Abstract
Plants have evolved different but interconnected strategies to defend themselves against herbivorous insects and microbial pathogens. We used an Arabidopsis/Pseudomonas syringae pathosystem to investigate the impact of pathogen-induced defense responses on cabbage looper (Trichoplusia ni) larval feeding. Arabidopsis mutants [npr1, pad4, eds5, and sid2(eds16)] or transgenic plants (nahG) that are more susceptible to microbial pathogens and are compromised in salicylic acid (SA)-dependent defense responses exhibited reduced levels of feeding by T. ni compared with wild-type plants. Consistent with these results, Arabidopsis mutants that are more resistant to microbial pathogens and have elevated levels of SA (cpr1 and cpr6) exhibited enhanced levels of T. ni feeding. These experiments suggested an inverse relationship between an active SA defense pathway and insect feeding. In contrast to these results, there was increased resistance to T. ni in wild-type Arabidopsis ecotype Columbia plants that were infected with P. syringae pv. maculicola strain ES4326 (Psm ES4326) expressing the avirulence genes avrRpt2 or avrB, which elicit a hypersensitive response, high levels of SA accumulation, and systemic acquired resistance to bacterial infection. Similar results were obtained with other ecotypes, including Landsberg erecta, Cape Verdi Islands, and Shakdara. When infected with Psm ES4326(avrRpt2) or Psm ES4326(avrB), nahG transgenic and npr1 mutant plants (which are more susceptible to virulent and avirulent P. syringae strains) failed to show the increased insect resistance exhibited by wild-type plants. It was surprising that wild-type plants, as well as nahG and npr1 plants, infected with Psm ES4326 not expressing avrRpt2 or avrB, which elicits disease, became more susceptible to T. ni. Our results suggest two potentially novel systemic signaling pathways: a systemic response elicited by HR that leads to enhanced T. ni resistance and overrides the SA-mediated increase in T. ni susceptibility, and a SA-independent systemic response induced by virulent pathogens that leads to enhanced susceptibility to T. ni.
Plants are frequently subjected to simultaneous insect herbivory and pathogen infection. They respond to these two different types of attackers with the induction of distinctive and overlapping subsets of secondary compounds or other defense responses involving antimicrobial or insecticidal activity. Although each type of interaction has been separately studied, the host response to the combined attack by insects and pathogens has received much less attention despite the abundance of reports indicating that pathogens and insects affect each other's performance on the host (Maleck and Dietrich, 1999; Paul et al., 2000). This study describes a model to study the three-way interactions between plants, insect herbivores, and microbial pathogens.
Plants respond to insect herbivory with a complicated arsenal of defensive responses, including the synthesis of insecticidal secondary metabolites, anti-feeding proteins, and/or volatile compounds to attract natural enemies of insect herbivores (Pare et al., 1998; Preston et al., 1999; Stotz et al., 1999). Wounding caused by insect herbivory induces a subset of plant defense responses, some of which are also activated by pathogen attack. A wound response pathway (elicited by insect feeding) has been extensively studied in tomato (Lycopersicon esculentum), tobacco (Nicotiana tabacum), Arabidopsis, and other species (Korth and Dixon, 1997; Felton et al., 1999; Reymond et al., 2000; Stotz et al., 2000). Activation of this pathway leads to the accumulation of jasmonic acid (JA) synthesized via the octadecanoic acid pathway and the production of anti-feeding proteinase inhibitors in local and distal parts of plants (Omer et al., 2000; Pechan et al., 2000). JA is an essential component mediating the signaling of this pathway, and exogenous addition of JA can increase the resistance of wild plants to insects in the field (Baldwin, 1998). In a similar manner, tomato (Howe et al., 1996) or Arabidopsis (McConn et al., 1997) mutants that block the JA-dependent pathway show increased sensitivity to insect feeding. Ethylene (Et) acts synergistically with JA for full induction of the wound response genes, and Et and JA may stimulate the biosynthesis of one another (O'Donnell et al., 1996; Penninckx et al., 1998; Alonso et al., 1999). Evidence that plants respond to insect-generated signals in addition to wounding has been obtained by transcription profiling of Arabidopsis leaves, which were wounded mechanically or were subjected to feeding by cabbage butterfly (Pieris rapae) larvae (Reymond et al., 2000).
Like insects, microbial pathogens induce a variety of host defense responses. One of the most effective defensive responses is elicited by so-called gene-for-gene relationships that involve the specific interaction between a pathogen avirulence (avr) gene product and a corresponding plant resistance (R) gene product (Flor, 1971). This type of plant-pathogen interaction is referred to as an incompatible interaction, and the pathogen expressing the avr gene is referred to as being avirulent. Among the approximate 150 R genes that have been identified in the completely sequenced Arabidopsis genome to date (The Arabidopsis Genome Initiative, 2000), RPS2 and RPM1 confer race-specific resistance to Pseudomonas syringae strains that express the avr genes avrRpt2 or avrB/avrRpm1, respectively (Kunkel et al., 1993; Yu et al., 1993; Grant et al., 1995). These two R genes have been widely used to examine the proposed ligand-receptor model of avr/R-gene interactions (Boyes et al., 1998; Leister and Katagiri, 2000; Nimchuk et al., 2000; Tao et al., 2000; Axtell et al., 2001), as well as the evolution of plant/pathogen interactions (Bergelson et al., 2001).
Plant defense responses activated upon avr/R recognition are often accompanied by a hypersensitive response (HR), which involves rapid programmed host cell death at the site of initial contact. The HR is mediated by a number of elicitors and secondary messengers, including reactive oxygen species and salicylic acid (SA; Grant et al., 2000; Heath, 2000; Klessig et al., 2000; McDowell and Dangl, 2000). Neighboring as well as distant host cells subsequently mount defense-related responses such as lignification and production of low-Mr antimicrobial compounds (e.g. phytoalexins) and pathogenesis-related (PR) proteins. The systemic activation of these defense responses, referred to as systemic acquired resistance (SAR), results in broad-spectrum resistance to many fungal, bacterial, and viral pathogens throughout the plant (Chester, 1933; Ross, 1961; Neuenschwander et al., 1995; Ryals et al., 1996). In many plants, the induction of SAR is preceded by the accumulation of SA, which has been shown to be necessary and sufficient for SAR induction in plants such as Arabidopsis. Exogenous application of SA elicits PR gene expression and enhanced pathogen resistance, whereas transgenic plants expressing a bacterial salicylate hydroxylase gene (nahG) that converts SA to catechol are deficient in SAR and are more susceptible to a variety of pathogens (Ward et al., 1991; Gaffney et al., 1993; Uknes et al., 1993; Delaney et al., 1994; Lawton et al., 1995).
In contrast to incompatible plant pathogen interactions that lead to HR and SAR, a compatible interaction resulting in disease can occur in the absence of a specific avr/R gene interaction. In compatible interactions, the pathogens are referred to as virulent, and the hosts as susceptible. Many of the same host responses involved in avr-R-mediated resistance also occur in compatible interactions, although they are activated more slowly or at a lower magnitude (Dixon and Harrison, 1990; Meier et al., 1993; Dixon et al., 1994; Ryals et al., 1996; Maleck et al., 2000). Genetic analysis in Arabidopsis has resulted in the most complete understanding of the similarities and differences in host responses to virulent and avirulent pathogens. A variety of Arabidopsis defense-related genes have been identified whose products appear to function specifically downstream of R-avr-recognition, specifically in defense responses that occur in compatible plant-pathogen interactions or nonspecifically in signal responses pathways that function in response to virulent and avirulent pathogens (Glazebrook, 2001).
The best-characterized Arabidopsis defense-related gene, NPR1 (nonexpressor of PR genes, also known as NIM1), which plays an important role in the response to virulent and avirulent pathogens, acts downstream of SA accumulation (Cao et al., 1994, 1997; Delaney et al., 1995; Ryals et al., 1997; Shah et al., 1997). npr1 mutant plants accumulate SA but have greatly reduced expression of the PR1, PR2, and PR5 genes and exhibit enhanced susceptibility to a variety of virulent and avirulent fungal and bacterial pathogens. EDS1 (enhanced disease susceptibility) is another well-studied defense-related gene that functions in response to virulent and avirulent pathogens (Parker et al., 1996; Aarts et al., 1998; Falk et al., 1999). PAD4 (phytoalexin deficient), on the other hand, encodes a product that only appears to function in response to virulent pathogens (Glazebrook and Ausubel, 1994; Glazebrook et al., 1997; Zhou et al., 1998). Like NPR1, EDS1 and PAD4, as well as several other Arabidopsis genes including EDS5 (Glazebrook et al., 1996; Rogers and Ausubel, 1997) are SID2 (SA induction deficient; Nawrath and Metraux, 1999) are involved in SA-mediated signaling.
When mutated, all of the genes described in the preceding paragraph result in an enhanced disease susceptibility phenotype. In contrast, Arabidopsis mutants that exhibit enhanced resistance to virulent and avirulent pathogens and that affect SA signaling pathways have also been isolated. cpr1 and cpr6 (constitutive expressor of PR genes) mutants exhibit constitutively high SA levels and PR gene expression (Bowling et al., 1994; Clarke et al., 1998), whereas acd (accelerated cell death; Greenberg and Ausubel, 1993; Greenberg et al., 1994; Rate et al., 1999) and lsd (lesions simulating disease; Dietrich et al., 1994) mutants exhibit spontaneous HR-like lesions in addition to constitutive SA and PR gene expression.
In addition to SA, JA and Et also play key roles in defending plants against microbial pathogens. A JA/Et-mediated pathway induces the accumulation of the antimicrobial peptides thionin and defensin, and appears to be particularly important in conferring Arabidopsis resistance to necrotrophic fungal pathogens (Penninckx et al., 1996; Bohlmann et al., 1998; Manners et al., 1998). SA-mediated signaling pathways and JA/Et-mediated pathways appear to be at least in part mutually antagonistic (Dong, 1998; Pieterse et al., 1998). For example, in the Arabidopsis cpr6 mutant, which has high constitutive SA levels, blocking the SA pathway by nahG, npr1, eds5, or pad4 resulted in enhanced expression of the JA/Et response gene PDF1.2 (encoding defensin; Clarke et al., 1998, 2000). On the other hand, SA and JA/Et pathways also appear to intersect, sharing the same regulatory components, because NPR1 has been shown to be required for SAR and a response called induced systemic resistance, which is a JA/Et-activated response elicited by nonpathogenic root-colonizing bacteria (Pieterse et al., 1998; Pieterse and Van Loon, 1999). In addition, there is evidence that in some cases, SA and JA can act synergistically to increase disease resistance (van Wees et al., 2000). Furthermore, high-throughput microarray analysis of the induction of selected Arabidopsis genes on activation of defense responses has revealed that a large set of Arabidopsis genes can be induced by SA or JA (Schenk et al., 2000).
Crosstalk between insect-plant interactions and pathogen-plant interactions has been recognized for a long time (Price et al., 1980; Jones, 1984; Doherty et al., 1988; Doares et al., 1995), consistent with the observations that insects activate JA/Et-mediated defense response pathways and that SA-mediated and JA/Et-mediated pathways can be antagonistic and/or synergistic. For example, transgenic tobacco plants compromised in SA-mediated SAR exhibited enhanced systemic resistance to larvae of Heliothis virescens, whereas plants with elevated phenylpropanoid levels associated with SA biosynthesis exhibited compromised induced insect resistance (Felton et al., 1999). In a similar manner, SA has been found to inhibit the JA-dependent insect defense pathway in tomato at several steps, including disrupting H+/K+ transport at the plasma membrane and inhibiting JA synthesis (Doherty et al., 1988; Peña-Cortés et al., 1993; Doares et al., 1995).
In the results described here, we use the well-studied pathosystem consisting of Arabidopsis and P. syringae pv. maculicola strain ES4326 (Dong et al., 1991) to study the effects of bacterially induced plant defenses on insect feeding. We take advantage of Arabidopsis mutants that are altered in defense against bacterial pathogens, and we examine the effects of infection by virulent and avirulent isolates of P. syringae on insect feeding. As an insect model, we have chosen Trichoplusia ni (cabbage looper; Lepidoptera:Noctuidae), a host plant generalist that feeds on a wide variety of plants, including Arabidopsis (Shorey et al., 1962; Grant-Peterson and Renwick, 1996; Jander et al., 2001). Our results indicate that virulent and avirulent pathogens have different effects on the induction of Arabidopsis defenses against T. ni feeding and, surprisingly, that virulent pathogens appear to inhibit Arabidopsis insect defenses in an SA-independent manner.
RESULTS
T. ni Feeding on Arabidopsis Defense-Related Mutants
To determine whether Arabidopsis mutants that have been generated to dissect the plant signal transduction pathways involved in pathogen defense also affect Arabidopsis-insect interactions, we tested several mutants with compromised or enhanced resistance to pathogens in a T. ni weight gain assay. In particular, we concentrated on a set of transgenic plants and mutants that exhibit enhanced resistance or susceptibility to a variety of bacterial pathogens and/or to obligate fungal pathogens such as Peronospora parasitica or Erysiphe orontii. Enhanced susceptibility mutants and transgenic lines fall into three broad classes: mutants and transgenics that have depleted SA levels (nahG transgenics; Delaney et al., 1994) or are deficient in SA biosynthesis [sid2(eds16); Nawrath and Metraux, 1999; Dewdney et al., 2000], mutants that appear to be deficient in signaling and/or that have low SA levels (pad4, eds1-2, eds5, eds15; Glazebrook et al., 1996; Rogers et al., 1996; Falk et al., 1999; Nawrath and Metraux, 1999; Dewdney et al., 2000), and a mutant that is unresponsive to SA (npr1; Cao et al., 1994; Delaney et al., 1995). The enhanced resistance mutants (cpr1, cpr6, and acd2; Greenberg and Ausubel, 1993; Bowling et al., 1994; Clarke et al., 1998) all have high levels of SA and increased resistance. In addition to expressing high levels of SA and defense-related gene transcripts, the acd2 mutant also forms spontaneous HR-like lesions that occur in the absence of bacterial infection (Greenberg and Ausubel, 1993; Greenberg et al., 1994).
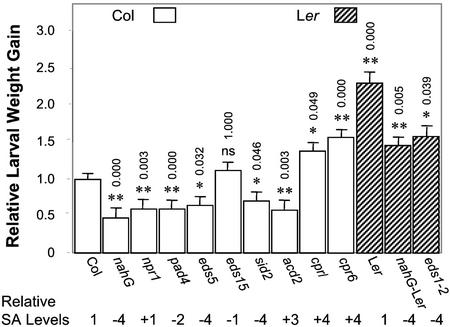
Larvae were weighed after 5 d of feeding (Fig. 1). For each experiment, the weights of the larvae feeding on various mutants or the Ler accession were normalized to the average weight of the larvae feeding on wild-type Col plants as described in “Materials and Methods.” Relative weight gain data from four sets of experiments were analyzed with ANOVA, and the means and ses obtained from this analysis are reported in Figure 1. Weight gain of larvae feeding on a Col nahG transgenic and on the Col npr1, eds5, pad4, sid2(eds16), and acd2 mutant plants was 30% to 50% less than that of larvae feeding on wild-type plants. There was no significant difference in larval weight gain when feeding on Col eds15 compared with wild-type Col.
Figure 1.
Larval weight gain of T. ni feeding for 5 d on Arabidopsis wild-type plants and defense-related mutants. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on wild-type Columbia (Col) plants. The bars represent the least square means (±ses, ANOVA) of relative larval weight gain from four independent experiments. Open bars and hatched bars correspond to the Col and Landsberg erecta (Ler) accessions, respectively. Relative SA levels refer to SA accumulation following infection with Psm ES4326(avrRpt2) or Erysiphe orontii in the case of the npr1, pad4, eds5, eds15, sid2(eds16), and eds1 mutants (Delaney et al., 1995; Zhou et al., 1998; Nawrath and Metraux, 1999; Dewdney et al., 2000; Feys et al., 2001), or the levels in uninfected plants in the case of the acd2, cpr1, and cpr6 mutants (Bowling et al., 1994; Greenberg et al., 1994; Clarke et al., 1998). Relative SA levels: −4, <0.25; −3, 0.25 to 0.5; −2, 0.5 to 0.75; −1, 0.75 to 1; 0, 1.0; +1, 1 to 1.5; +2, 1.5 to 2; +3, 2 to 3; +4, >3-fold wild-type (Wt) levels. The numbers above the bars represent P values adjusted using the Bonferroni method from multiple comparisons between Col wild-type and mutant plants. The P value for Ler shows the difference between Ler and Col wild-type plants. The P values for nahG-Ler and eds1-2 are for the comparisons with Ler wild-type plants. ns, Not significant; P > 0.05, * 0.01 < P < 0.05, **P < 0.01.
As described previously (Jander et al., 2001) and illustrated in Figure 1, T. ni larvae gain significantly more weight when feeding on the Ler accession compared with Col. Nevertheless, similar to results obtained with the Col plants, Ler nahG transgenic plants showed increased resistance to T. ni larval feeding compared with Ler wild type (37% less weight gain). In addition, the eds1 mutant (which has decreased levels of SA and is in the Ler background) also showed increased resistance (32% less weight gain).
In contrast to the nahG transgenic plants and most of the Arabidopsis mutants that are more susceptible to pathogen infection, the Col cpr1 and cpr6 mutant plants were significantly more susceptible to T. ni feeding, with 38% and 57% increased larval weight gain relative to wild-type plants, respectively. It is interesting that the acd2 mutant, which also exhibits constitutively elevated SA levels, was more resistant to T. ni feeding, similar to the nahG transgenics and eds mutants.
To summarize this series of experiments, the nahG transgenic plants and most of the eds mutants that exhibit compromised pathogen resistance or defects in SAR signaling also showed significant resistance to T. ni feeding. In a converse manner, the cpr mutants, which are more pathogen resistant and which exhibit a constitutive SAR response, were more susceptible to T. ni feeding. It is important to note that as illustrated in Figure 1, with the exception of acd2, there was a good correlation between previously reported SA levels in the mutants and transgenics and susceptibility/resistance to T. ni feeding. This correlation includes the eds15 mutant, which although compromised in pathogen resistance, has almost wild-type levels of SA (80%; Dewdney et al., 2000). In general, the mutants (and nahG transgenic plants) with low SA levels were more resistant to T. ni feeding, whereas mutants with high SA levels were more susceptible to feeding. These results, which are consistent with previously published data, suggest that SA response pathways are involved in the regulation of insect defenses in the plant (Felton et al., 1999; Thaler et al., 1999). An apparent exception is the acd2 mutant. These plants develop spontaneous HR-like lesions that lead to the production of high systemic levels of SA. Nevertheless, acd2 plants were resistant to T. ni feeding, similar to the eds, pad4, and npr1 mutants and the nahG transgenic plants, which have low SA or are compromised in SA signaling.
Correlation between the Extent of Leaf Defoliation and Larval Weight Gain
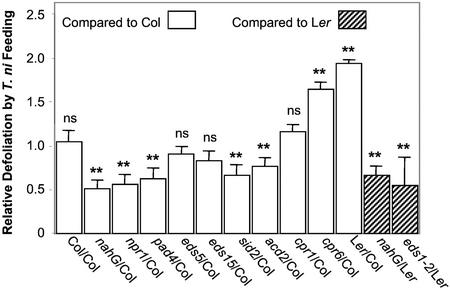
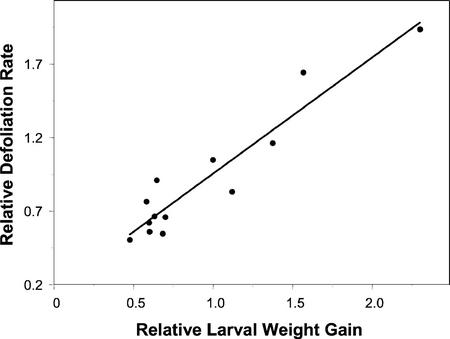
The change in T. ni larval weight gain observed in Figure 1 could be a consequence of a variety of different responses in the insect, including altered amounts of leaf ingested or efficiency of digestion or assimilation. To investigate further the different levels of T. ni larval weight gain when the caterpillars fed on different Arabidopsis ecotypes and mutants, we measured the amount of leaf area consumed after larvae fed on the plants for 3 d using a scoring system described in “Materials and Methods.” In this series of experiments, T. ni caterpillars were given a choice between the Col and Ler ecotypes or between wild-type and mutant plants planted side by side. As shown in Figure 2, T. ni larvae consumed significantly less leaf tissue of the nahG transgenic plants and of the npr1, pad4, sid2(eds16), acd2, eds1 5, and eds1 mutant plants compared with the relevant wild-type plants. In contrast, the larvae consumed significantly more of the cpr1 and cpr6 leaf tissue. A comparison of the data in Figures 1 and 2 showed that with the exception of the eds5 and eds15 mutants, there was good agreement between larval weight gain and the extent of defoliation (P value = 2.391e−7). Despite these two discrepancies, there was a strong positive correlation (r2 = 0.92) when the weight gain data were plotted against the defoliation data as shown in Figure 3.
Figure 2.
Relative defoliation rate of T. ni feeding on various Arabidopsis wild-type plants and defense-related mutants. The bars represent the means (±ses) of the relative defoliation rates obtained by comparing the extent of defoliation for each particular ecotype, transgenic, or mutant plant with the average extent of defoliation observed for the relevant wild-type plants that had been planted side by side with the particular experimental plants. The asterisks indicate the significance level, determined by permutation tests, of the defoliation differences between control plants (Col in the case of nahG, npr1, pad4, eds5, sid2, acd2, cpr1, cpr6, and Ler indicated by open bars, and Ler in the case of nahG-Ler and eds1-2 indicated by hatched bars) and experimental plants. ns, Not significant; P > 0.05, * 0.01< P < 0.05, ** P < 0.01.
Figure 3.
Scatterplot of relative defoliation rates against relative larval weight gains, showing a strong positive correlation between larval weight gains and defoliation rates (r2 = 0.92, P < 0.001).
Altered T. ni Weight Gain on Wild-Type Plants Challenged with Virulent and Avirulent P. syringae pv. maculicola Isolate ES4326 (Psm ES4326)
The experiments in the preceding sections utilizing Arabidopsis defense-related mutants and transgenic plants suggested that there is a negative correlation between pathogen and insect resistance in Arabidopsis. To investigate this relationship further, we investigated how infection of Arabidopsis plants by a bacterial pathogen affects insect feeding. We challenged wild-type Arabidopsis plants of different ecotypes with avirulent and virulent strains of the bacterial pathogen Psm ES4326. Psm ES4326 causes disease characterized by water-soaked lesions and chlorosis on a variety of Arabidopsis ecotypes (Dong et al., 1991; Whalen et al., 1991) and is not thought to elicit an SAR response. In contrast, derivatives of Psm ES4326 carrying the avrRpt2 or avrB avirulence genes [Psm ES4326(avrRpt2) or Psm ES4326(avrB)] elicit an HR and SAR on Arabidopsis ecotypes that carry the RPS2 or RPM1 resistance genes, respectively (Kiedrowski et al., 1992; Yu et al., 1993; Rogers and Ausubel, 1997).
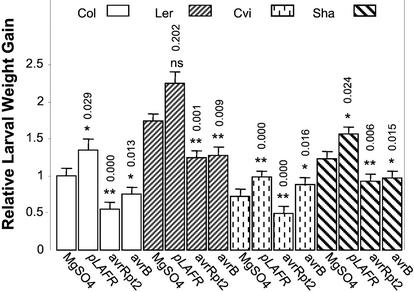
In most cases, T. ni larval feeding was inhibited on plants in which an HR was elicited by the interaction of avrRpt2 with RPS2 or avrB with RPM1 (Fig. 4). Larvae feeding on plants infected with Psm ES4326(avrRpt2) had 45%, 28%, 32%, and 25% decreased weight gain on the Col, Ler, Cape Verde Islands (Cvi), and Shakdara (Sha) ecotypes, respectively, compared with growth on plants mock-inoculated with 10 mm MgSO4. In a similar manner, infection with Psm ES4326(avrB) compromised larval growth by 25%, 26%, and 21%, respectively, on Col, Ler, and Sha. The observation that infection with avirulent pathogens (which elicit SAR) caused increased resistance to T. ni feeding was surprising given the data presented in Figure 1, which showed that the cpr1 and cpr6 mutants (which exhibit a constitutive SAR phenotype) were more sensitive to T. ni.
Figure 4.
Larval weight gain of T. ni feeding on various Arabidopsis ecotypes infiltrated with isogenic virulent and avirulent strains of Psm ES4326. As described in “Materials and Methods,” lower leaves were inoculated with 10 mm MgSO4 or with Psm ES4326(pLAFR3), Psm ES4326(avrRpt2), or Psm ES4326(avrB). Four days later, the inoculated leaves were removed and newly hatched T. ni larvae were placed on the upper leaves. Larval weight gain was measured after 5 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on wild-type Col plants inoculated with 10 mm MgSO4. The bars represent the least square means (±ses, ANOVA) of the relative larval weight gain data from three independent experiments. The numbers above each bar correspond to the Bonferroni adjusted P values from multiple t tests. For each ecotype, weight gain of T. ni larvae feeding on bacterial infected plants were compared with that on MgSO4-treated plants of the same ecotype. ns, Not significant; P > 0.05, * 0.01 < P < 0.05, ** P < 0.01.
It is interesting that in contrast to the other ecotypes, Cvi was not more resistant to T. ni feeding after Psm ES4326(avrB) infection (Fig. 4). However, this observation correlates with the fact that Cvi is a “natural” rpm1 mutant and does not respond to avrB (Debener et al., 1991; Stahl et al., 1999), and is consistent with the result that infection with Psm ES4326 not carrying avrRpt2 or avrB [Psm 4326(pLAFR3)] rendered plants of all ecotypes more susceptible to T. ni feeding (Fig. 4). Larvae gained 35%, 30%, 35%, and 27% more weight on Col, Ler, Cvi, and Sha infected with Psm ES4326(pLAFR3), respectively, compared with those feeding on plants mock infected with 10 mm MgSO4.
Larval Weight Gain on Arabidopsis R Gene Mutants Inoculated with Avirulent Pathogens
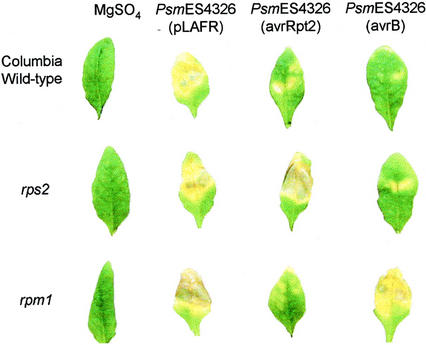
To confirm that the increased T. ni resistance of plants infected with avirulent pathogens is a direct result of the avr-R gene-for-gene-dependent defense response, we measured T. ni weight gain on Col rps2 and Col rpm1 mutant plants infected with Psm ES4326(avrRpt2) or Psm ES4326(avrB). The rationale for this experiment was that when R gene mutants are challenged with bacterial strains carrying the corresponding avr genes, the mutant plants cannot recognize the avr genes and fail to induce an HR and SAR. This is illustrated in Figure 5, which shows the symptoms of Arabidopsis wild-type plants as well as rps2 and rpm1 mutants infiltrated with Psm ES4326(avrRpt2), Psm ES4326(avrB), or Psm ES4326 not carrying avrRpt2 or avrB [Psm ES4326(pLAFR3)]. Psm ES4326(pLAFR3) caused water-soaked lesions in infected leaves. Wild-type Col plants infected with Psm ES4326(avrRpt2) or Psm ES4326(avrB) (or the Col rps2 mutant infected with Psm ES4326(avrB) or the Col rpm1 mutant infected with Psm ES4326(avrRpt2) exhibited an HR response and greatly reduced lesion formation. In contrast, Col rps2 plants infected with Psm 4326(avrRpt2) or Col rpm1 plants infected with Psm ES4326(avrB) did not exhibit an HR and developed water-soaked lesions similar to plants infected with Psm ES4326(pLAFR3) (Fig. 5).
Figure 5.
Disease symptoms that developed on wild-type ecotype Col plants or on rps2 or rpm1 mutant plants 4 d after they were inoculated with Psm ES4326(pLAFR3), Psm ES4326(avrRpt2), or Psm ES4326(avrB).
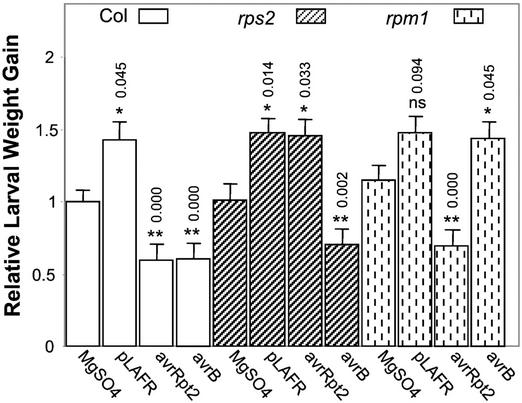
Regardless of the Arabidopsis Col genotype, infection with Psm ES4326(pLAFR3) resulted in increased T. ni larval weight gain compared with that on plants mock inoculated with 10 mm MgSO4 (Fig. 6), similar to the results shown in Figure 4. In all of the plant-pathogen interactions in which an HR was generated [Col infected with Psm ES4326(avrRpt2) or Psm ES4326(avrB), Col rps2 infected with Psm ES4326(avrB), or Col rpm1 infected with Psm ES4326(avrRpt2)], there was a 31% to 41% decrease in T. ni weight gain compared with those on plants that were mock inoculated with MgSO4. In contrast, in the plant-pathogen interactions in which water-soaked disease lesions were generated [Col infected with Psm ES4326(pLAFR3), Col rps2 infected with Psm ES4326 or Psm ES4326(avrRpt2), or Col rpm1 infected with Psm ES4326(pLAFR3) or Psm ES4326(avrB)], the larval weight gain was 26% to 46% more than the weight gain on mock-inoculated plants. These results indicate that increased insect resistance is a result of a defense response specifically induced in Arabidopsis by the interaction of avrRpt2 and RPS2 or avrB and RPM1.
Figure 6.
Larval weight gain of T. ni feeding on wild-type Arabidopsis plants and R gene mutant plants that were infiltrated with various strains of Psm ES4326. As described in “Materials and Methods,” lower leaves were inoculated with 10 mm MgSO4 or with Psm ES4326(pLAFR), Psm ES4326(avrRpt2), or Psm ES4326(avrB). Four days later, the inoculated leaves were removed and newly hatched T. ni larvae were placed on the upper leaves. Larval weight gain was measured after 5 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on mock-inoculated wild-type Col plants. The bars represent the means (±ses, ANOVA) of relative weight gain from four independent experiments. The numbers above each bar correspond to the Bonferroni adjusted P values from multiple t tests. The P values for bacteria-treated wild-type or R gene mutant plants are from multiple comparisons against mock-treated wild-type or R gene mutant plants, correspondingly. ns, Not significant; P > 0.05, * 0.01 < P < 0.05, ** P < 0.01.
T. ni Weight Gain on Arabidopsis Defense-Related Mutants Inoculated with Virulent and Avirulent Psm ES4326 Strains
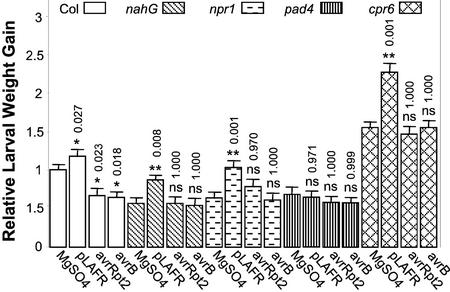
To identify potential crosstalk between plant defense responses and insect feeding behavior, the SAR compromised mutants pad4 and npr1, nahG transgenic plants, and the constitutive SAR mutant cpr6 were infected with Psm ES4326 ± avrRpt2 or avrB and were then fed to T. ni. It is interesting that all of these plant lines with the exception of pad4 were more susceptible to T. ni feeding after infection with Psm ES4326 not carrying avrRpt2 or avrB [Psm ES4326(pLAFR3)] compared with plants mock inoculated with 10 mm MgSO4 (Fig. 7), similar to the results obtained previously in Figures 4 and 6. On the other hand, infection with Psm ES4326(avrRpt2) or Psm ES4326(avrB) did not have an appreciable effect on T. ni feeding compared with mock-inoculated controls. Overall, the results suggest that increased larval weight gain on plants previously infected with a virulent pathogen is SA independent, whereas the decreased larval weight gain on plants previously infected with avirulent pathogens may be SA dependent.
Figure 7.
Larval weight gain of T. ni feeding on Arabidopsis defense-related mutants infiltrated with various strains of Psm ES4326. As described in “Materials and Methods,” lower leaves were inoculated with 10 mm MgSO4 or with Psm ES4326(pLAFR), Psm ES4326(avrRpt2), or Psm ES4326(avrB). Four days later, the inoculated leaves were removed and newly hatched T. ni larvae were placed on the upper leaves. Larval weight gain was measured after 5 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on mock-inoculated wild-type Col plants. The bars represent the means (±ses, ANOVA) of relative weight gain from four independent experiments. The numbers above each bar correspond to the Bonferroni adjusted P values from multiple t tests. The P values for bacteria-treated wild-type or defense-related mutant plants are from the multiple comparisons against mock-treated wild-type or defense-related mutant plants, respectively. ns, Not significant; P > 0.05, * 0.01 < P < 0.05, ** P < 0.01.
DISCUSSION
Many important plant diseases are transmitted from plant to plant by insect herbivores. Therefore, plant defensive systems are under constant selective pressure to optimize their response to microbial pathogens and insects in a concerted manner. Not surprisingly, diseased host plants represent a complex feeding niche for phytophagous insects, and both increased and decreased insect resistance has been reported for diseased plants (Harrison et al., 1980; Purcell, 1991; Hatcher and Paul, 2000). The present study used a genetic approach to help elucidate the complicated interplay between bacterial resistance and insect resistance in plants. A better understanding of these interactions in plant defensive systems is of theoretical and practical importance.
Taken on their own, the T. ni feeding experiments described in Figures 1 and 2 with Arabidopsis plants defective in the SA signaling pathway suggest that a component of the SA signal transduction cascade inhibits the induction of defenses against T. ni. Larvae consumed less leaf tissue and gained approximately 50% less weight on nahG plants, and 40% less weight on npr1, pad4, eds5, and sid2(eds16) plants. With the exception of npr1, the degree of resistance to T. ni feeding correlated with a decrease in induced SA levels in response to bacterial pathogens. Because the npr1 mutant, which has normal levels of SA but which does not respond to SA signaling, was also resistant to T. ni feeding, a component of the signal cascade downstream of NPR1 may be responsible for inhibiting defense responses against insects. The effect of SA levels on Arabidopsis resistance to insects can also be seen in the case of the cpr1 and cpr6 mutants, which exhibit constitutively elevated levels of SA. Larvae showed approximately 50% increase in weight gain on these mutants. These findings agree with previous reports of an inverse relationship between insect feeding and SA-mediated SAR induction (Doares et al., 1995; Niki et al., 1998; Felton et al., 1999; Thaler et al., 1999). Moreover, there was a strong positive relationship between larval weight gain and the amount of tissue consumed (Fig. 3). This result, on the one hand, strengthens the above-mentioned finding that an SA-dependent pathway inhibits insect resistance, and on the other justifies the utilization of larval weight gain for these experiments to assay plant resistance.
One caveat to the apparent correlation between SA levels in the various mutants and the extent of larval feeding is the fact that SA levels were not determined directly in our experiments, but were obtained from the literature. Moreover, many of these SA levels correspond to those observed in pathogen-infected plants, which are not directly relevant to the experiments shown in Figures 1 and 2 that did not involve prior pathogen infection. In the future, direct measurement of SA levels in plants undergoing T. ni feeding will help elucidate the precise role of SA signaling in mediating insect defense responses.
In contrast to the results that we obtained with the cpr1 and cpr6 mutants, which have a constitutive SAR response and are more sensitive to insect feeding, when we induced SAR with Psm ES4326 expressing avrRpt2 or avrB, we did not observe the expected result of increased weight gain by T. ni larvae (Fig. 4). In fact, T. ni larvae gained an average of 30% less weight when SAR was induced by inoculating the Col, Ler, or Sha ecotypes with Psm ES4326(avrRpt2) or Psm ES4326(avrB) than when feeding on mock-inoculated plants (Fig. 4). We verified that the enhanced resistance of these plants to T. ni feeding was due to avr-R gene interactions by showing that two R gene mutants in the Col background, rps2 and rpm1, were not more resistant to T. ni feeding when preinoculated with Psm ES4326(avrRpt2) or Psm ES4326(avrB), respectively (Fig. 6). Similarly, ecotype Cvi, which is a “natural” rpm1 mutant, did not exhibit enhanced resistance to T. ni feeding when infected with Psm ES4326(avrB) (Fig. 4).
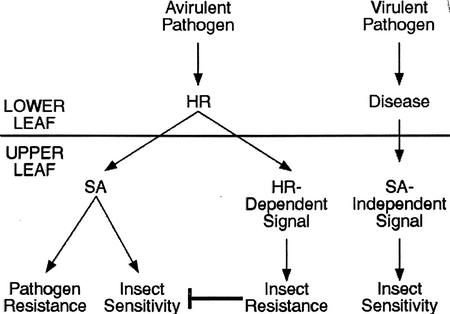
Our results are similar to those of Stout et al. (1999) who saw that SAR induced with benzothiadiazole inhibited plant resistance to Helicoverpa zea, whereas SAR induced by infection with P. syringae pv. tomato decreased the consumption of tomato plants by H. zea. One explanation for our observations and those of Stout et al. (1999) is that the HR elicited upon avr-R recognition overrides the SA-mediated increase in sensitivity to T. ni feeding. The finding that acd2 mutants were more resistant rather than more susceptible to insect feeding even though they have high systemic levels of SA supports this hypothesis. In contrast to cpr1 and cpr6, acd2 mutants exhibit spontaneous HR-like lesions in addition to constitutive SAR induction seen in the cpr mutants (Greenberg and Ausubel, 1993; Greenberg et al., 1994). Therefore, we conclude that a systemic signal in addition to SA is involved in Arabidopsis defenses against insects and that it is induced by the avr/R gene interaction (Fig. 8). The observation that the cpr1 and cpr6 mutants, which have constitutively high levels of SA, were more susceptible to T. ni feeding, makes it unlikely that the enhanced resistance to feeding elicited by infection with the avirulent Psm strains can be solely a consequence of the systemic accumulation of SA. In a nahG, npr1, pad4, or cpr6 genetic background, infection with an avirulent pathogen did not result in increased insect resistance (Fig. 7). This indicates that HR and SA may be involved in modulating the proposed systemic signal leading to insect resistance. The proposed systemic signal may be required for formation of the HR or it may be generated from the HR lesion itself (Fig. 8). It is interesting that this conclusion is consistent with a previous study that concluded that SA is not the mobile systemic signal that activates SAR (Vernooij et al., 1994).
Figure 8.
Model of the signaling leading to insect resistance or sensitivity after pathogen infection. Infection with an avirulent pathogen increases SA levels, which has been shown to cause insect sensitivity. However, the simplest interpretation of the data presented in this paper is that an unknown signal from an avirulent infection apparently overrides the SA signaling, thereby increasing insect resistance. This signal may partly depend on HR and SA. Infection with a virulent pathogen appears to result in another unidentified signal that systemically increases insect sensitivity in an SA-independent manner.
A second unexpected result from this work is the observation that infection of Arabidopsis with the virulent pathogen Psm ES4326(pLAFR3) led to an increase in T. ni weight gain. The feeding results from wild-type plants of four ecotypes tested (Col, Ler, Cvi, and Sha) showed that larvae gained about 30% more weight on diseased plants. In a similar manner, when rps2 or rpm1 mutants were inoculated with Psm ES4326(avrRpt2) or Psm ES4326(avrB), respectively, there was an increase in the weight gain of T. ni larvae feeding on these plants (Fig. 6). It seems unlikely that the increased susceptibility to T. ni feeding is a consequence of systemic SA-mediated signaling because virulent pathogens are not thought to induce systemic accumulation of SA and because, as discussed above, the induction of SAR by avirulent pathogens led to increased resistance to T. ni feeding. Nevertheless, the increased insect susceptibility to T. ni elicited by virulent pathogens must be a systemic response because the infected leaves were removed prior to the start of feeding. The data in Figure 7 support the conclusion that the proposed systemic signal elicited by virulent pathogens is not SA. That is, larvae feeding on mutants that affect SA signaling (npr1, pad4, and NahG) still showed increased weight gain after their host plants were infected with Psm ES4326(pLAFR3). Moreover, infection of the cpr6 mutant, which has high constitutive levels of SA, with Psm ES4326(pLAFR3), also resulted in a further increase in larval weight gain compared with a mock-infected cpr6 mutant. Thus, virulent pathogens seem to induce increased sensitivity to insects in a manner that is not tightly coupled to SA signaling (Fig. 8).
The above discussion assumes that the proposed systemic signal generated by virulent pathogens is of host origin, but we cannot rule out the possibility that the systemic signal is actually a bacterial product. Another explanation for the enhanced sensitivity to T. ni of plants infected with virulent pathogens is that the infected plant is depleted of resources due to fighting the bacterial infection and thus cannot mount an adequate defense against insect feeding. However, evidence against this latter explanation comes from the observation that infection of nahG and npr1 plants with virulent and avirulent pathogens causes similar disease symptoms (Cao et al., 1994; Bowling et al., 1997), even though only infection with the virulent pathogen resulted in increased larval weight gain.
Our studies on the three-way interactions between Arabidopsis, Psm ES4326, and T. ni show a complex pattern of susceptibility/resistance to T. ni larval feeding. Whereas mutants that express SAR constitutively exhibit enhanced sensitivity to insect feeding, the activation of an HR by an avirulent pathogen or the acd2 mutation results in a systemic increase in resistance to insect feeding. In contrast, infection with a virulent pathogen results in increased sensitivity of Arabidopsis to insect feeding. This increase in sensitivity appears to be due to a previously unreported systemic response that is independent of the SAR signaling pathway (Fig. 8). JA and Et are known to be involved in mediating insect defense responses (O'Donnell et al., 1996; Penninckx et al., 1998; Alonso et al., 1999). Furthermore, JA, along with HR and SA, has been shown to be induced by avr-R recognition (van Wees et al., 1999). It is possible that JA mediates insect resistance upon avr-R recognition, but it is unknown whether the level of JA induced is high enough to overcome the antagonistic effect of SA. Future work will determine whether the two signaling pathways proposed in Figure 8 are mediated by either or both of these molecules.
MATERIALS AND METHODS
Bacterial Strains and Media
The bacterial strain Pseudomonas syringae pv. maculicola (Psm) strain ES4326 has been described previously(Dong et al., 1991). Plasmid pLH12, a derivative of pLAFR3, carries avrRpt2 (Whalen et al., 1991), and plasmid pVB01 carries avrB (Innes et al., 1993a). P. syringae strains were grown at 28°C in King's B media (King et al., 1954) supplemented with 100 μg mL−1 streptomycin and 10 μg mL−1 tetracycline for strains carrying pLAFR3 and pLH12. Streptomycin (100 μg mL−1) and kanamycin (50 μg mL−1) were used for strains carrying pVB01.
Growth of Plants
Arabidopsis plants were grown in Metromix 200 soil (Scott, Marysville, OH) in a climate-controlled greenhouse (20°C ± 2°C, relative humidity 70% ± 5%) under natural light supplemented with 12 h per day of artificial light on a 12-h light cycle/12-h dark cycle. Flats were rotated every 3 to 4 d to minimize environmental variance. Arabidopsis ecotype Col plant lines used in these experiments included: a transgenic line expressing the bacterial nahG gene (Reuber et al., 1998), npr1 (Cao et al., 1994), pad4 (Glazebrook et al., 1996), eds5 (Glazebrook et al., 1996), eds15 (Dewdney et al., 2000), sid2 (also called eds16; Dewdney et al., 2000), cpr1 (Bowling et al., 1994), cpr6 (Clarke et al., 1998), and acd2 (accelerated cell death; Greenberg and Ausubel, 1993). Arabidopsis Col plants representing three different R gene genotypes were used: ecotype Col-0 wild-type (genotype RPS2/RPS2 RPM1/RPM1), rps2-101C (genotype rps2-101C/rps2-101C RPM1/RPM1; Yu et al., 1993), and rps3-1 (genotype RPS2/RPS2rpm1/rpm1; Debener et al., 1991; Innes et al., 1993b). Arabidopsis ecotype Ler plant lines used in these experiments included Ler wild type, a transgenic line expressing the bacterial nahG gene obtained from X. Dong, Department of Biology, Duke University, Durham, NC and eds1-2 (Parker et al., 1996). Other Arabidopsis ecotypes used were Cvi and Sha, both of which were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH).
Cabbage Looper (Trichoplusia ni) Larval Weight Gain Assay
T. ni eggs (Entopath Inc., Easton, PA) were incubated at 32°C to synchronize hatching. Newly emerged first instar larvae (less than 8 h old) were placed on the leaves of 5- to 6-week-old plants. Each pot contained one plant fed to one randomly assigned larva. Fine fabric cloth bags were used to cover each plant and the larva feeding on it. For each ecotype, transgenic, or mutant, 36 plants, all in separate pots in a single flat, were used in each feeding experiment. Flats were rotated periodically to minimize environmental fluctuations, and the flats containing the various ecotypes, transgenics, or mutants were randomly assigned positions in the greenhouse from experiment to experiment. Each feeding experiment was carried out independently at least three times. Larvae were collected after feeding for 6 d, dried at 80°C for 3 d, and weighed individually. It was assumed that the initial weights of all of the larvae were the same and were negligible compared with the final weights. Larval weight gain data are reported as means and ses normalized to values obtained from wild-type plants.
T. ni Weight Gain Assay on Infiltrated Plants
Prior to infiltration, bacterial strains were grown overnight in King's B, grown to mid-log phase, and resuspended in 10 mm MgSO4. The undersides of lower leaves were inoculated with suspensions of bacterial cells at a titer of 104 CFU cm−1 leaf area with a 1-mL syringe without a needle forcing the suspension through the stomata. Four days postinoculation, infected leaves were removed and the plants were used for subsequent insect feeding experiments. Cabbage looper larvae were grown as described above, and as in the weight gain assay, fine fabric cloth bags were used to cover each plant, and each plant was grown in a separate pot, 36 pots to a flat. In each experiment, 144 plants of a particular ecotype, transgenic, or mutant were used. For these experiments, one-quarter of the 36 plants in each flat were inoculated with MgSO4, one-quarter with Psm ES4326(pLAFR3), one-quarter with Psm ES4326(avrRpt2), and one-quarter with Psm ES4326(avrB). These variously inoculated plants were randomly assigned to particular rows in each flat. Flats were rotated periodically to minimize environmental fluctuations, and the flats containing the various ecotypes, transgenics, or mutants were randomly assigned positions in the greenhouse from experiment to experiment. Each infection/feeding experiment was carried out independently at least three times. Larvae were collected after feeding for 6 d, dried at 80°C for 3 d, and weighed individually. Larval weight gain data are reported as means and ses normalized to values obtained from wild-type plants.
Insect Defoliation Rating Scores
In flats that contained 36 pots, two plants of different ecotypes or one wild-type and one transgenic or mutant plant were planted side by side in a single pot. The plants were grown for 5 to 6 weeks. One-third instar T. ni larva was placed on the soil between the two plants in each pot. Each pot was then covered with a fine fabric bag to contain the plants and the larva. However, for experiments with cpr6 and acd2, water was used to separate the pots from each other rather than covering the plants with fabric bags. For each particular ecotype, transgenic, or mutant plant, approximately 30 pairs of such plants and relevant wild-type plants were used in each experiment in which both plants in the pot had grown well. Each experiment for each type of comparison was repeated at least three times. The loss of plant material was scored 3 d after T. ni larvae feeding began. The level of insect defoliation was ranked on a scale from 1 to 5, with a score of 1 indicating that less than 25% leaf tissue was left; 2, less than 50% leaf tissue was left; 3, approximately one-half of the leaf tissue was consumed; 4, less than 25% of leaf tissue was eaten; and 5, plants were essentially untouched. At least one-half of the scoring was carried out by a person who did not know which plants were being used in the particular experiment.
Statistical Analysis
All statistical analysis was carried out with the S-PLUS 4.0 software package (Insightful Co., Seattle). Weight gain data were normalized to the average weight gain of larvae feeding on untreated (in the experiments without inoculation) or mock-treated (in the experiments with inoculation) wild-type Col plants for each set of experiments. The relative weight gain data were analyzed with ANOVA that included experiment as a factor. The figures show the least square means and ses from ANOVA. Multiple t tests, with Bonferroni adjusted significance levels, were carried out to determine actual P values for differences between the different lines in the treatments. In the case of the defoliation data, for each particular ecotype, transgenic, or mutant plant, the defoliation scores were normalized to the average extent of defoliation observed for the relevant wild-type plants that had been planted side by side with the particular experimental plants. The relative defoliation scores were analyzed using permutation tests.
ACKNOWLEDGMENTS
We thank Jennifer Thaler, Anurag Agrawal, Julia Dewdney, Julie Stone, and Mary Wildermuth for helpful discussions and for critically reading the manuscript, and Junni Zhang and Samuel Kou for help with the statistical analysis.
Footnotes
This work was supported by the Department of Organismic and Evolutionary Biology (Harvard University Graduate Student Grant to J.C.) and by the National Institutes of Health (grant no. GM48707 to F.M.A.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010815.
LITERATURE CITED
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, Staskawicz BJ. Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding Pseudomonas syringae avrRpt2 avirulence gene. Mol Plant-Microbe Interact. 2001;14:181–188. doi: 10.1094/MPMI.2001.14.2.181. [DOI] [PubMed] [Google Scholar]
- Baldwin IT. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Bohlmann H, Vignutelli A, Hilpert B, Miersch O, Wasternack C, Apel K. Wounding and chemicals induce expression of the Arabidopsis thaliana gene THI2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett. 1998;437:281–286. doi: 10.1016/s0014-5793(98)01251-4. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clark JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–64. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Chester KS. The problem of acquired physiological immunity in plants. Quart Rev Biol. 1933;8:275–324. [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X. Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell. 1998;10:557–569. doi: 10.1105/tpc.10.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debener T, Lehnackers H, Arnold M, Dangl JL. Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1991;1:289–302. doi: 10.1046/j.1365-313X.1991.t01-7-00999.x. [DOI] [PubMed] [Google Scholar]
- Delaney T, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessman H, Ward E et al. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000;24:205–218. doi: 10.1046/j.1365-313x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ. Activation, structure and organization of genes involved in microbial defense in plants. Adv Genet. 1990;28:165–234. doi: 10.1016/s0065-2660(08)60527-1. [DOI] [PubMed] [Google Scholar]
- Dixon RA, Harrison MJ, Lamb CJ. Early events in the activation of plant defense responses. Annu Rev Phytopathol. 1994;32:479–501. [Google Scholar]
- Doares SH, Narvaez-Vasquez J, Coconi A, Ryan CA. Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 1995;108:1741–1746. doi: 10.1104/pp.108.4.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty HM, Selvendran RR, Bowles DJ. The wound response in tomato plants can be inhibited by aspirin and related hyrodxybenzoic acid. Physiol Mol Plant Pathol. 1988;33:377–384. [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XN. SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- Falk A, Feys B, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Korth KL, Bi JL, Wesley SV, Huhman DV, Mathews MC, Murphy JB, Lamb C, Dixon RA. Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory. Curr Biol. 1999;9:317–320. doi: 10.1016/s0960-9822(99)80140-7. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement for salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol. 2001;4:301–308. doi: 10.1016/s1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Use of Arabidopsis for genetic dissection of plant defense responses. Annu Rev Genet. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- Grant JJ, Yun BW, Loake GJ. Oxidative burst and cognate redox signaling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J. 2000;24:569–582. doi: 10.1046/j.1365-313x.2000.00902.x. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. Structure of the Arabidopsis RPM1 gene which enables dual-specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- Grant-Peterson J, Renwick JAA. Effects of ultraviolet B exposure of Arabidopsis thaliana on herbivory by two crucifer-feeding insects. Environ Entomol. 1996;25:135–142. [Google Scholar]
- Greenberg JT, Ausubel FM. Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 1993;4:327–341. doi: 10.1046/j.1365-313x.1993.04020327.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM. Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- Harrison M, Brewer J, Merrill L. Insect development in the transmission of bacterial pathogens. In: Harris KF, Maramorosch K, editors. Vectors of Plant Pathogens. New York: Academic Press; 1980. pp. 201–292. [Google Scholar]
- Hatcher PE, Paul ND. On integrating molecular and ecological studies of plant resistance: variety of mechanisms and breadth of antagonists. J Ecol. 2000;88:702–706. [Google Scholar]
- Heath M. Hypersensitive response-related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/a:1026592509060. [DOI] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes R, Bent A, Kunkel B, Bisgrove S, Staskawicz B. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syrnigae avirulence genes. J Bacteriol. 1993a;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, Bisgrove SR, Smith NM, Bent AF, Staskawicz BJ, Liu Y-C. Identification of a disease resistance locus in Arabidopsis that is functionally homologous to the RPG1 locus of soybean. Plant J. 1993b;4:813–820. doi: 10.1046/j.1365-313x.1993.04050813.x. [DOI] [PubMed] [Google Scholar]
- Jander G, Cui J, Nhan B, Pierce NE, Ausubel FM. The TASTY locus on Chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni. Plant Physiol. 2001;126:890–898. doi: 10.1104/pp.126.2.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CG. Microorganisms as mediators of plant resource exploitation by insect herbivores. In: Slobodchikoff CN, Gaud WS, Price PW, editors. A New Ecology: Novel Approaches to Interactive Systems. New York: Wiley and Sons; 1984. pp. 53–99. [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL. Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J. 1992;11:4677–4684. doi: 10.1002/j.1460-2075.1992.tb05572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of phycocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Klessig D, Durner J, Noad R, Navarre D, Wendehenne D, Kumar D, Zhou J, Shah J, Zhang S, Kachroo P et al. Nitric oxide and salicylic acid signaling in plant defense. Proc Natl Acad Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth K, Dixon R. Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant-Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- Leister RT, Katagiri F. A resistance gene product of the nucleotide binding site: leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 2000;22:345–354. doi: 10.1046/j.1365-313x.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- Maleck K, Dietrich RA. Defense on multiple fronts: How do plants cope with diverse enemies? Trends Plant Sci. 1999;4:215–219. doi: 10.1016/s1360-1385(99)01415-6. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgern T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IAMA, Vermaere K, Kazan K, Brown R, Morgan A, Maclean DJ, Curtis MD, Cammue BPA, Broekaert WF. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol. 1998;38:1071–1080. doi: 10.1023/a:1006070413843. [DOI] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL. Signal transduction in the plant immune response. Trends Biochem Sci. 2000;25:79–82. doi: 10.1016/s0968-0004(99)01532-7. [DOI] [PubMed] [Google Scholar]
- Meier BM, Shaw N, Slusarenko AJ. Spatial and temporal accumulation of defense gene transcripts in bean (Phaseolus vulgaris) leaves in relation to bacteria-induced hypersensitive cell death. Mol Plant-Microbe Interact. 1993;6:453–466. doi: 10.1094/mpmi-6-453. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Metraux J-P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenschwander U, Vernooij B, Friedrich L, Uknes S, Kessmann H, Ryals J. Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance? Plant J. 1995;8:227–233. [Google Scholar]
- Niki T, Mitsuhara I, Seo S, Ohtsubo N, Ohashi Y. Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 1998;39:500–507. [Google Scholar]
- Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;12:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ. Ethylene as a signal mediating the wound response of tomato plants. Science. 1996;274:1914–1917. doi: 10.1126/science.274.5294.1914. [DOI] [PubMed] [Google Scholar]
- Omer AD, Thaler JS, Granett J, Karban R. Jasmonic acid induced resistance in grapevines to a root and leaf feeder. J Econ Entomol. 2000;93:840–845. doi: 10.1603/0022-0493-93.3.840. [DOI] [PubMed] [Google Scholar]
- Pare PW, Alborn HT, Tumlinson JH. Concerted biosynthesis of an insect elicitor of plant volatiles. Proc Natl Acad Sci USA. 1998;95:13971–13975. doi: 10.1073/pnas.95.23.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ. Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell. 1996;8:2033–2046. doi: 10.1105/tpc.8.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul ND, Hatcher PE, Taylor JE. Coping with multiple enemies: an integration of molecular and ecological perspectives. Trends Plant Sci. 2000;5:220–225. doi: 10.1016/s1360-1385(00)01603-4. [DOI] [PubMed] [Google Scholar]
- Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell. 2000;12:1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents would-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux J-P, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux J-P, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Loon LC. Salicylic acid-independent plant defense pathways. Trends Plant Sci. 1999;4:52–58. doi: 10.1016/s1360-1385(98)01364-8. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, van Pett JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CA, Lewandowski C, Enyedi AJ, Baldwin IT. Tobacco mosaic virus inoculation inhibits wound-induced jasmonic acid- mediated responses within but not between plants. Planta. 1999;209:87–95. doi: 10.1007/s004250050609. [DOI] [PubMed] [Google Scholar]
- Price PW, Bouton CE, Gross P, McPheron BA, Thompson JN, Weis AE. Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Annu Rev Ecol Syst. 1980;11:41–65. [Google Scholar]
- Purcell AH. Interactions among plant pathogenic prokaryotes, plants, and insect vectors. In: Barbosa P, Krischik VA, Jones CJ, editors. Microbial Mediation of Plant-Herbivore Interactions. New York: Wiley Inter-Science; 1991. pp. 383–406. [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber TL, Plotnikova JM, Dewdney J, Rogers EE, Wood W, Ausubel FM. Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 1998;16:473–485. doi: 10.1046/j.1365-313x.1998.00319.x. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–720. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Volko S, Ausubel FM. Arabidopsis thaliana enhanced disease susceptibility (eds) mutants. In: Stacey G, Mullin B, Gresshoff PM, editors. Biology of Plant-Microbe Interactions. International Society of Molecular Plant-Microbe Interactions, St. Paul. 1996. pp. 47–56. [Google Scholar]
- Ross AF. Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology. 1961;14:329–339. doi: 10.1016/0042-6822(61)90318-x. [DOI] [PubMed] [Google Scholar]
- Ryals J, Weymann K, Lawton K, Friedrich L, Ellis D, Steiner H-Y, Johnson J, Delaney TP, Jesse T, Vos P et al. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell. 1997;9:425–439. doi: 10.1105/tpc.9.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner H-Y, Hunt MD. Systemic acquired resistance. Plant Cell. 1996;8:1809–1819. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- Shorey HH, Andres LA, Hale RL. The biology of Trichoplusia ni (Lepidoptera: Noctuidae): life history and behavior. Ann Entomol Soc Am. 1962;55:591–597. [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Dynamics of disease resistance polymorphism at the RPM1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Kroymann J, Mitchell-Olds T. Plant-insect interactions. Curr Opin Plant Biol. 1999;2:268–272. doi: 10.1016/S1369-5266(99)80048-X. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Pittendrigh BR, Kroymann J, Weniger K, Fritsche J, Bauke A, Mitchell-Olds T. Induced plant defense responses against chewing insects: Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 2000;124:1007–1018. doi: 10.1104/pp.124.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout MJ, Fidantsef AL, Duffey SS, Bostock RM. Signal interactions in pathogen and insect attack: systemic plant-mediated interactions between pathogens and herbivores of the tomato, Lycopersicon esculentum. Physiol Mol Plant Pathol. 1999;54:115–130. [Google Scholar]
- Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Fidantsef AL, Duff SS, Bostock RM. Trade-offs in plant defense against pathogens and herbivores: a field demonstration of chemical elicitors of induced resistance. J Chem Ecol. 1999;25:1597–1609. [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Uknes S, Winter AM, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward E, Ryals J. Biological induction of systemic acquired resistance in Arabidopsis. Mol Plant-Microbe Interact. 1993;6:692–698. [Google Scholar]
- van Wees SC, de Swart EA, van Pelt JA, van Loon LC, Pieterse CM. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:8711–8716. doi: 10.1073/pnas.130425197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Luijendijk M, Smoorenburg I, van Loon LC, Pieterse CM. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol. 1999;41:537–549. doi: 10.1023/a:1006319216982. [DOI] [PubMed] [Google Scholar]
- Vernooij B, Friedrich L, Morse A, Reist R, Kolditz-Jawhar R, Ward E, Uknes S, Kessmann H, Ryals J. Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell. 1994;6:959–965. doi: 10.1105/tpc.6.7.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Payne GP, Moyer MB, Williams SC, Dincher SS, Sharkey KC, Beck JJ, Taylor HT, Ahl-Goy P, Meins FJ et al. Differential regulation of β-1,3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 1991;96:390–397. doi: 10.1104/pp.96.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G-L, Katagiri F, Ausubel FM. Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol Plant-Microbe Interact. 1993;6:434–443. doi: 10.1094/mpmi-6-434. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream of salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]