Abstract
The ability of meristems to continuously produce new organs depends on the activity of their stem cell populations, which are located at the meristem tip. In Arabidopsis, the size of the stem cell domain is regulated by two antagonistic activities. The WUS (WUSCHEL) gene, encoding a homeodomain protein, promotes the formation and maintenance of stem cells. These stem cells express CLV3 (CLAVATA3), and signaling of CLV3 through the CLV1/CLV2 receptor complex restricts WUS activity. Homeostasis of the stem cell population may be achieved through feedback regulation, whereby changes in stem cell number result in corresponding changes in CLV3 expression levels, and adjustment of WUS expression via the CLV signal transduction pathway. We have analyzed whether expression of CLV3 is controlled by the activity of WUS or another homeobox gene, STM (SHOOT MERISTEMLESS), which is required for stem cell maintenance. We found that expression of CLV3 depends on WUS function only in the embryonic shoot meristem. At later developmental stages, WUS promotes the level of CLV3 expression, together with STM. Within a meristem, competence to respond to WUS activity by expressing CLV3 is restricted to the meristem apex.
The shoot apical meristem (SAM) of higher plants is formed during embryogenesis and gives rise to leaves and stem after germination (Steeves and Sussex, 1989). The side branches of angiosperms originate from axillary meristems that arise in the axils of leaves, whereas flowers are formed from secondary meristems that are initiated at the flanks of the SAM. The cells in these three types of meristems are arranged in three clonal layers (L1, L2, and L3). All organs and also floral meristems are produced at the flanks of the meristem dome in the peripheral zone. Loss of cells from the meristem during organ formation has to be compensated by divisions of cells in the central zone that act as pluripotent stem cells. When these stem cells divide, their daughter cells are displaced to the periphery, where they will be incorporated into organ primordia and eventually differentiate. Therefore, the ability of meristems to continuously produce new organs depends on the activity of their stem cell populations. Stem cells can be initiated repeatedly during plant development, whenever a new axillary or floral meristem is formed. The stem cells of floral meristems are not permanent, but lose their undifferentiated state when the inner set of floral organs is produced. Thus, stem cell identity may represent a flexible state that is subject to both positive and negative regulation.
A current model proposes that the size of the stem cell population in meristems is controlled by a negative feedback regulation between two pathways that promote or restrict stem cell number (Waites and Simon, 2000). The WUS (WUSCHEL) gene of Arabidopsis, encoding a nuclear-localized homeodomain protein, is expressed underneath the stem cell domain of shoot and floral meristems. In wus mutants, the cells in the central zone differentiate prematurely, indicating that WUS promotes the initiation and maintenance of stem cells at the apex (Laux et al., 1996; Mayer et al., 1998). However, WUS function is not required for meristem initiation per se because wus mutants still form axillary and floral meristems. The expression of WUS is controlled by the three CLAVATA genes (CLV1, 2, and 3) that act together in a signal transduction pathway and restrict stem cell fate (Brand et al., 2000; Schoof et al., 2000). The CLV3 gene is expressed in the putative stem cells at the apex of shoot, floral, and axillary meristems (Fletcher et al., 1999). The CLV3 protein is proposed to be secreted from the stem cells and to act as a signaling molecule that binds to and thereby activates a heterodimeric receptor complex, consisting of the CLV1 and CLV2 proteins (Trotochaud et al., 2000). A consequence of this receptor activation is the restriction of WUS expression to a small domain in the deeper regions of the meristem (Brand et al., 2000). Increased expression of CLV3 in transgenic plants causes a further repression of WUS expression, and a loss of stem cells (Brand et al., 2000). These results, together with studies on the formation of active CLV receptor complexes, indicate that the size of the stem cell population is controlled by the availability of CLV3 to regulate WUS via the CLV pathway (Brand et al., 2000; Trotochaud et al., 2000).
The genes that control the expression levels and pattern of CLV3 are not yet known, but several observations suggest that one important activator of CLV3 expression could be WUS itself. First, non-differentiating cells that express CLV3 accumulate in clv1, clv2, or clv3 mutants due to unrestricted WUS expression (Fletcher et al., 1999; Brand et al., 2000). Second, transgenic plants that express WUS from the CLV1 promoter in a larger domain within the meristem accumulate undifferentiated cells that express CLV3, resembling clv mutants (Schoof et al., 2000). Third, when CLV3 expression in the meristem is under control of a heterologous promoter (the promoter of the UFO gene), the meristem fails to maintain stem cells and CLV3 expression from its own promoter, indicating that CLV3 expression and stem cell identity are subject to the same positive control mechanisms (Brand et al., 2000). Homeostasis of the stem cell population is then achieved through negative feedback regulation, whereby any increase or decrease in stem cell number results in a corresponding change in CLV3 transcript levels, and an immediate adjustment of WUS expression via the CLV signal transduction pathway. A simple explanation for these observations is that WUS directly promotes both stem cell fate and CLV3 expression. Alternatively, WUS could only be required to maintain a stem cell population, and CLV3 expression in these stem cells is promoted by other factors.
A second candidate regulator of CLV3 expression is the STM (SHOOT MERISTEMLESS) gene of Arabidopsis. STM encodes a homeodomain protein that is expressed throughout shoot and floral meristems, but is excluded from organ primordia (Long et al., 1996). Plants homozygous for loss-of-function mutations in the STM gene fail to initiate a recognizable SAM during embryogenesis (Barton and Poethig, 1993). In weak stm mutants, a shoot meristem can be formed that arrests prematurely, indicating that STM is also required at later stages for the maintenance of a self-renewing stem cell population in meristems (Endrizzi et al., 1996). Although both STM and WUS are independently activated in the embryonic shoot meristem, each appears to require the function of the other at the seedling stage (Mayer et al., 1998).
As a first step to investigate the regulation of CLV3, we have studied the dependence of CLV3 expression on STM and WUS function because both genes are required for the formation and maintenance of stem cell populations in meristems.
RESULTS
Construction of a CLV3 Reporter Gene
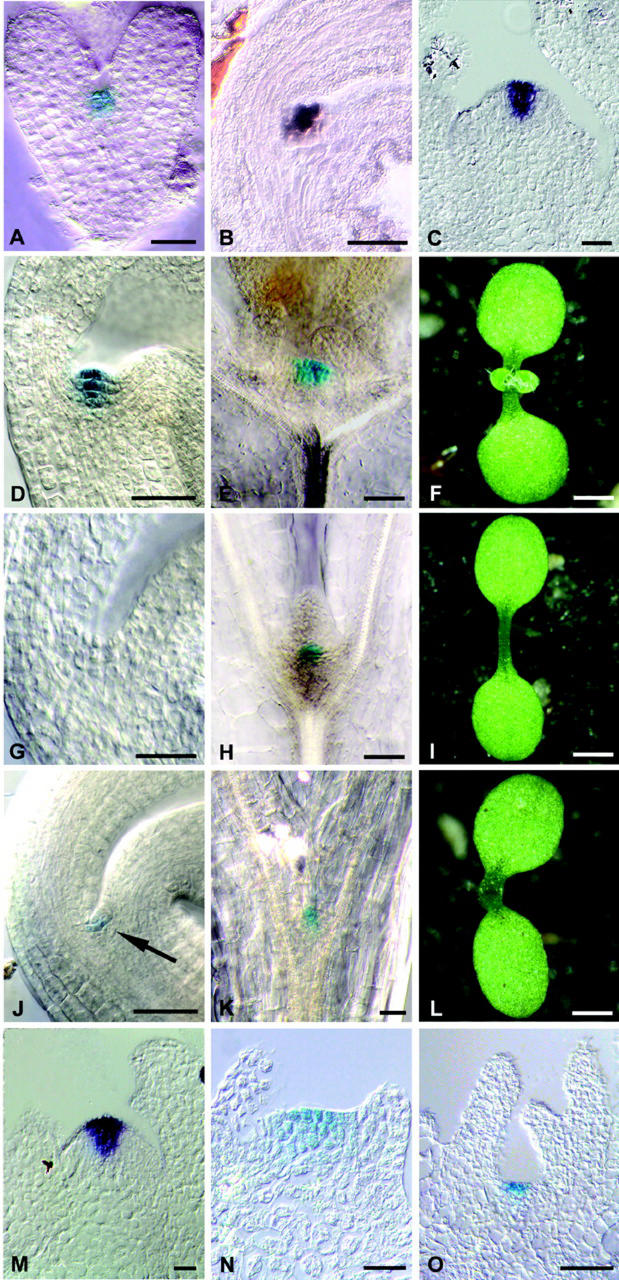
To facilitate expression studies with CLV3, we constructed a CLV3::GUS transgene that expresses the GUS (β-glucuronidase) reporter gene under control of CLV3 regulatory sequences. A transcriptional fusion of 1.5-kb genomic sequences immediately upstream of the translational start codon of CLV3 to a GUS reporter gene, pBU16D7, was not sufficient to express the GUS reporter in the pattern typical for CLV3. However, the weak clv3-3 allele carries a T-DNA insertion 175 bp downstream of the polyadenylation site, potentially disrupting or distancing an enhancer element (Fletcher et al., 1999). Therefore, we tested if additional sequences are required for CLV3 expression by inserting the 1.2-kb DNA sequences 3′ to the CLV3 translational stop codon downstream of the GUS gene in pBU16D7 to give pBU16 (CLV3::GUS). After plant transformation and selection of transgenic Arabidopsis, GUS activity in the plants was analyzed in whole-mount preparations and tissue sections. In wild-type Arabidopsis, CLV3 RNA is detected from the heart stage of embryogenesis onwards in the presumptive SAM (Fletcher et al., 1999). During further development, CLV3 remains expressed in the putative stem cells that are located in the central zone of the SAM, the axillary meristems, and in floral meristems. When CLV3::GUS transgenic plants were assayed for GUS activity, staining was first detectable at the heart stage of embryogenesis in the presumptive SAM (Fig. 1A). At later stages, a dark-blue GUS staining was visible in the central zone of the SAM, axillary, and floral meristems, coinciding with the CLV3 RNA pattern (Fig. 1, B–F). Occasionally, a faint GUS staining was observed in cells immediately adjacent to the central zone (Fig. 1D). Because RNA in situ hybridizations confirmed that the pattern of GUS RNA in CLV3::GUS plants coincided with the CLV3 RNA distribution (Fig. 1C), we assume that GUS staining in neighboring cells is due to the stability of the GUS protein in the descendants of CLV3-expressing cells. We concluded that the CLV3::GUS transgene can be used as a reliable reporter for CLV3 expression in planta.
Figure 1.
Dependence of CLV3 expression on WUS and STM. A, GUS-stained heart stage CLV3::GUS embryo. B, CLV3 expression in a bent cotyledon stage embryo, detected by in situ hybridization with a CLV3 probe. C, CLV3 expression in a CLV3::GUS inflorescence meristem, detected by in situ hybridization with a GUS probe. D, GUS-stained mature CLV3::GUS embryo; compare with B. E, GUS-stained wild-type seedling 10 d after germination (d.a.g.). F, Wild-type seedling 10 d.a.g. The first leaf pair is visible. G, GUS-stained mature CLV3::GUS/wus-1 embryo. CLV3 expression is not detectable. H, GUS-stained CLV3::GUS/wus-1 seedling 10 d.a.g. I, wus-1 seedling 10 d.a.g. J, GUS-stained mature CLV3::GUS/stm-11 embryo showing CLV3 expression (arrow). K, GUS-stained CLV3::GUS/stm-11 seedling 10 d.a.g. showing CLV3 expression. L, An stm-11 seedling 10 d.a.g. has formed cotyledons, but no SAM is visible. M, Wild-type CLV3::GUS axillary meristem 21 d.a.g. GUS RNA is detected by in situ hybridization. N, GUS-stained CLV3::GUS/wus-1 axillary meristem 21 d.a.g. O, GUS-stained CLV3::GUS/stm-11 axillary meristem 21 d.a.g. Scale bars in F, I, and L = 1 mm; in all other figures, scale bars = 20 μm.
Requirement of WUS Function for CLV3 Expression during Development
wus mutants fail to maintain a sufficient number of stem cells, resulting in a meristem arrest after formation of a few leaves (Laux et al., 1996). To test whether WUS activity is required not only for stem cell specification, but also for the early expression of CLV3 during embryogenesis, we introduced the CLV3::GUS reporter into wus-1 mutants. Plants homozygous for the CLV3::GUS transgene that carried the loss-of-function wus-1 allele were obtained, and after self-fertilization, mature embryos were assayed for reporter gene activity. Of 259 embryos analyzed, 58 (22.4%, expected 25%) were identified as wus-1 mutants by the lack of a SAM (Fig. 1, F and I). All wus-1 embryos failed to express CLV3::GUS, indicating that WUS function is required for the early activation of CLV3 expression in the embryo (Fig. 1G; Table I).
Table I.
CLV3∷GUS expression in embryos and seedlings
| Genotype | Embryo
|
Seedling
|
||||
|---|---|---|---|---|---|---|
| n | g.s. | % | n | g.s. | % | |
| wt | 414 | 405 | 96 | 207 | 198 | 96 |
| wus-1 | 58 | 0 | 0 | 106 | 83 | 78 |
| stm-11 | 86 | 6 | 7 | 59 | 15 | 25 |
| stm-5 | – | n.d. | – | 48 | 27 | 56 |
GUS staining of mature embryos or seedlings at 10 d.a.g. of the indicated genotypes, carrying the CLV3∷GUS reporter. The no. of seedlings that showed a GUS signal (g.s.), percentage of the total (%), and the total of individuals analyzed (n) are tabulated. n.d., Not determined.
During further development, wus-1 mutant seedlings initiate leaves, axillary meristems, and inflorescences with a reduced number of floral organs (Laux et al., 1996). Therefore, we tested wus-1 seedlings for expression of the CLV3::GUS marker, and found GUS-expressing cells at 9 to 10 d.a.g. (Fig. 1H; Table I). At this stage, a flattened SAM and two leaf primordia had been formed, and 83 of 106 wus-1 seedlings expressed the CLV3 reporter gene at low levels in a small group of cells in the center of the SAM. In later arising axillary meristems, strong GUS staining was observed in the central zone in a pattern comparable with wild-type meristems (Fig. 1, M and N). These results indicate a decreasing dependence of CLV3 expression in WUS function during development: WUS is required to establish CLV3 expression in the embryo, and for the up-regulation of CLV3 in the SAM after germination. In axillary meristems, establishment of CLV3 expression is WUS independent, and therefore may be controlled by other factors that could act redundantly with WUS.
WUS Promotes CLV3 Expression at the Meristem Tip
To test whether WUS is sufficient to promote CLV3 expression, we designed a transgene that allows the induction of WUS activity in the whole plant. A translational WUS-GR fusion was designed that is expressed under control of the CaMV (cauliflower mosaic virus) 35S promoter in transgenic plants (WUS-GR). Nuclear entry of the fusion protein now depends on the addition of the synthetic glucocorticoid hormone dexamethasone (Dex; Lloyd et al., 1994). The WUS-GR transgene was transformed into Arabidopsis plants carrying the CLV3::GUS reporter gene. The expression of the WUS-GR transgene was confirmed by RNA in situ hybridization (data not shown). If the presence of the WUS protein in the nucleus is sufficient to activate CLV3 expression, we would expect GUS activity throughout Dex-treated plants.
Untreated transgenic plants were phenotypically wild type and showed GUS-staining patterns typical for CLV3. However, seedlings that were treated with Dex during germination formed broad but flat shoot meristems that showed an intense GUS staining that was restricted to the apical cell layers of the SAM (Fig. 2A, compare with Fig. 1D).
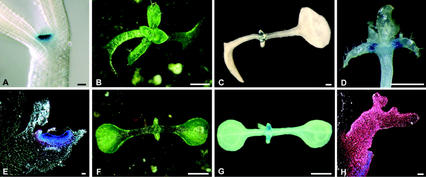
Figure 2.
Induction of CLV3 expression by WUS. A, GUS-stained CLV3::GUS/WUS-GR seedling 2 d.a.g., induced with Dex during germination. B, CLV3::GUS/WUS-GR seedling 24 d.a.g., induced with Dex at 4 d.a.g. C, GUS-stained seedling, as in B. D, GUS-stained CLV3::GUS/WUS-GR seedling 28 d.a.g., induced with Dex at 4 d.a.g. The cotyledons have been removed and GUS staining is visible in the leaf axils. E, Longitudinal section through a GUS-stained leaf, as in D; strong GUS expression is found in the apical cell layers of the laterally expanded axillary meristem. F, CLV3::GUS/WUS-GR/STM-GR seedling 24 d.a.g., induced with Dex 4 d.a.g. (compare with B). G, GUS-stained seedling, as in F; GUS expression is found in leaves. H, Section through a GUS-stained primary leaf of G, viewed with dark-field illumination. GUS signal (red) is found in all leaf cells. Scale bars in A, E, and H = 20 μm; in B through D, F, and G, scale bars = 1 mm.
When seedlings were sprayed with Dex 4 d.a.g., the cotyledons curled downwards and leaves that initiated during further development failed to expand (Fig. 2B). In addition to the staining in the shoot meristem, strong GUS staining was now found in axillary meristems that had formed on the adaxial side of leaves close to their base at 21 d.a.g. (Fig. 2. C–E). Compared with wild type (Fig. 1M), these axillary meristems did not form a typical meristematic dome, but appeared to be laterally expanded, thus resembling the SAM in Dex-treated WUS-GR plants. Activity of the CLV3 reporter was highest in the meristematic cells of the apical cell layers, and decreased toward the deeper layers and the periphery, where initiation of new organ primordia was observed. Although leaf development was affected by the activation of the WUS-GR transgene, we never observed any GUS staining in leaf tissues. Thus, WUS expression is not sufficient to activate CLV3 expression on its own, and additional localized factors may be required to cooperate with WUS in the activation of target genes.
Dependence of CLV3 Expression on STM
To test whether STM function is required for expression of CLV3, we introduced the CLV3::GUS transgene into plants carrying the loss-of-function allele stm-11 (Long et al., 1998). After self-fertilization, these plants segregated 25% stm-11 mutant embryos, which were identified by the lack of a visible SAM. Plants homozygous for CLV3::GUS were identified in the next generation, and used for the expression studies. Although mature wild-type embryos showed the typical CLV3::GUS staining in the SAM, most stm-11 mutant embryos did not express the CLV3::GUS reporter (Table I). However, in six of 86 stm-11 mutant embryos analyzed (7% of the total), we observed weak GUS staining in two to four cells between the cotyledons where the SAM is formed in wild-type embryos, indicating that STM function is not absolutely required for CLV3 expression in the embryo (Fig. 1J).
At the seedling stage, stm-11 mutants form partially fused cotyledons (Fig. 1L) and, even in the absence of an embryonic SAM, malformed leaves can be initiated at the cotyledon bases (Long and Barton, 1998; data not shown). stm-11 mutants were then tested for activity of CLV3::GUS 10 d.a.g., and GUS-positive cells were detected in the basal region of the cotyledons in 15 of 59 mutant seedlings (25% of the total; Table I; Fig. 1K). CLV3::GUS expression was also found in the axillary meristems of stm-11 mutants 21 d.a.g. (Fig. 1O).
To exclude effects due to the genetic background of stm-11, we also analyzed CLV3 expression in another strong stm mutant, stm-5 (Endrizzi et al., 1996). Under our growth conditions, stm-5 mutants are phenotypically weaker than stm-11. In plants homozygous for the stm-5 allele, GUS expression was observed in 27 of 48 seedlings (56% of the total) at 10 d.a.g. (Table I).
Taken together, our observations indicate that STM is not generally required for CLV3 expression in embryos and young seedlings. However, CLV3 expression levels are reduced in stm mutants, suggesting a role for STM in the up-regulation of CLV3 expression.
Ectopic STM Expression Is Not Sufficient to Activate CLV3 in Non-Meristematic Tissues
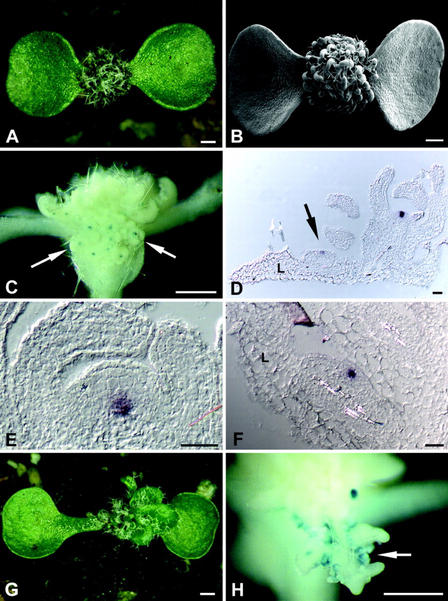
To further analyze the role of STM in the regulation of CLV3, we constructed a transgene that allows ectopic expression of STM. Previous studies have shown that transgenic plants expressing STM under control of the constitutive CaMV35S promoter are severely stunted with a highly disorganized shoot meristem, and arrest development at an early seedling stage (Williams, 1998). To control ectopic STM activity during development, we fused the STM coding region with the hormone-binding domain of the rat glucocorticoid receptor. This STM-GR fusion protein is constitutively expressed from the CaMV35S promoter throughout the plant. The STM-GR transgene was introduced into Arabidopsis plants carrying the CLV3::GUS reporter. If STM can act on its own to promote CLV3 expression, we expected to observe blue GUS staining in all tissues after STM activation by Dex addition.
The expression of the STM-GR transgene was confirmed by reverse transcriptase (RT)-PCR and RNA in situ hybridization (data not shown). Without Dex application, transgenic lines were indistinguishable from the wild type and expressed the CLV3 reporter gene in a normal pattern. Plants that were treated with Dex during germination carried more rounded cotyledons and small, lobed leaves with ectopic stipules along the leaf margin and fully differentiated trichomes (Fig. 3, A and B). On the adaxial (upper) leaf surface, ectopic meristems originated that again initiated new leaves, resulting in the formation of a dense array of small leaves and meristems. In some plants, ectopic meristems were also found on the surface of the cotyledons. Strong expression of the CLV3 reporter gene was detected in the SAM and ectopic meristems, in a pattern comparable with CLV3 expression in wild-type shoot meristems (Fig. 3, C and D). Occasionally, weak GUS staining was observed in the vasculature of the cotyledons and in stipules. Sensitive fluorometric GUS assays of leaf samples revealed an increase in GUS activity above background levels not before 5 d after Dex treatment (not shown). One explanation for this temporal delay in CLV3 activation is that STM does not act as a direct activator of CLV3. Instead, STM promotes the formation of new meristems, where CLV3 expression may be activated by other meristem-specific genes, e.g. WUS. We analyzed the expression pattern of WUS and CLV3 in STM-GR plants by RNA in situ hybridization. WUS was expressed in a small group of cells underneath the CLV3 expression domain, comparable with the expression pattern in wild-type meristems (Fig. 3, D–F). To test if WUS function is required for CLV3 expression, we introduced the STM-GR transgene into wus loss-of-function mutants. After crossing the STM-GR transgene into wus-1 mutants and self-fertilization, homozygous wus-1 seedlings were identified in the next generation and sprayed with Dex to activate STM. The leaves that were initiated during the first 3 weeks after Dex application were lobed and carried ectopic stipules, resembling leaves of STM-GR plants (Fig. 3, G and H). However, ectopic meristems were only occasionally formed, indicating that WUS function supports their induction by STM. CLV3::GUS expression was observed occasionally in ectopic stipules, but not in other non-meristematic tissues of these plants (Fig. 3H).
Figure 3.
Induction of CLV3 expression by STM. A, STM-GR seedling 24 d.a.g., induced with Dex at 4 d.a.g.; a dense array of leaves with new meristems is formed. B, Scanning electron micrograph of a seedling, as in A. C, GUS-stained CLV3::GUS/STM-GR seedling, as in A; ectopic meristems are initiated on the adaxial leaf surface, GUS signal is found in the center of these meristems (arrow). D, Section through a leaf of a STM-GR seedling, as in A. Expression of CLV3 is detected by RNA in situ hybridization in the central zone of ectopic meristems (arrow). E, Section through the SAM of a STM-GR seedling, as in A. WUS expression, detected by RNA in situ hybridization, is found in the inner regions of the meristem, similar to wild type. F, Section through an axillary meristem of a STM-GR seedling, as in A. The domain of WUS expression resembles the expression in the SAM. E and F, No ectopic expression of WUS is observed. G, CLV3::GUS/STM-GR/wus-1 seedling 24 d.a.g.; Dex induction commenced 7 d.a.g. when the seedlings could be phenotypically identified as wus mutants. H, GUS-stained seedling as in G; GUS signal was detected in ectopic stipules that occasionally formed on the leaf margins (arrow). Note the absence of ectopic meristems. L, Leaf. Scale bars in A through C, G, and H = 500 μm; in D through F, scale bars = 20 μm.
Co-Expression of WUS and STM Induces CLV3 Expression in Leaves
Because ectopic expression of either WUS or STM was not sufficient to activate CLV3 expression in non-meristematic regions, we then analyzed the consequence of misexpressing both genes together. After crossing of STM-GR transgenic plants to WUS-GR plants, the resulting F1 plants were allowed to self-fertilize, and seedlings of the F2 generation were induced with Dex 4 d.a.g. Within 2 d after Dex application, seedlings carrying only the STM-GR transgene can be distinguished from WUS-GR seedlings by their cotyledon shape. We identified STM-GR/WUS-GR seedlings as a new phenotypic class, characterized by upward curling of the cotyledons (Fig. 2F, compare with Figs. 2B and 3A). During the next 20 d after Dex treatment, small leaves were initiated that failed to expand. In contrast to STM-GR or WUS-GR plants, expression of the CLV3 reporter gene was detected throughout the leaves of the STM-GR/WUS-GR plants, indicating that both STM and WUS function are required and sufficient to activate CLV3 expression in non-meristematic cells (Fig. 2, G and H).
WUS Can Activate CLV3 within the Same Cells
In the wild type, a small group of cells underlying the stem cell domain expresses both WUS and STM. However, CLV3 RNA is not found in these cells, but in a separate domain at the tip of the meristem. The spatial separation of the WUS expression domain from the stem cell domain expressing CLV3 suggests that either the WUS protein itself, or a WUS-dependent signal, is transmitted to the cells at the apex of the meristem (Mayer et al., 1998). Is this non-cell autonomy of WUS required for CLV3 activation?
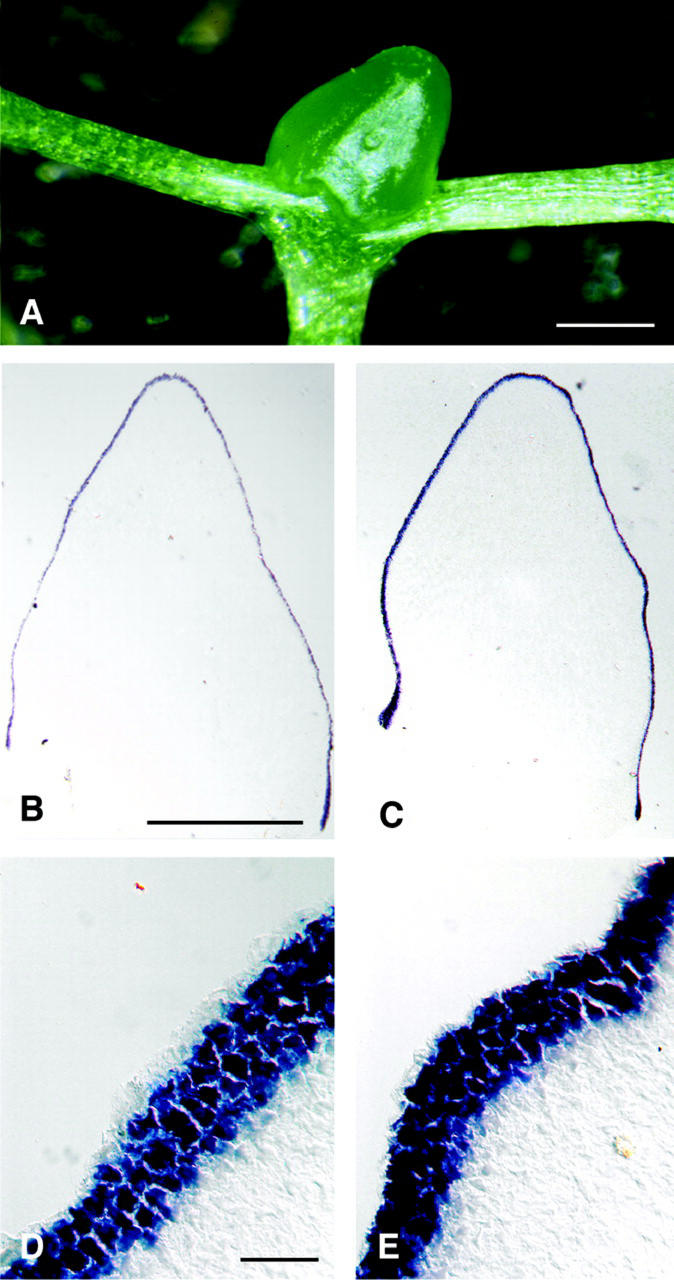
We tried to circumvent a non-cell-autonomous function of WUS by expressing WUS directly in the stem cell domain under control of the CLV3 promoter. We replaced the GUS reporter gene in pBU16 with a WUS cDNA, and the resulting transgene CLV3::WUS was introduced into wild-type Arabidopsis. Four weeks after germination, transgenic plants had formed an apparently normal hypocotyl and cotyledons. Whereas non-transgenic control plants had already initiated six visible leaves, the CLV3::WUS transgenic plants produced only large domes of cells between the cotyledons (Fig. 4A). These apices resembled enlarged meristems that lacked organ primordia. The outer cell layers consisted of small, meristematic cells that covered an inner mass of large and vacuolated cells. Interestingly, we never observed any vascular tissue in these aberrant meristems. RNA in situ hybridization revealed that CLV3 was now expressed not only in a few cells at the meristem apex, but at very high levels throughout the outer three cell layers, thus covering the apical dome (Fig. 4, C and E). WUS RNA was found only in cells that expressed CLV3 (Fig. 4, B and D), but not in its normal expression domain underneath the stem cells. This could be explained if WUS activity in the stem cell domain can directly induce CLV3 expression, resulting in a strong activation of the CLV signal transduction pathway, and repression of WUS transcription from its own promoter. Undifferentiated cells were still confined to the three outer meristem layers of the CLV3::WUS transgenic plants, indicating that WUS can promote stem cell fate in the same cells where it is expressed. Importantly, WUS fails to induce CLV3 expression in deeper regions of the meristem. We conclude that non-cell autonomy of WUS is not a prerequisite for CLV3 activation in the meristem.
Figure 4.
Cell-autonomous induction of CLV3 by WUS in the stem cell domain. A, CLV3::WUS seedling 28 d.a.g.; a large apical dome is formed between the cotyledons, but organs are missing. B, In situ detection of WUS RNA in sections through the apical dome of a CLV3::WUS seedling. WUS RNA is found only in three apical cell layers, but not in the deeper regions of the meristem. D, High magnification view of the outer cell layers in B. C, As in B, but detection of CLV3 RNA. CLV3 and WUS are expressed in the same cells. E, High magnification of C. Scale bars in A and B = 500 μm; scale bars in D = 10 μm.
DISCUSSION
Pluripotent stem cells serve as a source for new cells to compensate for cell loss from the meristem during organ formation. Homeostasis of the shoot meristem requires a continuous adjustment of stem cell number, which can be achieved through the antagonistic activities of WUS and the CLV signal transduction pathway. A current model proposes that WUS induces both stem cell fate and CLV3 expression; the size of the stem cell population is then controlled by a simple feedback system, where CLV3 activates a signal transduction pathway that results in a down-regulation of WUS expression, corresponding to CLV3 levels (Brand et al., 2000; Schoof et al., 2000).
Our analysis of CLV3 expression in wild-type and wus mutant embryos showed that WUS function is required for activation of CLV3 expression during embryogenesis. This could suggest that WUS is involved in the transcriptional control of CLV3. However, the lack of CLV3 expression could also be an indirect consequence of wus mutant embryos failing to initiate the cells that normally express CLV3. Activity of WUS is not confined to the embryonic SAM because inflorescence and floral meristems of wus mutants do not maintain permanent stem cells and abort organ formation prematurely (Laux et al., 1996). Importantly, the ability of wus mutants to initiate meristems at all reflects the activity of additional regulators that promote meristem function. We found that CLV3 is expressed in the abnormal shoot meristems of wus mutant seedlings and also in later arising axillary meristems in a pattern comparable with wild type, indicating that WUS is required for CLV3 expression only during embryogenesis. During seedling development, expression of CLV3 in the SAM and axillary meristems is promoted by additional factors that act partially redundantly with WUS to control stem cell fate.
Misexpression of WUS in the whole plant did not result in a widespread up-regulation of CLV3 expression. Instead, increased CLV3 expression was observed only in the SAM and axillary meristems, and within these meristems, high levels of CLV3 expression were found in the apical cell layers. This observation indicates that only cells at the tip of established meristems are competent to respond to WUS expression or to a WUS-derived signal (an alternative, but less likely explanation would be that CLV3 expression is actively repressed in all other cells). Consistent with a requirement for additional activators, Lenhard et al. (2001) reported that misexpression of WUS in whorls 2 and 3 of flowers, under control of the AP3 promoter, was insufficient to activate CLV3 expression in developing organ primordia.
In wild type, the STM gene is expressed in the entire meristem, but down-regulated at sites of organ formation (Long et al., 1998). To test the role of STM in the regulation of CLV3 expression, we first analyzed CLV3 expression in stm mutant embryos and seedlings. Although stm mutants are not able to form a functional SAM, we detected low levels of CLV3 expression in embryos, seedlings, and axillary meristems. In all cases, the patterns of CLV3 expression resembled those found in wild-type plants. At least part of this residual CLV3 expression could be due to WUS activity because WUS is initially expressed at a position corresponding to the shoot meristem in stm mutant embryos (Mayer et al., 1998). However, no WUS RNA was detected in the apices of stm seedlings (Mayer et al., 1998), suggesting that neither STM nor WUS are required to define the expression domain of CLV3.
Ectopic expression of STM resulted in the formation of lobed leaves and new, functional meristems on the leaf surface that expressed CLV3 in a wild-type pattern. In addition, CLV3 was occasionally expressed in the vasculature of some leaves and in ectopic stipules. The gain-of-function phenotype of an STM-related homeobox gene, KNAT1, shows a close association between ectopic meristems and leaf veins (Chuck et al., 1996). Thus, expression of CLV3 in vasculature tissue of STM-GR plants may reflect an increased capacity of vein tissue for meristem formation. Stipules originate during leaf formation in pairs at the leaf base, and several mutant backgrounds or transgenes that cause misexpression of knox genes in leaves result in the co-induction of ectopic stipules along the leaf margin and ectopic meristems, which has led to an interpretation of stipules as an indicator for meristem-like activity (Ori et al., 2000). However, expression of the CLV3::GUS reporter in STM-GR plants was not observed throughout leaves or hypocotyls, indicating that STM expression is not generally sufficient to activate CLV3 expression in non-meristematic tissues. Within ectopic meristems, we found both WUS and CLV3 expression in a wild-type pattern, suggesting that STM promotes CLV3 expression only indirectly by establishing a meristem-specific program of gene expression. We were not yet able to resolve whether the expression of CLV3 in ectopic meristems of STM-GR plants depends on WUS because the number of ectopic meristems that initiated in a wus mutant background was insufficient for a detailed analysis.
Co-expression of STM-GR and WUS-GR resulted in GUS staining throughout leaf tissue, implying that activity of both homeobox genes is necessary and sufficient to activate CLV3. The STM gene product itself, or target genes that are controlled by STM, therefore may be required to cooperate with WUS to promote CLV3 expression. This would explain why expression of WUS in the AP3 domain was not sufficient to control CLV3 expression in floral organs, where STM transcripts are down-regulated (Lenhard et al. , 2001). Notably, the levels of CLV3 expression in the leaves of STM-GR/WUS-GR plants were lower than in wild-type meristems. Given that co-expression of STM and WUS suffices to activate basic levels of CLV3 expression, we may speculate on what accounts for the separation of the CLV3 expression domain (at the tip of the meristem) from the WUS domain (in deeper cell layers) in meristems. One possible explanation is that WUS acts only non-cell-autonomously in meristems, and causes the production or activation of a signal that is perceived by competent cells at the meristem apex. When we expressed WUS under control of the CLV3 promoter, thus obliterating any requirement for a signaling process, the number of non-differentiating cells increased dramatically compared with wild type. This can be explained if WUS can promote stem cell fate and activate CLV3 expression cell autonomously at the meristem apex. As a consequence, WUS expression levels controlled by the CLV3 promoter increase further, thus creating a positive feedback loop of gene expression. WUS expression from its own promoter in deeper regions of the meristem is then expected to be switched off due to increased CLV3 dependent signaling via the CLV pathway (Brand et al., 2000). If WUS can act cell autonomously and no signaling process is required, the lack of CLV3 expression within the WUS expression domain could be explained with a requirement for localized cofactors to cooperate with WUS, allowing for CLV3 expression only at the apex. Further support for this view comes from the observation that constitutive expression of WUS throughout the plant activates high levels of CLV3 expression only in the apical cell layers of meristems. However, we found that STM, together with WUS, can activate CLV3 expression at low levels in leaves. This could indicate that the regulation of CLV3 expression also involves negative control mechanisms that are acting only in meristems.
In conclusion, we have shown here that only early CLV3 expression in the embryonic SAM depends on WUS function. During later stages of development, neither WUS nor STM are required to establish the pattern of CLV3 expression in the central zone of the SAM and axillary meristems. The role of WUS is now confined to increase the levels of CLV3 expression, together with STM. Within a meristem, only cells at the meristem apex can express CLV3, indicating that localized (and as yet unidentified) factors or a specific cellular competence are required to respond to WUS activity.
MATERIALS AND METHODS
Plant Material and Genetics
The Arabidopsis Columbia ecotype was used for Agrobacterium tumefaciens-mediated vacuum transformation (Clough and Bent, 1998). Transgenes were introduced into mutant backgrounds by crosses or by direct transformation. The stm-5, stm-11, and wus-1 mutants used in this study were described previously (Endrizzi et al., 1996; Laux et al., 1996; Long et al., 1998).
GUS Staining
We used the GUS staining protocol of Sieburth and Meyerowitz (1997) with minor modifications. Plants were incubated in a staining solution [50 mm NaPO4, 5 mm K3Fe(CN)6, 5 mm K4Fe(CN)6, and 10 mm X-Gluc] at 37°C for at least 3 h. The tissue was then fixed, cleared, and embedded in paraffin or visualized as whole mount. Sections (7 μm) were cut from the embedded tissue.
Molecular Methods
In situ hybridizations were performed as described previously (Brand et al., 2000). To construct the CLV3::GUS marker gene, we isolated the CLV3 regulatory sequences by PCR amplification from vector gE5.5CLV3 (Brand et al., 2000) carrying a genomic DNA fragment comprising the CLV3 gene. The 1.5-kb DNA promoter fragment 5′ to the transcriptional start of the CLV3 gene was amplified using the 3′primer 551 up-(5′CCCCCTGCAGAGAG-AAAGTGACTGAGTGAG3′), introducing a PstI site, and the 5′primer forward-(GTAAAAGGACGGCCAG) that binds to vector sequences. The enhancer fragment 1.2 kb downstream of the transcriptional stop was amplified with the 5′primer 553dow-(5′AAAAGCGGCCGCCCTAATCTCTTGTTGCTTTAA3′), introducing a NotI site, and the 3′primer 554dow-(5′CCCCGAGCTCTATGTGTGTTTTTTCTAAACAA3′), which introduces a SacI site. Both fragments were cloned into the pGreen vector (Hellens et al., 2000) using the EcoRI/PstI and NotI/SacI sites to give pBU14. The uidA (GUS) coding sequence was cloned between these two fragments into the SmaI site of pBU14 to give pBU16 (CLV3::GUS). Transgenic plants are selected by Basta resistance.
To obtain a CaMV35S::STM-GR transgene, we amplified a STM cDNA by RT-PCR using the following primers: STM5′-(5′GGGGTCTAGAGATGGAGAGTGGTTCCAACAGCA3′), which introduces an XbaI site 5′ to the translation start codon; and STM3′-(5′GGGGGGATCCGCAAGCATGGTGGAGGAGATGTGA3′), which replaces the stop codon with a BamHI site. The correct PCR products were cloned into the XbaI and BamHI sites of the pBIΔGR vector (Lloyd et al., 1994), in frame with the hormone-binding domain of the glucocorticoid receptor to give pRS4 (STM-GR). Transgenic plants were selected for kanamycin resistance.
The CaMV35S::WUS-GR transgene was constructed by PCR amplification of a WUS cDNA using the primers WUS5′-(5′TAGAGGATCCTATGGAGCCGCCACAGCATCAG3′) and WUS3′-(5′TTCAGGATCCTCGTTCAGACGTAGCTCAAGAG3′). The WUS5′primer introduces a BamHI site 5′to the translation start codon. The WUS3′ primer replaces the stop codon with a BamHI site. PCR products were cloned into the BamHI site of the pBIΔGR vector to give pMG10 (WUS-GR). Transgenic plants were selected for kanamycin resistance. Dex inductions were carried out by spraying plants with a solution containing 1 μm Dex (Sigma D8893).
To express the WUS coding region under control of CLV3 regulatory sequences, we isolated a WUS cDNA by RT-PCR using the primers WUSBam-(5′CCCAGGATCCAACACACATG-GAGCCGCCA3′) and WUSSpe-(5′AAAGACTAGTGGCGTAAGAGCTAGTTCAG3′), which introduce BamHI and SpeI sites, respectively, and inserted it into the BamHI and SpeI sites of pBU14 to give pMG7 (CLV3::WUS). Transgenic plants were selected for Basta resistance.
All plasmid constructs were verified by restriction digests and DNA sequencing to rule out any amplification errors. A minimum of 50 transgenic plants was obtained for each construct.
ACKNOWLEDGMENTS
We thank Philip M. Mullineaux and Roger P. Hellens (John Innes Centre, Norwich, UK) for providing the pGreen vector before publication, Alan M. Lloyd (University of Texas, Austin) for donating pBIΔGR, and John Chandler (Institut für Entwicklungsbiologie, Köln, Germany) for discussions.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. SI677/1–1, SFB243, and SFB572).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001867.
LITERATURE CITED
- Barton MK, Poethig RS. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wildtype and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Long JA, Barton MK. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Trotochaud AE, Jeong S, Clark SE. CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science. 2000;289:613–617. doi: 10.1126/science.289.5479.613. [DOI] [PubMed] [Google Scholar]
- Waites R, Simon R. Signaling cell fate in plant meristems: three clubs on one tousle. Cell. 2000;103:835–838. doi: 10.1016/s0092-8674(00)00186-0. [DOI] [PubMed] [Google Scholar]
- Williams RW. Plant homeobox genes: Many functions stem from a common motif. Bioessays. 1998;20:280–282. doi: 10.1002/(SICI)1521-1878(199804)20:4<280::AID-BIES2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]