Abstract
Phosphatidylglycerol is a ubiquitous phospholipid that is also present in the photosynthetic membranes of plants. Multiple independent lines of evidence suggest that this lipid plays a critical role for the proper function of photosynthetic membranes and cold acclimation. In eukaryotes, different subcellular compartments are competent for the biosynthesis of phosphatidylglycerol. Details on the plant-specific pathways in different organelles are scarce. Here, we describe a phosphatidylglycerol biosynthesis-deficient mutant of Arabidopsis, pgp1. The overall content of phosphatidylglycerol is reduced by 30%. This mutant carries a point mutation in the CDP-alcohol phosphotransferase motif of the phosphatidylglycerolphosphate synthase (EC 2.7.8.5) isoform encoded by a gene on chromosome 2. The mutant shows an 80% reduction in plastidic phosphatidylglycerolphosphate synthase activity consistent with the plastidic location of this particular isoform. Mutant plants are pale green, and their photosynthesis is impaired. This mutant provides a promising new tool to elucidate the biosynthesis and function of plastidic phosphatidylglycerol in seed plants.
Phosphatidylglycerol (PG) is one of the most common phosphoglycerolipids found in nature. It is the only major phospholipid present in the thylakoid membranes of plant chloroplasts (Marechal et al., 1997) and the only phospholipid in cyanobacteria, which strikingly resemble plant chloroplasts in their lipid composition (Murata and Nishida, 1987). A large body of correlative and direct evidence suggests that PG is critical for the structural and functional integrity of the thylakoid membrane. Thus, the presence of specific molecular species of PG in photosynthetic membranes correlates well with low-temperature-induced photoinhibition and chilling sensitivity of plants and cyanobacteria (Murata et al., 1992; Somerville, 1995). Light-harvesting pigment-protein complexes of PSII are specifically enriched in PG (Murata et al., 1990; Tremolieres et al., 1994). Moreover, PG is crucial for the in vitro trimerization of the major peripheral light-harvesting pigment-protein complexes (Nussberger et al., 1993; Hobe et al., 1994; Kühlbrandt, 1994) and the dimerization of the reaction/center core pigment-protein complexes of PSII (Kruse et al., 2000). It is also an integral component of the PSI reaction center (Jordan et al., 2001) and is required for the in vitro reconstitution of the light-harvesting pigment-protein complexes of PSI (Schmid et al., 1997). Thylakoid membranes treated with phospholipase A2 are PG depleted and are inhibited in their photosynthetic electron transport activities (Jordan et al., 1983; Siegenthaler et al., 1987). Furthermore, the anionic lipid PG interacts with the transit peptide of chloroplast precursor proteins during protein import into chloroplasts (van't Hof et al., 1993).
PG-deficient auxotrophic mutants of the cyanobacterium Synechocystis sp. PCC6803 that are severely impaired in photosynthesis have recently been isolated (Hagio et al., 2000; Sato et al., 2000). Two Arabidopsis mutants, pho1 and ats1(act1), are known that show, among other phenotypes, a decreased relative amount of PG (Härtel et al., 1998a; Kunst et al., 1988; Poirier et al., 1991). However, because both mutants are affected in multiple aspects of lipid metabolism, their usefulness for the investigation of specific PG functions in photosynthetic membranes of seed plants is limited.
Two enzymatic reactions are specific to PG biosynthesis in plants, the formation of phosphatidylglycerolphosphate (PGP) from CDP-diacylglycerol and glycerol-3-phosphate catalyzed by the PGP synthase (EC 2.7.8.5), and the subsequent dephosphorylation by PGP phosphatase (EC 3.1.3.27; Moore, 1982; Andrews and Mudd, 1986; Kinney, 1993). Labeling studies indicate that enzymatic activity involved in PG biosynthesis is associated with the inner mitochondrial membrane, the endoplasmic reticulum, and the chloroplast (Moore, 1982; Kinney, 1993, and refs. therein), suggesting that at least three isoforms of PGP synthase and PGP phosphatase exist in plant cells. None of them has been purified to homogeneity and studied in vitro. Here, we describe a chemically induced, PG-deficient mutant of Arabidopsis, pgp1, with a mutation in one isoform of PGP synthase.
RESULTS
Isolation of a New PG-Deficient Mutant of Arabidopsis
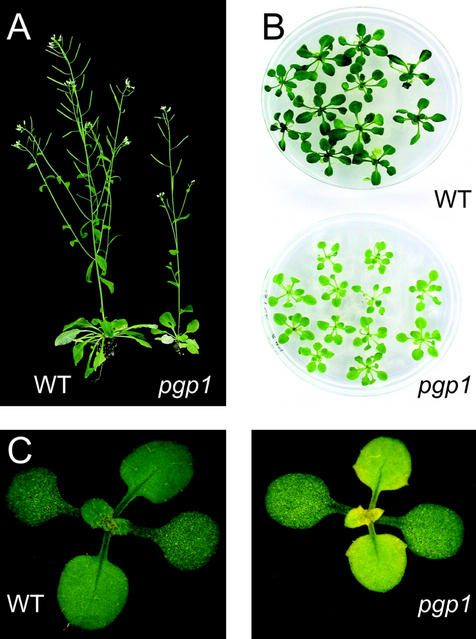
To isolate mutants of Arabidopsis deficient in the biosynthesis of complex lipids, we designed a brute force screening procedure based on thin layer-chromatographic analysis of lipid extracts from leaves of a chemically mutagenized Arabidopsis M2 population. Screening 3,000 M2-plants from 20 independent batches for lipid alterations by thin layer chromatography (TLC) of lipid extracts, we were not only able to isolate a galactolipid-deficient dgd1 mutant as previously described (Dörmann et al., 1995) but also two mutant lines with a slight reduction in PG content. One of these mutant lines was dark green, small, and resembled the pho1 mutant in appearance, whereas one mutant was pale green in color. Because of phosphate deprivation of the pho1 mutant in which the primary defect is in the translocation of phosphate from the root to the shoot system (Poirier et al., 1991), the relative content of phospholipids including PG is reduced, whereas the content of non-phosphorous lipids is increased (Essigmann et al., 1998; Härtel et al., 1998a). Unlike the pale green mutant, the dark green mutant was reduced in leaf phosphate content, and Yves Poirier confirmed by genetic complementation analysis that it carries a mutant allele of the pho1 locus (Y. Poirier, personal communication). The pale green mutant, designated pgp1 and back-crossed three times, is shown in Figure 1. Its growth was slightly reduced and, particularly under photoautotrophic growth conditions, developing young leaves in the center of the mutant rosette were yellowish in color (Fig. 1C). TLC of lipid extracts from expanded leaves followed by quantitative analysis of lipid composition of the pgp1 mutant showed that the PG content was reduced by approximately 30% (Table I). For comparison, we also included the analysis of the pho1 and the ats1(act1) mutants. In all three cases the reduction in PG content was similar. However, contrary to pgp1 and ats1(act1), the pho1 mutant showed an increase in the relative amounts of the sulfolipid sulfoquinovosyldiacylglycerol and the galactolipid digalactosyldiacylglycerol, a phenotype characteristic for pho1 (Härtel et al., 1998a).
Figure 1.
Growth and morphology of Arabidopsis wild type and pgp1 mutant. A, Six-week-old plants grown for 10 d on agar-solidified Murashige and Skoog medium with 1% (w/v) Suc followed by photoautotrophic growth on soil. B, Three-week-old plants grown on agar-solidified Murashige and Skoog medium with 1% (w/v) Suc. C, Three-week-old plants grown on soil. Note the yellow tissues in the center of the mutant rosette.
Table I.
Leaf lipid composition of Arabidopsis wild type and mutantsa
| WT | pgp1 | ats1(act1) | pho1 | |
|---|---|---|---|---|
| mol% | ||||
| MGDG | 48.5 ± 1.3 | 45.9 ± 3.3 | 41.0 ± 0.9 | 48.4 ± 1.1 |
| PG | 8.4 ± 0.6 | 5.6 ± 0.8 | 5.5 ± 0.6 | 4.9 ± 0.3 |
| DGDG | 16.3 ± 0.8 | 14.7 ± 1.9 | 13.3 ± 1.7 | 28.6 ± 0.3 |
| SQDG | 1.2 ± 0.4 | 1.0 ± 0.3 | 1.1 ± 0.4 | 5.8 ± 0.3 |
| PI | 1.8 ± 0.5 | 1.5 ± 0.4 | 0.9 ± 0.4 | 0.7 ± 0.3 |
| PE | 9.1 ± 0.3 | 13.1 ± 1.2 | 13.8 ± 0.7 | 4.3 ± 1.2 |
| PC | 14.7 ± 1.4 | 18.3 ± 2.4 | 24.4 ± 0.7 | 7.3 ± 0.7 |
Values represent means of three measurements. Plants were 25 to 30 d old. DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; SQDG, sulfoquinovosyldiacylglycerol.
The primary defect in the ats1(act1) mutant is a reduction in the activity of the plastidic glycerol-3-phosphate:acyl-ACP acyltransferase (Kunst et al., 1988). As a consequence, the plastid pathway of lipid assembly (Heinz and Roughan, 1983; Browse et al., 1986; Browse and Somerville, 1991) in this plant is shut down, and molecular species of monogalactosyldiacylglycerol containing 16:3 fatty acids (16 carbons, three double bonds) are strongly reduced. This phenotype was not visible in the fatty acid composition of monogalactosyldiacylglycerol of pgp1, which had an overall fatty acid composition similar to wild type (data not shown). Furthermore, morphology and growth of ats1(act1) are similar to wild type, unlike that observed for pgp1 (Fig. 1). This phenotypic analysis suggested that pgp1 was not a new allele of pho1 or ats1(act1), but a new locus affecting the biosynthesis of PG as was confirmed by genetic mapping (see below).
The pgp1 Locus Maps in the Proximity of a Putative PGP Synthase Gene on Chromosome 2
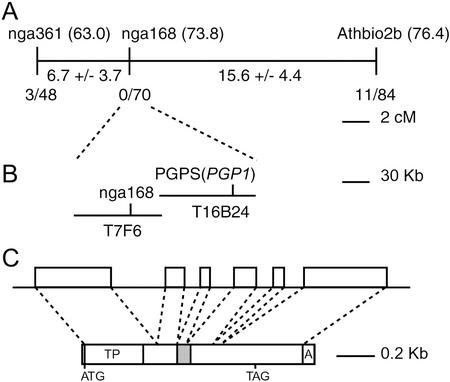
To narrow down the search for a candidate gene affected in pgp1, genetic mapping of the mutation leading to PG deficiency in pgp1 was employed. For this purpose we crossed pgp1, which is in the Col-2 genetic background, to a PGP1 wild-type line in the Ler-0 genetic background. In the F1 generation, all plants were phenotypically indistinguishable from wild type with regard to PG content based on semiquantitative TLC analysis with an example shown in Figure 2A (top, left two lanes). However, in the F2 generation of 233 plants screened, 54 (23%) plants were recovered with a reduced amount of PG, as shown for the homozygous pgp1 mutant in Figure 2A, whereas lipid extracts from all other plants were indistinguishable from wild-type extracts. Without exception, only these 54 plants were also reduced in growth. The F3 progeny of these plants no longer segregated, suggesting that they represent the homozygous mutant fraction. The nearly three to one segregation observed for the F2 plants was in agreement with a single nuclear recessive mutation that is responsible for the PG deficiency in the pgp1 mutant. A small mapping population of 42 homozygous pgp1 mutants derived from this cross was used to map simple sequence polymorphic markers (SSLPs; Bell and Ecker, 1994) relative to the pgp1 locus. Only markers on chromosome 2 at approximately 70 centimorgans showed linkage to pgp1. Recombination frequencies and calculated map distances for this region are shown in Figure 3A. Two minimally overlapping bacterial artificial chromosomes (BACs) sequenced by The Arabidopsis Genome Initiative (2001), one containing the most tightly linked SSLP marker nga168 as derived from The Arabidopsis Information Resource Web page (http://www.Arabidopsis.org), are shown in Figure 3B. The nucleotide sequence of T16B24 (GenBank accession no. AC004697) contains an open reading frame (BAC locus T16B24.7; nucleotides 78, 883–880, 690; gene At2g3920) with a translated amino acid sequence (GenBank accession no. AAC28995) annotated as CDP-diacylglycerol glycerol-3-phosphate 3-phosphatidyltransferase (PGP synthase). The predicted intron-exon structure for this gene is shown in Figure 3C and was later confirmed after the isolation of the cDNA (see below). Searching the amino acid sequence (Altschul et al., 1990) of PGP synthase from Escherichia coli (GenBank accession no. P06978) against all amino acid sequences of Arabidopsis confirmed this open reading frame on chromosome 2 as putative PGP synthase and revealed two additional putative PGP synthase genes of Arabidopsis, one on chromosome 3 (BAC locus T15C9.30; gene At3g55030) and one on chromosome 4 (BAC locus T4B21.19; gene At4g04870). Targeting signal analysis (Emanuelsson et al., 2000) suggested that, of all three putative PGP synthases, only the one encoded on chromosome 2 contained a putative chloroplast transit peptide.
Figure 2.
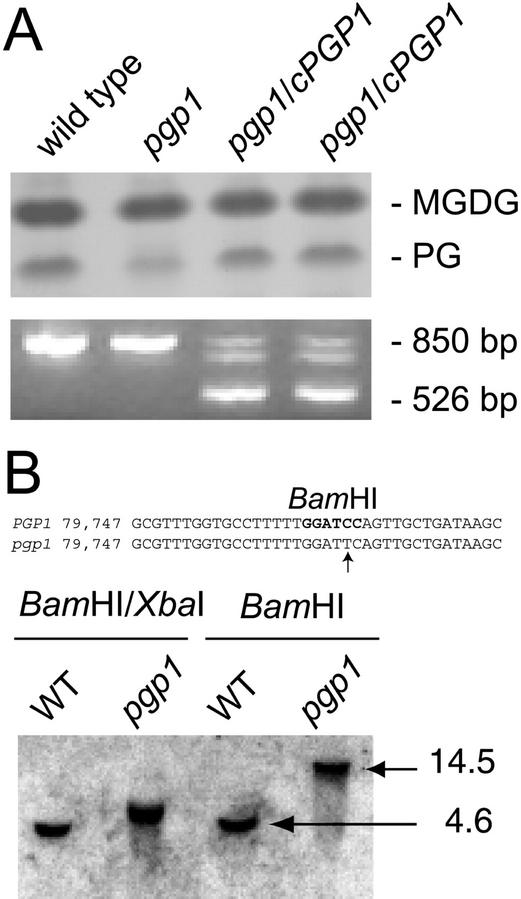
Lipid phenotype of pgp1 and molecular defect. Sections of a lipid chromatogram (top panel) and a DNA gel of PCR products (bottom panel) of wild type, the pgp1 mutant, and two independent transgenic pgp1 lines transformed with the PGP1 wild-type cDNA (pgp1/cPGP1) are shown. Lipids were analyzed by TLC and stained by exposure to iodine. Monogalactosyldiacylglycerol (MGDG), serving as loading control, and PG are shown. B, Restriction length polymorphism in the pgp1 mutant locus. Sequence comparison of the wild-type (PGP1) and mutant (pgp1) locus showing the mutated BamHI site (top). The nucleotide number refers to the GenBank accession number of the BAC clone T16B24 (accession no. AC004697). Genomic Southern blot of the wild type and the pgp1 mutant probed with the PGP1 gene (bottom). Restriction digests are indicated. Numbers indicate the size (in kilobases) of the fragments marked by arrows.
Figure 3.
The PGP1 locus on chromosome 2 of Arabidopsis. A, Genetic map showing SSLP markers (positions on map), experimentally determined map distances, and recombinant chromosomes/total chromosomes analyzed for each marker. B, BACs from the genome sequencing project and markers. C, Structure of the PGP1 gene (top) and cDNA (bottom). The gray box represents the exon carrying the mutation in pgp1. A, Poly(A) tail; ATG, start codon; TAG, stop codon; TP, transit peptide.
The pgp1 Mutant Is Recognizable by a Distinct RFLP in BAC Locus T16B24.7
To determine the exact molecular defect in the pgp1 mutant, a detailed analysis of the presumed PGP synthase locus on chromosome 2 was conducted in the mutant and the wild type. By chance, we discovered during the restriction analysis of cloned fragments derived from the mutant that the T16B24.7 equivalent locus in the pgp1 mutant is missing a BamHI restriction site resulting in a RFLP. Figure 2B shows a comparison of wild-type and pgp1 genomic DNA restricted with BamHI/XbaI or just BamHI and probed with a 987-bp genomic DNA fragment from the beginning of the open reading frame to the BamHI site present in the wild type (GenBank accession no. AC004697; nucleotides 78,777–79,764). In both digests, the size of the hybridizing fragment is increased in the pgp1 sample as predicted from the genomic sequence for the loss of a specific BamHI site (GenBank accession no. AC004697; nucleotide 79,764). Sequence analysis of the entire coding region of the respective gene isolated by PCR from wild-type and mutant genomic DNA confirmed a mutation altering the BamHI site (Fig. 2B).
Expression of a cDNA Derived from Wild-Type Locus T16B24.7 Restores PG Biosynthesis in the pgp1 Mutant Background
To obtain conclusive evidence that the deficiency in PG biosynthesis in the pgp1 mutant is indeed caused by the observed mutation in the T16B24.7 locus, the respective wild-type cDNA was isolated by reverse transcriptase (RT)-PCR and cloned into a T-DNA binary vector. This construct was used to transform pgp1 mutant plants by vacuum infiltration. Several hygromycin B resistant transformants were isolated, and their lipid composition was examined by semiquantitative TLC. Lipid extracts from two independent transformants are shown in Figure 2A (top). Their lipid composition and their macroscopic appearance was indistinguishable from wild type. The presence of the transgene construct was confirmed by PCR as shown in Figure 2A (bottom). The genomic sequence gives rise to a diagnostic fragment of 850 bp, whereas the transgene appears as 525-bp fragment. The intermediate size fragment was isolated and analyzed, but its sequence was unrelated to PGP1 or any of its paralogs (data not shown). Thus, introduction of the wild-type cDNA corresponding to T16B24.7 into the mutant background rescued lipid and morphological phenotypes, thereby confirming that the observed mutant phenotypes are caused by a single mutation in the T16B24.7 locus.
BAC Locus T16B24.7 Encodes a Fully Functional PGP Synthase in the Wild-Type But Not in the pgp1 Mutant
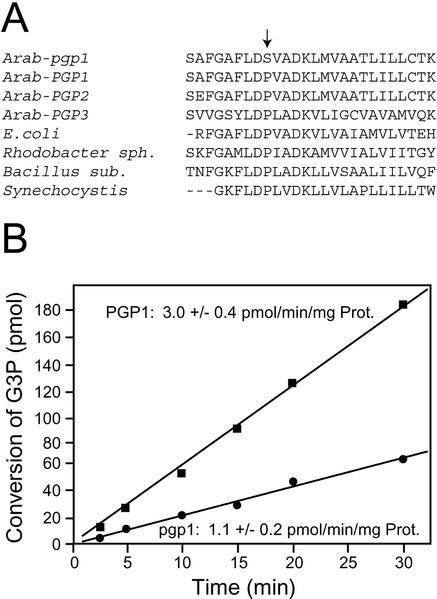
The mutation in pgp1 that abolished the BamHI site mentioned above is located in the third exon of the gene (Fig. 3C). In accordance, the predicted protein carries a Pro-170 to Ser mutation. The alignment of the predicted protein with the two other Arabidopsis paralogs and known bacterial PGP synthases (Fig. 4A) revealed that this Pro residue is part of a conserved active site motif for CDP-alcohol phosphotransferases (Williams and McMaster, 1998). At the beginning of this study, no experimental data were available for any of the three putative PGP synthase genes of Arabidopsis. To confirm that the wild-type locus T16B24.7 indeed encodes a fully functional PGP synthase, the respective full-length wild-type cDNA (GenBank accession no. AB048535) was isolated as described in “Materials and Methods.” In addition, the corresponding cDNA from the pgp1 mutant was cloned by RT-PCR. The 1,210-bp-long full-length wild-type cDNA encodes a 296-amino acid protein with a molecular mass of 32.2 kD. To test PGP synthase activity for the respective wild-type and mutant proteins, we expressed the cDNA for the predicted mature proteins (with the 93 N-terminal amino acids missing) in the E. coli mutant YA5512. This mutant is deficient in PGP synthase activity (Asai et al., 1989) and carries a Thr-60 to Pro replacement in the coding region of the pgsA gene (Usui et al., 1994). Consequently, this strain has very low residual PGP synthase activity leading to PG deficiency. In addition, the content of cardiolipin is very low because PG is a precursor for this lipid. Total lipids extracted from lines of YA5512 transformed with vector control (pQE32), the wild-type cDNA (pQE32-PGP1), or mutant cDNA (pQE32-pgp1) expression constructs were quantified. Both wild-type and mutant constructs partially restored the lipid composition of the E. coli mutant, but the pgp1 mutant construct did so to a lesser extent suggesting reduced activity (data not shown). Based on this result, it was concluded that PGP1 encodes a functional PGP synthase. Because mutant and wild proteins are expressed to the same extent in E. coli as confirmed by gel electrophoresis of total proteins (data not shown), the result suggested that the activity of the pgp1 mutant is impaired, but not abolished. This was directly confirmed by enzymatic assays under saturating substrate conditions in the linear range of the assay of crude protein extracts from the two E. coli strains as shown in Figure 4B. Three independent clones were tested for each construct and averaged. The specific activity of the mutant protein was reduced to approximately one-third of wild-type activity. The PGP synthase activity was not detectable in the crude protein extracts from YY5512 or YY5512 hosting the empty vector pQE32 (data not shown).
Figure 4.
Active site mutation of PGP synthase in the pgp1 mutant. A, Alignment of different Arabidopsis PGP1 paralogs (PGP2 and PGP3) and bacterial PGP synthase orthologs. GenBank accession numbers (Arabidopsis gene numbers) for the respective protein sequences are: Arab-PGP1, AAC28995 (At2g3920); Arab-PGP2, CAB82698 (At3g55030); Arab-PGP3, CAB80852 (At4g04870); E. coli, P06978; Rhodobacter sphaeroides, AAC44003; Bacillus subtilis, I39950; and Synechocystis sp., S76208. The arrow indicates the Pro (Pro-170) to Ser change in the pgp1 mutant. B, Reduced specific activity of the PGP1-Ser-170 mutant protein. Total activity in extracts of the E. coli YA5512 PGP synthase-deficient mutant expressing wild-type PGP1 cDNA (squares) or mutant pgp1 cDNA (circles) was determined. The two graphs represent single representative experiments. Equal amounts of protein were used. In addition, the mean specific activity (±se) for three independent PCR constructs is provided. The PGP synthase activities in YA5512 and YA5512 containing pQE32 were not detectable.
Plastidic PGP Synthase Activity Is Strongly Reduced in the pgp1 Mutant
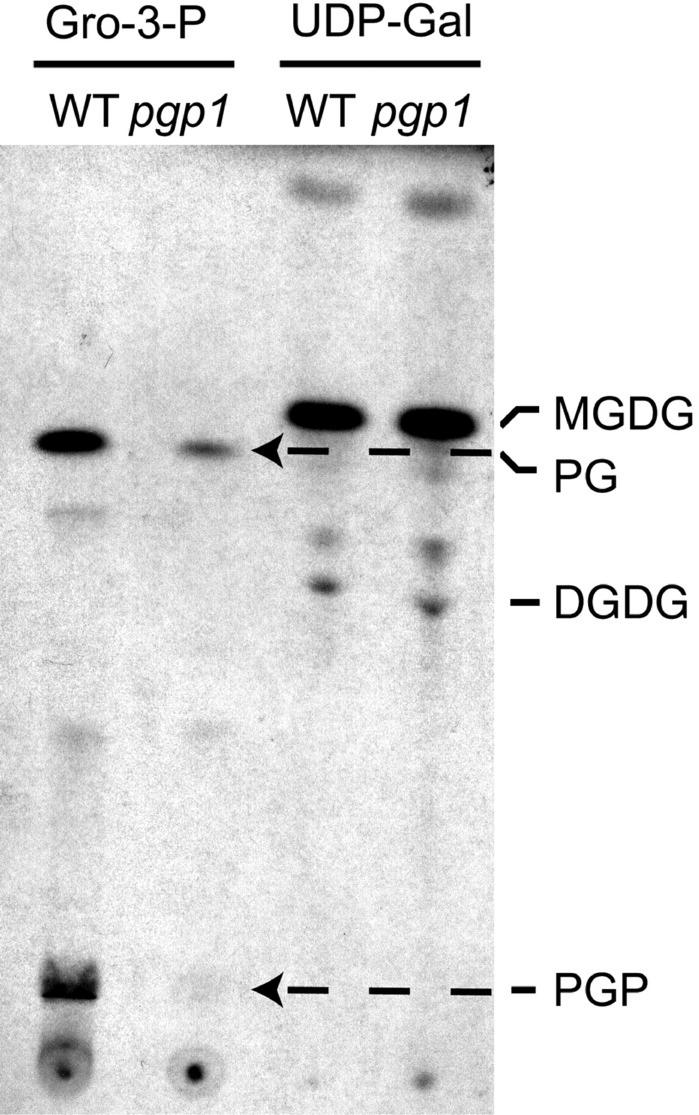
To determine the effect of the Pro-170 to Ser mutation in the pgp1 mutant, PGP-synthase activity was determined in isolated chloroplasts. When extracts from isolated wild-type chloroplasts were incubated with CDP-diacylglycerol and labeled glycerol-3-phosphate in the dark, two major lipids were labeled, PGP and PG, which was presumably derived from PGP because of the action of PGP phosphatase (Fig. 5). In chloroplast extracts from the pgp1 mutant, the incorporation of label into PGP was nearly abolished, and the labeling of PG was markedly reduced (Fig. 5). In comparison, incorporation of labeled UDP-Gal into galactolipids was not affected in the pgp1 mutant (Fig. 5). The labeling pattern was more complex when the PGP synthase assay was performed in the light (data not shown). Fatty acid biosynthesis and de novo lipid biosynthesis are apparently stimulated in the light and lead to the incorporation of label from glycerol-3-phosphate into the diacylglycerol moieties of other lipids. We, therefore, quantified the PGP synthase activity in dark-incubated chloroplasts by determining the amount of label in the total lipid extracts derived from the assay mixture. In the pgp1 mutant, the chloroplastic PGP synthase activity was reduced to approximately 19% of wild-type activity from 24.4 ± 4.4 to 4.7 ± 1.7 μmol glycerol-3-phosphate min−1 mg−1 chlorophyll.
Figure 5.
Chloroplastic PGP synthase and galactolipid biosynthetic activities in wild type and pgp1 mutant. Isolated and ruptured chloroplasts were either incubated with labeled glycerol-3-phosphate (Gro-3-P) and CDP-diacylglycerol (PGP synthase assay, left two lanes) or with labeled UDP-Gal (galactolipid biosynthesis, right two lanes). An autoradiograph of a thin layer chromatogram of labeled lipid extracts is shown. Identified lipids were digalactosyldiacylglycerol (DGDG), monogalactosyldiacylglycerol (MGDG), phosphatidylglycerol (PG), and phosphatidylglycerolphosphate (PGP).
Photosynthesis Is Impaired in the pgp1 Mutant
Although the loss of total PG in leaves of the pgp1 mutant does not exceed 30%, the growth of the mutant was reduced, and the mutant plants appeared pale green consistent with an impairment in photosynthesis (Fig. 1). This initial observation was confirmed by quantification of the pigment content of fully expanded leaves (Table II). Chlorophyll a and b as well as carotenoid amounts were reduced in the mutant/and the ratio of chlorophyll a/b was increased. This result points to an alteration in the structure of the photosynthetic apparatus in the mutant and explains the pale green color of pgp1 leaves. To investigate the functionality of the photosynthetic apparatus in the pgp1 mutant, we used noninvasive chlorophyll fluorescence measurements. This technology is well established and was successfully used to investigate photosynthetic competency and light utilization of other Arabidopsis lipid mutants (Härtel et al., 1997, 1998a, 1998b). Measurements under steady-state conditions at a photosynthetic photon fluence density comparable with that used for the growth of the plants revealed a marked decrease in the quantum yield of linear electron transport through PSII (ΦPSII) and in the efficiency of open PSII units (Fv′/Fm′; Table II). A small (approximately 6%) but significant decrease in the maximum photochemical efficiency of PSII (Fv/Fm) was observed for the mutant after a 1-h dark adaptation period. For comparison, the initial Fv/Fm ratios in the ats1(act1) and pho1 mutant were 0.79 ± 0.01 and 0.78 ± 0.02, respectively. Taken together, these alterations in the pigment composition and photosynthetic light energy utilization in the pgp1 mutant underline the importance of PG for the proper structure and function of photosynthetic membranes in seed plants.
Table II.
Pigment content (mg g−1 fresh wta and chlorophyll fluorescenceb of wild-type and mutant fully expanded leaves
| WT | pgp1 | |
|---|---|---|
| Chlorophyll a | 1.48 ± 0.08 | 1.17 ± 0.11 |
| Chlorophyll b | 0.49 ± 0.03 | 0.36 ± 0.03 |
| Chlorophyll a+b | 1.97 ± 0.11 | 1.55 ± 0.14 |
| Chlorophyll a/b | 3.01 ± 0.02 | 3.29 ± 0.07 |
| Carotenoids | 0.25 ± 0.02 | 0.19 ± 0.02 |
| Chlorophyll/carotenoid | 7.77 ± 0.26 | 7.99 ± 0.27 |
| Fv/Fm (n = 25) | 0.79 ± 0.02 | 0.74 ± 0.01 |
| φPSII (n = 10) | 0.55 ± 0.01 | 0.49 ± 0.01 |
| F′v/F′m (n = 10) | 0.69 ± 0.01 | 0.60 ± 0.01 |
Samples were taken from fully expanded leaves of 25 to 30-d-old plants harvested after 8 h of light exposure. Plants were grown under a 16-h-light/8-h-dark regime at a PPFD of 75 μmol m−2 s−1 in soil. Pigment values represent means (±se) of six independent determinations.
Data were acquired at a PPFD of 75 μmol m−2 s−2.
DISCUSSION
The majority of PG in green plant tissues is localized in the chloroplasts, and the biosynthesis of PG in green plants has been studied in greatest detail using intact or broken chloroplasts from pea (Pisum sativum) leaves (Andrews and Mudd, 1986). Based on pulse-chase labeling experiments, it was proposed that PG biosynthesis proceeds in chloroplasts as worked out for bacteria and yeast (Saccharomyces cerevisiae). First, CDP-diacylglycerol is formed from CTP and phosphatidic acid through the action of CDP-diacylglycerol synthase (EC 3.1.3.4). Plant cDNAs encoding this enzyme have been recently identified and expressed in E. coli (Kopka et al., 1997). Although this reaction is required for PG biosynthesis, it is not specific for this process, because CDP-diacylglycerol also serves as precursor in the biosynthesis of other phospholipids (Kinney, 1993). The second step of PG biosynthesis involves the transfer of glycerol-3-phosphate onto CDP-diacylglycerol giving rise to PGP and CMP. This reaction is catalyzed by PGP synthase (EC 2.7.8.5). Third, PGP is hydrolyzed to PG and inorganic phosphate by PGP phosphatase (EC 3.1.3.27). Although the latter two reactions had been observed in pea chloroplasts (Andrews and Mudd, 1986), no further biochemical analysis of the respective proteins has been performed. A mutant deficient in one of the two enzymes involved in plastidic PG biosynthesis would serve at least two purposes. First, it would permit the study of PG function in plastid membranes, and second, it would provide experimental evidence that the pathway described above indeed represents the major route of PG biosynthesis in plastids. Based on our analysis, pgp1 is just such a mutant, because it severely affects plastidic PGP synthase activity. The experimental evidence for this conclusion can be summarized as follows: first, pgp1 maps within 1 centimorgan of BAC T16B24.7 annotated as chloroplast-targeted PGP synthase; second, complementation analysis employing a PGP synthase-deficient E. coli mutant confirmed the gene product of BAC locus T16B24.7 is indeed a PGP synthase; third, introduction of wild-type cDNA corresponding to T16B24.7 into the mutant background rescued lipid and morphological phenotypes; fourth, a mutation in the third exon of T16B24.7 in pgp1 gives rise to an RFLP at the DNA level, and at the protein level to a Pro-170 to Ser substitution in a highly conserved motif characteristic for CDP-alcohol phosphotransferase; and fifth, PGP synthase activity in isolated chloroplasts of pgp1 is markedly reduced, consistent with the other findings. Taken together, these observation provided an unambiguous logical link between the molecular defect and the biochemical phenotype in pgp1, as well as a firm gene-product relationship between T16B24.7 and the encoded PGP synthase.
Although the Pro-170 to Ser mutation in pgp1 affects a highly conserved Pro in the active site (Williams and McMaster, 1998), the activity of the recombinant mutant protein was only reduced by approximately two-thirds. Thus, the mutation in pgp1 is “leaky.” Although the relative amount of PG was only reduced by 30% in the pgp1 mutant, this biochemical defect led to a visible phenotype. More detailed analysis revealed that pigment composition and photosynthetic light utilization are affected in this mutant, thereby explaining the slow growth and the pale green appearance of pgp1. Chlorophyll b is exclusively located in the light-harvesting pigment-protein complexes of PSII and PSI. Therefore, a decrease of total chlorophyll content associated with an increase in the chlorophyll a/b ratios in the pgp1 mutant (Table II) indicates both a decrease in the total number of photosystems and a stronger decline in light-harvesting antenna complexes relative to the chlorophyll a-containing reaction center/core complexes. Chlorophyll fluorescence measurements on intact leaves revealed that the chlorophyll loss was associated with a decline in the quantum efficiency of PSII photochemistry. In particular, ΦPSII and Fv′/Fm′ were markedly reduced in the mutant. Taken together, the chlorophyll fluorescence and pigment data for the pgp1 mutant most likely reflect the importance of PG in photosynthesis of seed plants. This is consistent with previous observations on cyanobacterial mutants that are deficient in PG biosynthesis (Hagio et al., 2000; Sato et al., 2000). These cyanobacterial mutants have an absolute requirement for PG supplementation in their medium, and deficiencies in photosynthetic competence correlate in severity with the amount of PG in cell membranes, which can be manipulated in this system through external supply. What is surprising about the pgp1 mutant in comparison with ats1(act1) is the fact that in the latter the mutant ion does not show a striking effect on growth and a less severe effect on photosynthesis in spite of a similar reduction in overall PG content (Kunst et al., 1988). Although we currently do not know the basis for this difference, we expect that the detailed analysis of the pgp1 mutant and a future reexamination of the ats1(act1) mutant may provide clues toward a deeper understanding of PG biosynthesis and function in plant chloroplasts.
While this work was under review, Müller and Frentzen (2001) published the expression of the PGP1 cDNA in yeast and demonstrated a mitochondrial localization of the enzyme in this heterologous system. Although the data presented here clearly show that PGP1 is the major chloroplastic isoform of PGP synthase, we currently cannot rule out that PGP1 is also associated with mitochondria in Arabidopsis cells.
MATERIALS AND METHODS
Plant Material
Surface-sterilized seeds of Arabidopsis (ecotype Col-2 or Landsberg erecta [Ler]-0) mutant, the pgp1 mutant that had been back-crossed multiple times, ats1(act1) mutant (Kunst et al., 1988), and pho1 mutant (Poirier et al., 1991) were germinated on 0.8% (w/v) agar-solidified Murashige and Skoog (1962) medium supplemented with 1% (w/v) Suc. The seedlings were kept on agar for 10 d before the transfer to pots containing a standard soil mixture (equal parts of Bacto Soil [Michigan Peat Company, Houston], medium vermiculite, and perlite) drenched with one-half-strength Arabidopsis nutrient solution (Estelle and Somerville, 1987). Plants were grown in growth chambers (AR-75L, Percival Scientific Inc., Boone, IA) under light of a photosynthetic photon flux density (400–700 nm) of 75 μmol m−2 s−1. A 14-h light/10-h dark regime was applied. The day/night temperature was controlled at 23°C/18°C.
Genetic and Molecular Procedures
The pgp1 mutant was isolated by TLC of lipid extracts of individual M2 plants as previously described for the isolation of the dgd1 mutant (Dörmann et al., 1995). The pgp1 mutant locus was mapped in a small F2 population (42 homozygous pgp1 plants) derived from a cross of pgp1 (Col-2) to a standard Ler (Ler-0) line employing single sequence polymorphic markers evenly spaced on the Arabidopsis genome map (http://www.Arabidopsis.org) under conditions described by Bell and Ecker (1994). At least four markers per chromosome were tested. Standard sequence searching tools (Altschul et al., 1990) were used at the National Center for Biological Information Web site (http://www.ncbi.nlm.nih.gov) to identify putative PGP synthase genes in the Arabidopsis genomic sequence (The Arabidopsis Genome Initiative, 2001). References to the respective DNA and amino acid sequences are provided as GenBank accession numbers throughout the text. New sequences were obtained at the Michigan State University Genomics Facility by automated sequencing.
For Southern-blot analysis, Arabidopsis DNA was extracted from 4-week-old plants (Rogers and Bendich, 1994). The digested DNA was separated on a 1% (w/v) agarose gel and transferred onto a Hybond N+ membrane (Amersham Pharmacia Biotech, Piscataway, NJ). Blots were hybridized at 65°C in standard hybridization solution (250 mm sodium phosphate buffer, 7% [w/v] SDS, 1 mm EDTA, 1% [w/v] bovine serum albumin, and 10 mg mL−1 autoclaved herring sperm DNA). Blots were repeatedly washed for 15 min in 2× SSC and 0.1% (w/v) SDS.
Cloning of the Mutant and Wild-Type cDNAs and Expression in Escherichia coli
The PGP1 cDNA was amplified in three steps from an Arabidopsis λgt11 cDNA library derived from leaves. An internal fragment of the Arabidopsis PGP1 cDNA was amplified by PCR using the primers 5′-CGCTGCAGGTCTGGCTTCGTTAAT-3′ and 5′-CGCTGCAGCTACTTCATTAGTACTTT-3′. The 5′-terminal region of the cDNA was obtained by the 5′-RACE method (5′-Full RACE Core Set, TaKaRa, Shiga, Japan) and the 3′-terminal region by amplification using an internal primer and oligo(dT). The amplified cDNAs were subcloned into pCR2.1 (Original TA Cloning Kit, Invitrogen, Carlsbad, CA), and their nucleotide sequences were determined. The sequence fragments were combined using appropriate restriction sites to obtain the full-length cDNA as deposited in GenBank (accession no. AB048535).
The E. coli mutant YA5512 defective in PGP synthase (Asai et al., 1989; Usui et al., 1994) was provided by K. Matsumoto (Saitama University, Japan). A region of the Arabidopsis PGP1 cDNA encoding the presumed mature form of PGP synthase (first 93 amino acids removed) was amplified by PCR using the primers 5′-AAGGTACCCTTCACCGCCTCCGT-3′ and 5′-AACTGCAGCTACTTCATTAGTACTTTCCA-3′. The corresponding pgp1 mutant sequence was cloned by RT-PCR using the same set of primers. For this purpose, total leaf RNA was isolated from approximately 20-d-old plants according to the method by Logemann et al. (1987). Poly(A+) mRNA was purified using an oligotex kit from Qiagen (Valencia, CA) according to the instructions. RT-PCR reactions were performed using the Omniscript RT-PCR system from Qiagen. The PCR product was cloned into the ligation-ready vector pPCR-Script Amp SK+ (Stratagene, La Jolla, CA). The presence of the C to T mutation in pgp1 cDNA was confirmed by DNA sequencing. The amplified DNA fragments from the wild type and the mutant were digested with KpnI and PstI and ligated into the KpnI and PstI sites of the expression vector pQE32 from Qiagen. The resulting plasmids expressing the wild type (pQE32-PGP1) and mutated protein (pQE32-pgp1) were used to transform E. coli YA5512. The constructs were confirmed by sequencing. YA5512 was also transformed with the vector pQE32 for control purposes. The transformants were grown at 37°C in Luria-Bertani medium supplemented with 100 μg mL−1 ampicillin. A 3-mL overnight culture was centrifuged, and the cells were washed and resuspended in 10 mL of the same medium with antibiotic. The culture was incubated at 37°C for 3 h, isopropyl-1-thio-β-galactoside was added to a final concentration of 0.4 mm, and the culture was further incubated for 3 h at 37°C. Cells were collected by centrifugation (10 min, 5,000g). The resulting cell pellet was resuspended in 50 mm Tris, pH 7.2, and 20% (v/v) glycerol and stored at −80°C. For enzyme assays, the cells were disrupted by one freeze/thaw cycle and ultrasonification (three times for 20 s). The sonicated suspension was centrifuged at 15,000g, and the supernatant (cell-free extract) was used for the assay of PGP synthase activity as described below.
Complementation Analysis
The full-length coding sequence of wild-type PGP synthase was amplified by RT-PCR using the primer 5′-AAGGTACCATGCTCAGATCCGGTCTGGCT-3′ and 5′-AACTGCAGCTACTTCATTAGTACTTTCCA-3′ as described above for the pgp1 mutant sequence. This fragment was ligated into the Stratagene pPCR-Script Amp SK+ vector, and the nucleotide sequence was determined. The insert was excised, with KpnI taking advantage of the site in the vector and in one of the primers, and was subsequently cloned into the respective site of the binary vector pBINAR-Hyg (Dörmann and Benning, 1998). Arabidopsis pgp1 (Col-2) plants were transformed in planta (Bechtold and Pelletier, 1998). Transformants were selected on agar-solidified Murashige and Skoog medium containing 1% (w/v) Suc and 70 μg mL−1 hygromycin B. The lipid phenotype was determined by thin layer chromatographic analysis of lipid extracts (see below). The presence of the transgene construct was confirmed by PCR using the primers 5′-AAGGTACCATGCTCAGATCCGGTCTGGCT-3′ and 5′-ACCATAAGCTTATCAGCAACTGGATTCAA-3′.
Lipid Analysis
Rosette leaves were immediately frozen in liquid nitrogen upon harvesting, and lipids were extracted as previously described (Dörmann et al., 1995). Bacterial cells were extracted as described by Benning and Somerville (1992). Lipid extracts were analyzed on activated ammonium sulfate-impregnated silica gel TLC plates using a solvent system of acetone:toluene:water (91:30:7, v/v). Lipids were visualized with iodine vapor and identified by cochromatography with lipid extracts of known composition. For quantitative analysis, individual lipids were isolated from TLC plates and used to prepare fatty acid methyl esters. The methyl esters were quantified by GLC using myristic acid as the internal standard (Rossak et al., 1997).
Chloroplast Isolation and PGP Synthase Assay
Chloroplasts were isolated according to Joy and Mills (1987) from 4-week-old Arabidopsis plants grown on agar-solidified Murashige and Skoog medium supplied with 1% (w/v) Suc. The equivalent of 5 g of rosette leaves was typically used in a single preparation. The chloroplast PGP synthase activity was measured by the incorporation of radiolabled glycerol-3-phosphate into the chloroform-soluble product PGP according to Mudd and coworkers (1987). The reaction mixture contained 25 mm HEPES (pH 7.3), 2 mm MgCl2, 0.1 mm CDP-diacylglycerol, 0.5 mm [14C]dl-glycerol 3-phosphate (1 μCi 150 nmol−1), 0.05% (v/v) Triton X-100, and chloroplast suspension equivalent to 150 μg of chlorophyll in a final volume of 150 μL. The reaction mixture for the determination of galactolipid synthase activities in isolated chloroplasts contained the same buffer except that 0.5 μCi of UDP-[14C] Gal (325 mCi mmol−1; Amersham) was added instead of CDP-diacylglycerol and labeled glycerol-3-phosphate. Furthermore, this assay was performed in a total volume of 120 μL containing chloroplasts equivalent to 150 μg of chlorophyll. The incubations were performed at 27°C for 60 min and were terminated by the addition of 0.5 mL of 0.1 m HCl in methanol. The lipids were extracted with 2 mL of chloroform and 3 mL of 1 m MgCl2. The chloroform phase was evaporated under a stream of nitrogen and then redissolved in 0.2 mL of chloroform. A 25-μL aliquot was taken for the determination of radioactivity by scintillation counting. The remainder was used for lipid analysis by TLC as described above.
Chlorophyll Fluorescence Measurements and Pigment Analysis
In vivo room temperature chlorophyll fluorescence was monitored with plants, which were dark adapted for 1 h as described (Härtel et al., 1998c). The fluorescence parameters used in this study are as previously defined (Genty et al., 1989; van Kooten and Snel, 1990). Pigments were determined as described by Lichtenthaler (1987).
Footnotes
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–98ER20305 to C.B.), by the Michigan State University Center for Novel Plant Products (grant to C.B.), by a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Science, Sport and Culture (grant no. 12640635 to H.W.), and by a Program for Promotion of Basic Research Activities for Innovative Biosciences grant (to H.W.) from the Bio-oriented Technology Research Advancement Institution (Tokyo). M.H. was supported by a fellowship from the Japanese Society for the Promotion of Science.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002725.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews J, Mudd JB. Phosphatidylglycerol synthesis in pea chloroplasts. Plant Physiol. 1986;79:259–265. doi: 10.1104/pp.79.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2001;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Asai Y, Katayose Y, Hikita C, Ohta A, Shibuya I. Suppression of the lethal effect of acidic-phospholipid deficiency by defective formation of the major outer membrane lipoprotein in Escherichia coli. J Bacteriol. 1989;171:6867–6869. doi: 10.1128/jb.171.12.6867-6869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thalianaplants by vacuum infiltration. In: Salinas J, Martinez-Zapater JM, editors. Arabidopsis Protocols, Methods in Molecular Biology. Vol. 82. Totowa, NJ: Humana Press; 1998. pp. 259–266. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174:2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J, Somerville C. Glycerolipid biosynthesis: biochemistry and regulation. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the “16:3” plant Arabidopsis thaliana. Biochem J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C. The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J. 1998;13:641–652. doi: 10.1046/j.1365-313x.1998.00067.x. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Essigmann B, Güler S, Narang RA, Linke D, Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thalianawith an altered morphology. Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- Genty B, Briantais JM, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochem Biophys Acta. 1989;990:87–92. [Google Scholar]
- Hagio M, Gombos Z, Varkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H. Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol. 2000;124:795–804. doi: 10.1104/pp.124.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Essigmann B, Lokstein H, Hoffmann-Benning S, Peters-Kottig M, Benning C. The phospholipid-deficient pho1 mutant of Arabidopsis thalianais affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biochim Biophys Acta. 1998a;1415:205–218. doi: 10.1016/s0005-2736(98)00197-7. [DOI] [PubMed] [Google Scholar]
- Härtel H, Lokstein H, Dörmann P, Grimm B, Benning C. Changes in the composition of the photosynthetic apparatus in the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiol. 1997;115:1175–1184. doi: 10.1104/pp.115.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtel H, Lokstein H, Dörmann P, Trethewey RN, Benning C. Photosynthetic light utilization and xanthophyll-cycle activity in the galactolipid deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiol Biochem. 1998b;36:407–417. [Google Scholar]
- Härtel H, Reinhardt I, Grimm B. Relationship between energy-dependent fluorescence quenching and xanthophyll-cycle-pigments in transgenic chlorophyll-deficient tobacco grown under different light intensities. J Photochem Photobiol B Biol. 1998c;43:136–145. [Google Scholar]
- Heinz E, Roughan G. Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 1983;72:273–279. doi: 10.1104/pp.72.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobe S, Prytulla S, Kühlbrandt W, Paulsen H. Trimerization and crystallization of reconstituted light-harvesting chlorophyll a/b complex. EMBO J. 1994;13:3423–3429. doi: 10.1002/j.1460-2075.1994.tb06647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan BR, Chow W-S, Baker AJ. The role of phospholipids in the molecular organisation of pea chloroplast membranes: effect of phospholipid depletion on photosynthetic activity. Biochim Biophys Acta. 1983;725:77–86. [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Δ resolution. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- Joy KW, Mills WR. Purification of chloroplasts using silica sols. Methods Enzymol. 1987;148:179–199. [Google Scholar]
- Kinney AJ. Phospholipid head groups. In: Moore TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 259–284. [Google Scholar]
- Kopka J, Ludewig M, Müller-Röber B. Complementary DNAs encoding eukaryotic-type cytidine-5′-diphosphate-diacylglycerol synthases of two plant species. Plant Physiol. 1997;113:997–1002. doi: 10.1104/pp.113.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse O, Hankammer B, Konczak C, Gerle C, Morris E, Radunz A, Schmid GH, Barber J. Phosphatidylglycerol is involved in the dimerization of photosystem II. J Biol Chem. 2000;275:6509–6514. doi: 10.1074/jbc.275.9.6509. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Structure and function of the plant light-harvesting complex, LHC-II. Curr Opin Struct Biol. 1994;4:519–528. [Google Scholar]
- Kunst L, Browse J, Somerville C. Altered regulation of lipid biosynthesis in a mutant Arabidopsisdeficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc Natl Acad Sci USA. 1988;85:4143–4147. doi: 10.1073/pnas.85.12.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic membranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- Marechal E, Block MA, Dorne A-J, Joyard J. Lipid synthesis and metabolism in the plastid envelope. Physiol Plant. 1997;100:65–77. [Google Scholar]
- Moore TS. Phospholipid biosynthesis. Annu Rev Plant Physiol. 1982;33:235–259. [Google Scholar]
- Mudd JB, Andrews JE, Sparace SA. Phosphatidylglycerol synthesis in chloroplast membranes. Methods Enzymol. 1987;148:338–345. [Google Scholar]
- Müller F, Frentzen M. Phosphatidylglycerolphosphate synthases from Arabidopsis thaliana. FEBS Lett. 2001;509:298–302. doi: 10.1016/s0014-5793(01)03163-5. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Murata N, Fujimura Y, Higashi S. Glycerolipids in various preparations of photosystem II from spinach chloroplasts. Biochim Biophys Acta. 1990;1019:261–268. [Google Scholar]
- Murata N, Ishizaki-Nishizawa O, Higashi S, Hayashi H, Tasaka Y, Nishida I. Genetically engineered alteration in the chilling sensitivity of plants. Nature. 1992;356:710–713. [Google Scholar]
- Murata N, Nishida I. Lipids of blue-green algae (cyanobacteria) In: Stumpf PK, editor. The Biochemistry of Plants. Vol. 9. New York: Academic Press; 1987. pp. 315–347. [Google Scholar]
- Nussberger S, Dörr K, Wang DN, Kühlbrandt W. Lipid-protein interactions in crystals of plant light-harvesting complex. J Mol Biol. 1993;234:347–356. doi: 10.1006/jmbi.1993.1591. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsisdeficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–8. [Google Scholar]
- Rossak M, Schäfer A, Xu N, Gage DA, Benning C. Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch Biochem Biophys. 1997;340:219–230. doi: 10.1006/abbi.1997.9931. [DOI] [PubMed] [Google Scholar]
- Sato N, Hagio M, Wada H, Tsuzuki M. Requirement of phosphatidylglycerol for photosynthetic function in thylakoid membranes. Proc Natl Acad Sci USA. 2000;97:10655–10660. doi: 10.1073/pnas.97.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid VHR, Cammarata KV, Bruns BU, Schmidt GW. In vitro reconstruction of the photosystem I light-harvesting complex LHCI-730: Heterodimerization is required for antenna pigment organization. Proc Natl Acad Sci USA. 1997;94:7667–7672. doi: 10.1073/pnas.94.14.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler P-A, Smutny J, Rawler A. Involvement of distinct populations of phosphatidylglycerol and phosphatidylcholine molecules in photosynthetic electron-flow activities. Biochim Biophys Acta. 1987;891:85–93. [Google Scholar]
- Somerville C. Direct tests of the role of membrane lipid composition in low-temperature-induced photoinhibition and chilling sensitivity in plants and cyanobacteria. Proc Natl Acad Sci USA. 1995;92:6215–6218. doi: 10.1073/pnas.92.14.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolieres A, Dainese P, Bassi R. Heterogenous lipid distribution among chlorophyll-binding proteins of photosystem II in maize mesophyll chloroplasts. Eur J Biochem. 1994;221:721–730. doi: 10.1111/j.1432-1033.1994.tb18785.x. [DOI] [PubMed] [Google Scholar]
- Usui M, Sembongi H, Matsuzaki H, Matsumoto K, Shibuya I. Primary structures of the wild-type and mutant alleles encoding the phosphatidylglycerophosphate synthase of Escherichia coli. J Bacteriol. 1994;176:3389–3392. doi: 10.1128/jb.176.11.3389-3392.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooten O, Snel JFH. The use of fluorescence nomenclature in plant stress physiology. Photosynth Res. 1990;25:147–150. doi: 10.1007/BF00033156. [DOI] [PubMed] [Google Scholar]
- van't Hof R, van Klompenburg W, Pilon M, Kozubek A, de Korte-Kool G, Demel RA, Weisbeek PJ, de Kruijff B. The transit sequence mediates the specific interaction of the precursor of ferredoxin with chloroplast envelope membrane lipids. J Biol Chem. 1993;268:4037–4042. [PubMed] [Google Scholar]
- Williams JG, McMaster CR. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiaecholinephosphotransferase. J Biol Chem. 1998;273:13482–13487. doi: 10.1074/jbc.273.22.13482. [DOI] [PubMed] [Google Scholar]