Abstract
SPY (SPINDLY) encodes a putative O-linked N-acetyl-glucosamine transferase that is genetically defined as a negatively acting component of the gibberellin (GA) signal transduction pathway. Analysis of Arabidopsis plants containing a SPY::GUS reporter gene reveals that SPY is expressed throughout the life of the plant and in most plant organs examined. In addition to being expressed in all organs where phenotypes due to spy mutations have been reported, SPY::GUS is expressed in the root. Examination of the roots of wild-type, spy, and gai plants revealed phenotypes indicating that SPY and GAI play a role in root development. A second SPY::GUS reporter gene lacking part of the SPY promoter was inactive, suggesting that sequences in the first exon and/or intron are required for detectable expression. Using both subcellular fractionation and visualization of a SPY-green fluorescent protein fusion protein that is able to rescue the spy mutant phenotype, the majority of SPY protein was shown to be present in the nucleus. This result is consistent with the nuclear localization of other components of the GA response pathway and suggests that SPY's role as a negative regulator of GA signaling involves interaction with other nuclear proteins and/or O-N-acetyl-glucosamine modification of these proteins.
GAs are endogenous plant growth regulators that have been studied for over 70 years. Until relatively recently, most of this research has concentrated on determining the physiological role of various GAs, defining the GA biosynthetic pathway in plants and fungi, and developing practical uses for GAs and chemical inhibitors of GA biosynthesis in agriculture. Over the last decade, considerable progress has also been made in understanding how plants are able to perceive the level of endogenous GAs and the mechanism by which the GA signal is transduced (Thornton et al., 1999a; Lovegrove and Hooley, 2000; Sun, 2000; Richards et al., 2001). This research has been made possible by advances in molecular genetic techniques in model systems such as Arabidopsis, rice (Oryza sativa), and the aleurone layer of cereal grains. In Arabidopsis, several negatively acting components of the GA response pathway have been characterized in some detail, including SPY (SPINDLY; Jacobsen and Olszewski, 1993; Jacobsen et al., 1996), and two members of the GRAS family (Pysh et al., 1999), RGA (REPRESSOR OF ga1-3) and GAI (GA INSENSITIVE; Peng et al., 1997; Silverstone et al., 1998). The cloning of GAI has led to the identification of orthologous genes from other species such as the wheat (Triticum aestivum) rht homeo-alleles that are the genetic basis of the “green revolution” (Peng et al., 1999a). Other potential GA-signaling proteins include SHI (SHORT INTERNODES), SLY (SLEEPY), and PKL (PICKLE) in Arabidopsis (Steber et al., 1998; Fridborg et al., 1999; Ogas et al., 1999), and GAMyb in barley (Hordeum vulgare; Gubler et al., 1999). A role for heterotrimeric G proteins has also been suggested based on work with inhibitors in wild oat (Avena sativa) aleurones (Jones et al., 1998) and analysis of the d1 mutant of rice (Ashikari et al., 1999; Fujisawa et al., 1999; Ueguchi-Tanaka et al., 2000). Several other second messengers that play a role in the process have also been identified (Lovegrove and Hooley, 2000).
RGA and GAI are thought to be nuclear-localized transcriptional regulators that act as repressors of GA signal transduction (Silverstone et al., 2001). At present, the identity of the genes regulated by RGA and GAI is not known, but expression of an RGA/GAI homolog from rice, OsGAI, in yeast (Saccharomyces cerevisiae) cells suggests that RGA and GAI are likely to be transcriptional activators or co-activators that control the expression of other negatively acting components of GA response (Ogawa et al., 2000). The RGA and GAI genes appear to be expressed throughout the plant and loss-of-function rga and gai mutations increase GA response in hypocotyls, rosette leaves, and internodes (Peng et al., 1997; Silverstone et al., 1997a, 1999b). Consistent with previous models that activation of GA signaling involves inhibition of a repressor of GA response (Harberd et al., 1998; Sun, 2000; Richards et al., 2001), recent work shows that GA treatment causes degradation of RGA (Silverstone et al., 2001).
In contrast, the available evidence suggests that SPY is a cytosolic O-linked GlcNAc transferase (OGT; Thornton et al., 1999b; Roos and Hanover, 2000). As a consequence, SPY is thought to act as a repressor of GA signaling by posttranslationally O-GlcNAc modifying as yet unknown target proteins. Although there is strong evidence to suggest that SPY is required for normal GA response in both Arabidopsis (Jacobsen et al., 1996) and barley (Robertson et al., 1998), the phenotypes of spy mutants also suggests that SPY may have additional roles in plant development (Swain et al., 2001). A complex role for SPY is consistent with proposed functions of animal OGTs, which are also thought to modify proteins involved in a wide range of cellular functions (Comer and Hart, 2000).
Although limited experiments to examine the localization of SPY mRNA by in situ hybridization in seedlings and developing flowers has detected SPY mRNA in these tissues (Jacobsen et al., 1997), the expression throughout development has not been characterized. Moreover, it is not known if SPY expression is regulated during development or by environmental or hormonal signals. Although the sequence of the SPY protein contains no obvious localization signals, suggesting that it is localized in the cytosol, the localization of SPY has also not been determined.
In this paper, the expression of the SPY gene and the cellular localization of the SPY protein are examined. SPY expression was determined using a SPY::GUS reporter gene, and both subcellular fractionation and visualization of a SPY-green fluorescent protein (GFP) fusion protein were used to determine the localization of SPY. Based on the pattern of SPY expression, a new role for the SPY and GAI loci in root development is identified.
RESULTS
SPY Is Expressed throughout the Plant
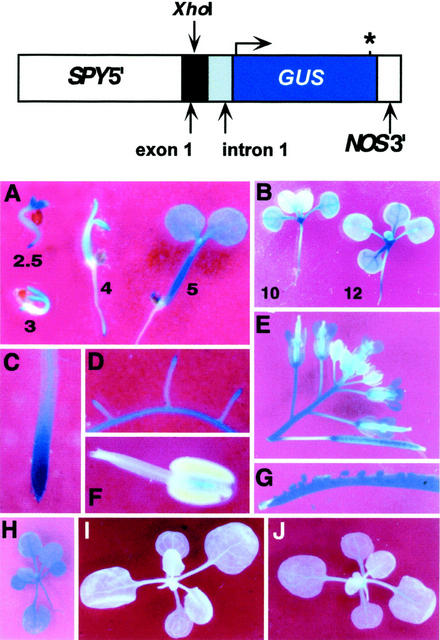
Two reporter genes that place the expression of β-glucuronidase (GUS) under the control of sequences from upstream of the start of SPY translation were constructed and used to characterize the SPY promoter and its activity. The first exon of SPY is untranslated and by comparing the promoter activity of reporter genes that either contained (SPY::GUS1) or lacked (SPY::GUS2) the complete first intron and exon sequences, the role of these sequences in controlling SPY expression was tested. A map of SPY::GUS1 is shown in Figure 1. SPY::GUS2 was identical to SPY::GUS1 except that it does not contain the SPY region 3′ to the XhoI site present in exon 1. No GUS activity was detected in any transgenic plant containing SPY::GUS2, indicating that sequences in the first exon and/or intron are required for detectable levels of expression (data not shown).
Figure 1.
Analysis of SPY::GUS1 expression. The expression of the SPY gene during plant development was examined using the SPY::GUS1 reporter gene, which expresses GUS under the control of the SPY promoter. SPY5′ is SPY genomic sequence from an HindIII site 2,361 bp upstream of the 5′ end of exon 1, all of exon 1 (which is not translated), intron 1, and the first 16 nucleotides of exon 2 just before the SPY start codon. An asterisk represents the stop codon. For A through D, I, and J, the GUS staining reaction was allowed to proceed at 25°C, whereas other images were stained at 37°C to increase the intensity of the staining (see “Materials and Methods”). The numbers in A and B indicate seedling age in days. The plant in H is 3 weeks old. The plants in I and J were treated with either 2 μL of ethanol or 2 μL of ethanol containing 20 μg of GA3, respectively, and stained 24 h after treatment.
The pattern of GUS expression in plants carrying SPY::GUS1 matched the localization of SPY mRNA as determined by in situ hybridization (Jacobsen et al., 1997). For example, both methods of detection revealed SPY expression throughout seedlings with more intense signal in the shoot apex and root tips. In addition, expression of the SPY cDNA under the control of the promoter used in SPY::GUS1 rescues spy mutants (Swain et al., 2001). Therefore, we believe that the pattern of GUS expression obtained with this construct reflects the expression pattern of the SPY gene and have further characterized SPY expression by determining the GUS expression pattern.
Eleven independent lines carrying SPY::GUS1 in the No-O background were identified. Based on preliminary examination of the GUS staining of the 11 lines, two representative lines, 702 and 711, each containing a single transgene locus, were selected for more detailed analysis. Both lines gave the same results and data from line 702 are shown in Figure 1. In general, GUS activity was detected in all organs of the plant and at all stages of the life cycle; however, some developmental regulation was apparent. GUS activity was detected 1 d after germination in the radicle just before its emergence from the seed (data not shown). At 2.5 and 3 d after germination, expression in the young seedling was highest in the cotyledons and the root tip. At 3, 4, and 5 d, expression was also detectable in the hypocotyl. At 10 d of age, GUS activity in the first pair of true leaves was reduced relative to the rest of the seedling. Two days later, this difference disappeared and the intensity of staining was again fairly similar throughout the aboveground portion of the plant, with a higher staining intensity in the vegetative apex (Fig. 1B). This developmental regulation of GUS activity was not detected in leaves developing at later nodes (note staining in youngest visible leaf in Fig. 1, B and H). Older plants (Fig. 1H) also displayed uniform GUS staining throughout the vegetative organs, but this staining was less intense (the intensity of the staining of the plant shown in Fig. 1H was enhanced relative to the seedling shown in Fig. 1I by staining it at a higher temperature; see “Materials and Methods”). In older seedlings, expression was observed throughout the root, particularly at the tip of the primary root (Fig. 1C) and in lateral roots (Fig. 1D). GUS staining was also observed in trichomes and senescing leaves (data not shown), and in inflorescence internodes, flowers (anther connective, sepals, and carpels, Fig. 1, E and F). Expression was observed in the seeds and carpels of fully elongated siliques (Fig. 1, E and G). Lower expression was also observed in expanding siliques (Fig. 1E) and in the developing seeds in these siliques when they were cut open to allow the GUS substrate to penetrate (data not shown). Expression was also detected in the embryo of maturing seeds (after the endosperm disappeared) when No-O flowers were pollinated with pollen from line 711 (data not shown).
The Effects of Hormones and Temperature on the Expression of the SPY::GUS1 Reporter Gene
To investigate possible transcriptional regulation of SPY, various treatments and growth regulators were applied to 702 and 711 plants. Plants were germinated on Murashige and Skoog plates without growth regulators (see “Materials and Methods”) and at 5 d of age were transferred to new plates containing 10−5 m naphthaline acetic acid (an auxin), 10−5 m benzyl amino purine (a cytokinin), or 10−5 m abscisic acid (ABA). Other plants were transferred to Murashige and Skoog plates and placed in the dark at 4°C, 22°C, or 30°C. Control plants were transferred to fresh plates and kept at 22°C in the light. Seedlings were stained for GUS activity after 19 h of treatment. No differences in GUS staining were observed between control plants and plants receiving any of the treatments.
Because SPY is thought to be a negative regulator of GA signal transduction, we also examined whether the SPY::GUS1 gene responded to GA3 treatments. Lines 702 and 711 in the No-O background were analyzed by treating 3-week-old seedlings with either 20 μg of GA3 in 2 μL of ethanol or 2 μL of ethanol only (control). After 24 h, whole seedlings were stained for GUS activity and GA-treated seedlings were found to stain slightly more intensely than control plants, particularly in the shoot apex (Fig. 1, I and J). This experiment was repeated several times with similar results on each occasion. To quantify this difference, GUS activity in seedlings representing four independent lines in the No-O background, including 702 and 711, was quantified. Although we observed a slight (70%) increase in GUS activity in response to continuous growth in the presence of 10−6 m GA3, we were not able to repeat this result when the same SPY::GUS1 construct was introduced in the Columbia background (data not shown). We were also unable to observe any effect of the ga1, gai, and spy-4 mutations on SPY::GUS1 activity in the Columbia background (data not shown).
In conclusion, the SPY::GUS1 reporter gene suggests that SPY is expressed throughout the life of the plant and in most plant organs examined, and there is at most only minor regulation at the transcriptional level by the treatments investigated.
SPY Is Present in the Nucleus and Cytosol
Two approaches were used to determine the cellular localization of the SPY protein. In the first approach, a construct expressing full-length SPY-GFP fusion protein under the control of the SPY promoter was introduced into plants and the localization of GFP was determined (Fig. 2). In the second approach, SPY was detected on protein blots containing proteins from different subcellular fractions (Fig. 3).
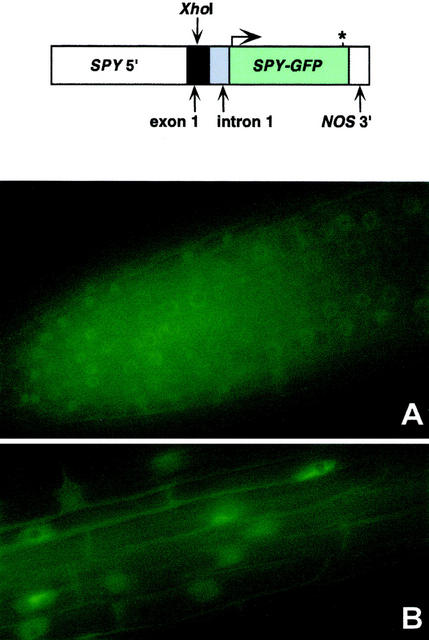
Figure 2.
SPY-GFP is present predominantly in the nucleus. Localization of SPY-GFP in the roots of SPY::SPY-GFP plants. An asterisk represents the stop codon. A, SPY-GFP is expressed in roots. B, GFP is localized predominantly to the nucleus of cells in the zone of elongation.
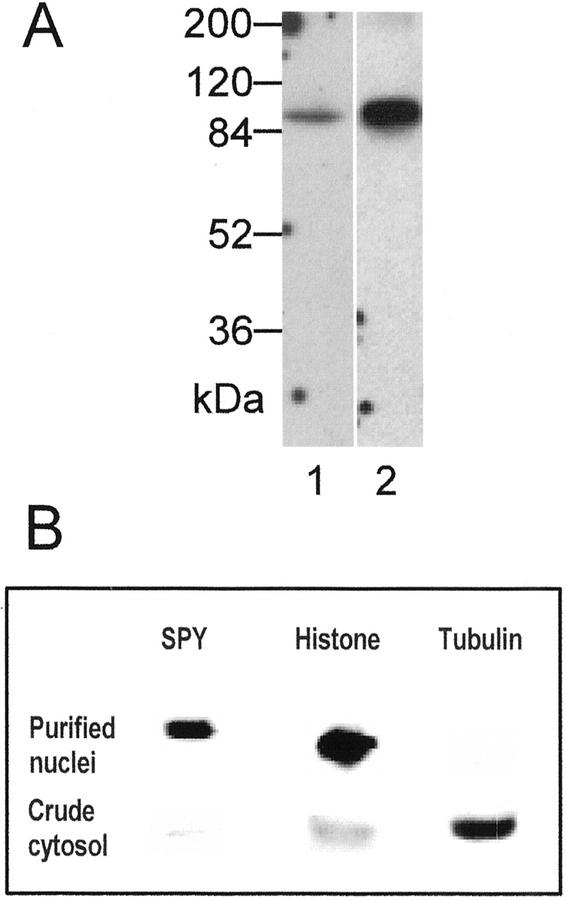
Figure 3.
SPY is present predominantly in the nucleus. A, Anti-SPY antibody was used to detect SPY in protein extracts from Arabidopsis seedlings (lane 1) and from cauliflower (Brassica oleracea var. botrytis) heads (lane 2). Each lane contained 50 μg of protein. SPY was detected using two different antisera (see “Materials and Methods”) with similar results. B, Identical blots, each containing nuclear and crude cytosolic protein preparations from cauliflower inflorescence, were probed with anti-SPY, antihistone, and antitubulin antibodies. Each lane contained approximately 35 μg of protein.
Before initiating experiments to localize the SPY-GFP protein, the functionality of the fusion protein was assessed by determining if expression of this protein in spy-3 seeds restored sensitivity to paclobutrazol, a GA biosynthesis inhibitor. T2 seeds from 10 independently generated spy-3 lines carrying the SPY-GFP transgene were scored for resistance to kanamycin, the marker linked to the SPY-GFP transgene, and resistance to paclobutrazol (Table I). In every line, germination on paclobutrazol was reduced and the ratios of resistance to kanamycin (kanr):sensitivity to kanamycin (kans) and sensitivity to paclobutrazol (Paclos):resistance to paclobutrazol (Paclor) seeds were similar, indicating that the SPY-GFP transgene encodes a protein with SPY activity.
Table I.
The SPY-GFP transgene encodes a protein with SPY activity
| Linesb | No. of Seedlings
|
Segregation Ratio | χ2c | No. of Seedlingsa
|
Segregation Ratio | χ2 | ||
|---|---|---|---|---|---|---|---|---|
| Kanr | Kans | Paclos | Paclor | |||||
| WT | – | – | – | – | 24 | 0 | – | – |
| spy-3 | – | – | – | – | 2 | 18 | – | – |
| 2b | 40 | 7 | 3:1 | 1.92 | 41 | 11 | 3:1 | 0.31 |
| 3a | 33 | 12 | 3:1 | 0.05 | 40 | 11 | 3:1 | 0.24 |
| 4a | 19 | 14 | 3:1 | 14.00 | 18 | 14 | 3:1 | 4.50 |
| 5a | 53 | 21 | 3:1 | 0.34 | 26 | 8 | 3:1 | 0.03 |
| 6b | 30 | 10 | 3:1 | 0.00 | 27 | 11 | 3:1 | 0.24 |
| 7a | 43 | 13 | 3:1 | 0.07 | 29 | 9 | 3:1 | 0.02 |
| 9a | 30 | 0 | 15:1 | 1.88 | 38 | 3 | 15:1 | 0.08 |
| 9b | 36 | 4 | 15:1 | 0.90 | 17 | 2 | 15:1 | 0.56 |
Paclor seeds germinate in the presence of 35 mg L−1 paclobutrazol while Paclos seeds do not.
The kanamycin and paclobutrazol sensitivity of seeds of different lines; wild-type Columbia (WT), spy-3, and spy-3 segregating for a SPY-GFP transgene (2b–9b) were determined.
P = 0.050 when χ2 = 3.84.
The SPY-GFP transgene is driven by the same promoter as SPY::GUS1, and the expression pattern in roots, where autofluorescence was low enough for the protein to be easily detected, was similar to the SPY::GUS1 expression pattern (Figs. 1C and 2A). In root cells, the majority of the GFP fluorescence is from the nucleus, although some is also from the cytoplasm (Fig. 2B). In comparisons between plants carrying the SPY-GFP transgene and untransformed controls, the fluorescence from the cytosol of the transgenic plant was clearly stronger and distinguishable from that of the control, indicating that the cytosolic SPY-GFP fluorescence of the transgenic plant was not attributable to autofluorescence from the cell wall (data not shown). In other parts of the plant, autofluorescence prevented GFP localization. Examination of SPY-GFP seedlings 0, 15, 30, 45, and 60 min after treatment with 10−4 m GA3 or 1.5 × 10−6 m ABA did not detect any effect of these treatments on either the localization or abundance of SPY-GFP. We also failed to observe any change in GFP activity 24 and 48 h after transfer to Murashige and Skoog plates containing 10−4 m paclobutrazol (data not shown).
We have found that SPY from Arabidopsis plants is difficult to detect by western-blot analysis, presumably because it is rare. Because immunolocalization experiments (T. Thornton and N. Olszewski, unpublished data) and the expression pattern of SPY::GUS1 (Fig. 1B) suggested that SPY might be more abundant in apices, we attempted to detect SPY from cauliflower inflorescences. A western blot containing total soluble cauliflower inflorescence proteins and a protein sample from Arabidopsis seedlings, in which SPY had been concentrated and enriched by precipitation with ammonium sulfate, was probed with affinity-purified anti-SPY antibodies that recognize recombinant SPY protein expressed in both Escherichia coli and insect cells. A single protein of the expected size of SPY was detected in both samples (Fig. 3A), suggesting that these antibodies recognize cauliflower SPY and that it is more abundant in the inflorescence of cauliflower than in Arabidopsis plants.
To confirm that SPY is localized to the nucleus, duplicate western blots containing proteins from purified cauliflower nuclei and total soluble proteins were probed with antibodies against SPY, histone, and tubulin (Fig. 3B). Consistent with the SPY-GFP localization to the nucleus, SPY was most abundant in the purified nuclei. The nuclei proteins do not react strongly with antitubulin antibodies, indicating that the nuclei are not highly contaminated with cytosolic proteins. SPY protein was also detected in the soluble protein fraction, suggesting that SPY is also present in the cytosol. However, this protein preparation also contained histones, indicating that this fraction was contaminated with nuclear proteins. Consequently, we were not able to determine if SPY is also present in the cytosol of cauliflower cells.
SPY Is Required for Normal Leaf and Root Growth
Detailed examination of the phenotype of several spy mutants has been used previously to examine the role of SPY in plant development (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996). Nevertheless, it is likely that additional roles for SPY remain to be discovered, especially because more recent work suggests that SPY may play a role in plant development beyond its role in GA signaling (Swain et al., 2001). Because the expression of a gene in a particular organ or developmental stage is consistent with a physiological role for the corresponding protein, the pattern of SPY expression was compared with known roles for SPY. Two aspects of SPY expression suggested possible new functions for SPY. In contrast to other leaves, a change in SPY::GUS1 expression during development was readily detectable in the first pair of true leaves (Fig. 1B). To test the hypothesis that SPY may have a unique role in these leaves, the first pair of leaves was examined for defects in development. The distance across the first leaf pair of spy plants was smaller than those of wild type (WT; Table II), confirming that SPY is essential for the normal development of these leaves, but no additional defects in leaf development were detected. Previous analysis of severe spy mutants late in vegetative development revealed that they possess smaller rosettes than WT plants (Swain et al., 2001). Therefore, although SPY expression varies during the growth of the first leaf pair, these leaves exhibited no phenotypes that are not observed in leaves where SPY expression is constant throughout development.
Table II.
Early leaf growth is altered in spy mutants
All in the Columbia background.
Distance tip to tip for the first pair of true leaves at 11 d of age. At this stage, the young leaves were nearly horizontal. This measurement was used because of the difficulty in distinguishing between the first and second true leaf at this developmental stage.
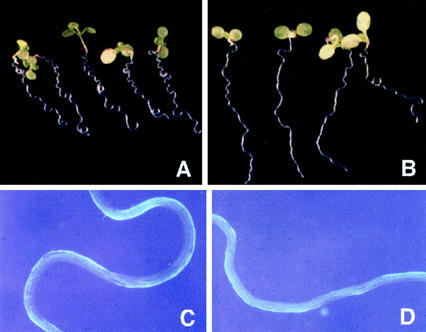
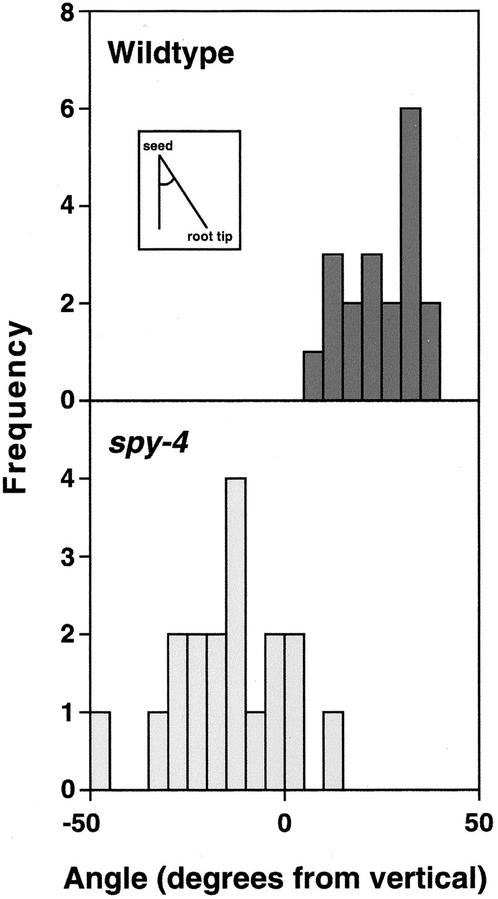
The SPY::GUS1 and SPY::SPY-GFP reporter constructs also reveal that SPY is expressed in roots (Figs. 1 and 2). Initial inspection of WT and spy mutant roots did not reveal any obvious differences in morphology or growth. To examine root development more carefully, roots were allowed to grow along the surface of an agar plate inclined at an angle of about 30° from the vertical. Due to the nutation of the root as it elongates, WT roots grew aslant of the vertical and the root also exhibited a wavy phenotype. Because it has been reported that this phenotype is difficult to detect in the Columbia background (Rutherford and Masson, 1996), we used spy mutants in the Wassilewskija (Ws) and Landsberg erecta (La-er) backgrounds. As shown in Figure 4, spy-4 roots behave very differently from WT Ws roots under these conditions, demonstrating that root growth is altered in the mutant plants. The wavy pattern was less pronounced in spy-4 roots (compare Fig. 4, C with D) and the mean angle the roots deviated from the vertical was reversed and significantly different (P < 0.001) from WT (Fig. 5; Table III). To confirm that this phenotype is due to loss-of-SPY function and not some previously unrecognized mutation in the spy-4 plants, the growth of spy-5 roots was also examined and found to exhibit similar abnormal root growth (P < 0.001; Table III).
Figure 4.
spy-4 roots grow abnormally on slanted plates. WT and spy-4 seedlings were germinated on slanted plates on 1% (w/v) agar as described in “Materials and Methods” to compare root growth and development. A, WT Ws seedlings. B, spy-4 (Ws background) seedlings. C, Enlarged view of WT roots showing uniform twisting pattern. D, Enlarged view of spy-4 roots showing abnormal twisting pattern.
Figure 5.
SPY is required for normal “root waving.” Seedlings were grown on slanted plates as for Figure 4, and the angle of the root tip from the vertical (relative to the highest part of the plate) determined as shown in the inset. The data show the root angle observed from the front of the plates. The frequency value represents the number of seedlings in each class.
Table III.
Root waving is altered in the spy and gai mutants
| Genotype | Agara | Angleb |
|---|---|---|
| % | degrees | |
| Ws | 1.0 | 28.1 ± 8.9 |
| spy-4 | 1.0 | −15.7 ± 5.0 |
| Ws + 1 × 10−5m GA3 | 1.0 | 32.0 ± 2.3 |
| La-er | 1.0 | 27.5 ± 2.8 |
| spy-5 | 1.0 | 6.7 ± 4.2 |
| Ws | 0.6 | 40.7 ± 3.1 |
| spy-4 | 0.6 | −6.8 ± 6.5 |
| La-er | 0.6 | 21.0 ± 2.3 |
| gai | 0.6 | 29.0 ± 1.4 |
% Agar used to solidify the medium.
Angle calculated as described in Figure 4.
In an attempt to determine whether the altered angle of root growth of spy mutants represents a physiological role for GAs, two experiments were performed. In the first experiment, we were unable to mimic the spy-4 phenotype by growing WT Ws roots on plates containing 10−5 m GA3 (Table III). In the second experiment, the effect of the gai mutation, which reduces GA response (Peng et al., 1997, 1999b), was examined. When grown on plates solidified with 1.0% (w/v) agar, no differences between WT La-er and gai were detected (data not shown). However, when plates solidified with 0.6% (w/v) agar were used instead, gai roots grew at a slightly but significantly greater (P < 0.001) angle than WT La-er roots (Table III). On 0.6% (w/v) plates, the difference between WT Ws and spy-4 was still apparent (Table III), suggesting that spy-4 and gai have opposite effects on this phenotype.
DISCUSSION
Sequences in the First Intron or Exon of the SPY Gene Are Required for Promoter Activity
The SPY transcript has a 350-bp untranslated leader at the 5′ end, and the start codon of the SPY open reading frame (ORF) is located at position 27 of the second exon (Fig. 1). The genomic region between 3,032 and 11 bp upstream of the start of translation is likely to encompass the full SPY promoter. Driving the expression of the SPY ORF with this promoter rescues spy mutants (Swain et al., 2001), and the seedling expression pattern of SPY::GUS1 (Fig. 1), which is driven by this promoter, accurately reflects the pattern of SPY expression as determined by in situ hybridization in young seedlings and flowers (Jacobsen et al., 1997).
In contrast, the promoter region of the SPY::GUS2 gene, which in comparison with the SPY::GUS1 gene lacked the 3′ most 255 bp of the first exon, the entire first intron (320 bp), and 16 bp of the second exon, was inactive, indicating that this region contains sequences that are essential for promoter activity. A role for introns in the regulation of gene expression has now been recognized for a number of plant genes (e.g. Sieburth and Meyerowitz, 1997; Silverstone et al., 1997b). In contrast to the apparent importance of intron 1, there is no evidence to suggest that the other introns are required because driving expression of either the SPY or the SPY-GFP ORFs with the promoter used in SPY::GUS1 can rescue spy mutants (Swain et al., 2001; Table I).
SPY Acts throughout the Plant
Analysis of the spy mutant phenotype has provided the most compelling evidence that SPY is a negatively acting component of GA response (Jacobsen and Olszewski, 1993; Jacobsen et al., 1996; Swain et al., 2001). In particular, this hypothesis is supported by the interaction between the ga1 and spy mutations. The GA1 locus encodes copalyl diphosphate synthase, the first enzyme in the GA biosynthetic pathway (Sun and Kamiya, 1994). Mutant ga1 plants are severely GA deficient and exhibit a range of phenotypes throughout the plant and throughout the life cycle, including poor seed germination, reduced vegetative growth, delayed flowering, and abnormal flower development and male sterility (Koornneef and van der Veen, 1980). The ability of spy mutants to partially or fully suppress the various ga1 mutant phenotypes suggests that loss of SPY activity either increases GA responsiveness or partially abolishes the requirement for GA. The expression of the SPY gene has been analyzed using the SPY::GUS1 reporter gene (Fig. 1). The reporter gene is expressed essentially constitutively throughout the life of the plant and in all plant organs examined. This expression pattern is consistent with the observed phenotypes of ga1 spy double mutants (Silverstone et al., 1997a; Swain et al., 2001) and with other spy mutant phenotypes. For example, the SPY locus was originally isolated in a genetic screen for mutants able to germinate in the presence of paclobutrazol, an inhibitor of GA biosynthesis (Jacobsen and Olszewski, 1993), and we found that SPY is expressed in the emerging radicle (Fig. 1).
To determine whether the expression of SPY in roots (Figs. 1 and 2) reflects a role for SPY in root development, the root growth of spy mutants was examined. Mutant roots exhibit alterations in the angle that they grow along a slanted surface and the waving pattern that they exhibit during this growth (Figs. 4 and 5). These phenotypes could result from an alteration(s) in the nutation of the root tip, thigmomorphogenesis, the elongation rate, and/or the ability of the root to adhere to the surface. Abnormal root waving has also been observed for mutants with defects in a range of developmental processes other than GA response (e.g. Rutherford and Masson, 1996; Mullen et al., 1998). The spy root phenotype could not be induced in WT plants by exogenous GA application (Table III), although in this experiment GA was applied to the entire root, whereas SPY expression is not evenly distributed along the length of the root but appears to be highest in the root tip (Figs. 1 and 2). However, the gai mutation, which results in reduced GA response (Peng et al., 1997, 1999b), has a root phenotype opposite to that observed in spy mutants (Table III), suggesting that GA response is involved in this phenomenon. Other studies have suggested a role for GAs in root elongation growth (e.g. Yaxley et al., 1999), and the GA1 gene is expressed in Arabidopsis root tips (Silverstone et al., 1997b), consistent with a need for root cells to be able to respond to GAs.
Transcriptional Regulation of SPY
Although we found some evidence for developmental regulation of SPY, we observed no evidence for regulation in response to treatment of plants with naphthalene acetic acid, benzyl amino purine, ABA, or in response to dark, heat, or cold. The ga1, gai, and spy-4 mutations also did not obviously alter SPY expression in the Columbia background. Although a slight (less than 2-fold) induction of SPY::GUS1 expression by GA3 was observed in the No-O background (Fig. 1, I and J), this induction was not detected in the Columbia background. Hence, it appears that if SPY is transcriptionally regulated by GA, it is at most relatively minor. The potential for transcriptional regulation of SPY by GA is, however, consistent with the slight induction of RGA and GAI (Silverstone et al., 1998) and of OsGAI (Ogawa et al., 2000) in response to applied GA. The possibility that GA response is attenuated by GA action is similar to models of feedback and feedforward regulation of GA metabolism (Coles et al., 1999; Xu et al., 1999) and suggests that both the endogenous GA level and sensitivity to GA may be under homeostatic control.
Given that SPY now appears to have multiple roles in plant growth and development, it seems less likely that significant regulation of SPY activity will occur by transcriptional control because changing the amount of SPY protein will not be selective for a single pathway. Instead, it is more likely that regulation of SPY occurs at the protein level with SPY-interacting proteins or posttranslational modification regulating SPY's substrate specificity and/or activity. This model has also been proposed for the regulation of animal OGTs (Kreppel and Hart, 1999; Lubas and Hanover, 2000).
SPY Is Present in the Nucleus and Cytosol
Both the localization of SPY-GFP (Fig. 2) and subcellular fractionation experiments (Fig. 3) demonstrate that the majority of the SPY protein is present in the nucleus. It also appears likely that some SPY protein is present in the cytoplasm. How SPY becomes nuclear localized is unclear because, unlike animal OGT (Kreppel et al., 1997; Lubas et al., 1997), it does not contain any obvious nuclear localization signals (NLS; Jacobsen et al., 1996). Nuclear localization could occur instead as a result of an interaction between SPY and proteins containing an NLS. This model is supported by the presence of tetratricopeptides in SPY, which in other proteins function as protein-protein interaction domains (Blatch and Lässle, 1999). Interactions with any of several other components of the GA response pathway could also localize SPY to the nucleus. Both RGA and the closely related GAI protein contain a consensus NLS motif (Silverstone et al., 1998), as does a RGA/GAI homolog from rice, OsGAI (Ogawa et al., 2000). GFP fusion proteins with RGA and OsGAI are nuclear localized in onion (Allium cepa) epidermal cells (Silverstone et al., 1998; Ogawa et al., 2000). GAMyb is a transcriptional activator and presumably is also nuclear localized (Gubler et al., 1999). Although we do not know if SPY from plants is O-GlcNAc modified, SPY produced in insect cells is (T. Thornton and N. Olszewski, unpublished data). Because O-GlcNAc addition can serve as an NLS for animal proteins (Snow and Hart, 1998), O-GlcNAc modification of SPY may potentially serve as a NLS in plants.
The presence of SPY in the nucleus suggests that its role as a negative regulator of GA signaling involves interaction with other nuclear proteins and/or O-GlcNAc modification of these proteins. Because missense mutations affecting either the tetratricopeptide or OGT catalytic domain affect GA signaling (Jacobsen et al., 1996; T.-S. Tseng and N. Olszewski, unpublished data), we believe that the latter possibility is more likely. In animals, both RNA polymerase II and a number of transcription factors are known to be O-GlcNAcylated and this modification is believed to be regulatory. Therefore, SPY's action as a negative regulator of GA signaling could involve the modification of transcription factors such a RGA, GAI, or GAMyb, or, alternatively, SPY could be localized to the promoters of GA-regulated genes and then modify the basal transcription machinery as it interacts with the promoter.
MATERIALS AND METHODS
Growth Conditions and Plant Material
All seeds were stratified for 3 d at 4°C under dim light to aid germination. All plants were grown under an 18-h long-day photoperiod of 120 μmol m−2 s−1 consisting of white fluorescent light with a temperature of 22°C (day) and 20°C (night). For the root assays, seeds were placed on the surface of 1× Murashige and Skoog plates sealed with medical (porous) tape and stratified as usual. When transferred to a growth room, the plates were placed at an angle of 30° from the vertical with the seeds toward the light source and at the top of the plate. The majority of roots grew on the surface of the agar. Roots that grew into the agar did not exhibit the root waving phenotype and were not measured. Values are shown as the mean ± se, and Student's t test was used to determine the statistical significance of differences between genotypes. To allow comparison of leaf size with WT Columbia and spy-3, the spy-4 mutation, originally generated by a T-DNA insertion in the Ws background, was backcrossed into the Columbia genetic background six times. The ga1 and gai lines were each backcrossed into the Columbia genetic background three times before combining with SPY::GUS1 lines in the Columbia background. Upon request, all novel materials described here will be made available for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this paper that would limit their use in noncommercial research purposes.
Constructs and Determination of SPY Gene Expression
All constructs were generated using standard molecular techniques. All SPY::GUS1 lines described in detail here contain a single locus, based on segregation of kanamycin resistance on the T-DNA containing SPY::GUS1 (Fig. 1). The SPY::GUS1 construct contained genomic sequence from a HindIII site 2,361 bp upstream of the 5′ end of exon 1, all of exon 1 (324 bp) and intron 1 (320 bp), and 16 nucleotides from the 5′ end of exon 2 so that the most 3′ nucleotide of this promoter corresponds to a position 11 nucleotides upstream of the SPY start codon in exon 2. SPY::GUS2 was identical to SPY::GUS1 except that the region of the SPY gene 3′ of the XhoI site, which cuts at position 69 in exon 1, was not included.
An additional reporter gene, SPY::SPY-GFP (Fig. 3), expressing a full-length SPY-GFP fusion protein under the control of the SPY promoter, was also constructed using standard methods. The SPY ORF from a full-length SPY cDNA was amplified by PCR using the following primers: 5′-AGCTGGCTGGGAATACTC-3′ and 5′-ATGCGGCCGCCATGGAGCTAGTGGAGTCCATTCTC-3′. The PCR product was subcloned into the pCR 2.1-TOPO plasmid (Invitrogen, Carlsbad, CA). The StuI and NotI fragment of the TOPO construct was used to replace the same restriction fragment of construct F (Swain et al., 2001). The new construct F was then digested with SalI and NcoI to isolate the full-length SPY promoter (as for SPY::GUS1) and cDNA. The SalI-NcoI fragment was ligated along with a NcoI-EcoRI DNA fragment from a 35S-sGFP-TYG-nos construct (Chiu et al., 1996; J. Sheen, personal communication) into pOCA28.
Binary vectors containing the different reporter genes were transformed into Agrobacterium tumefaciens strain C58C1 (pMP90) and/or AGL1 (Lazo et al., 1991). WT Arabidopsis ecotype Columbia and spy-3 were transformed by vacuum infiltration (Ye et al., 1999) using C58C1 (pMP90). Root explants of the ecotype No-O were transformed by the method of Valvekens et al. (1988) using AGL1.
GUS activity was determined as described in Jefferson et al. (1987) with the addition of 2 mm ferri- and ferro-cyanide, and quantified fluorometrically as described by Hull and Devic (1995). For Figure 1, A through D, I, and J, the reaction was allowed to proceed for 24h at 25°C, whereas other images were stained for 24h at 37°C to increase the intensity of the staining. GFP was localized in 10-d-old SPY::SPY-GFP plants that had been grown in the presence of kanamycin using an E800 microscope (Nikon, Tokyo) with a 470- to 490-nm excitation filter and a 520- to 580-nm barrier filter. Images were captured with a Cool Cam system. To determine the effects of GA and ABA on the localization and abundance of SPY-GFP, seedlings were transferred to media containing either no added hormone, 100 μm GA3, or 1.5 μm ABA and visually scored 0, 15, 30, 45, and 60 min after transfer. The effects of paclobutrazol on SPY-GFP localization and abundance were scored 24 and 48 h after transfer to 100 μm paclobutrazol. The kanamycin sensitivity of seeds was scored by germinating surface-sterilized seeds on medium solidified with 0.6% (w/v) phytoagar (Sigma, St. Louis), and containing 1× Murashige and Skoog salts (Sigma), 1% (w/v) Suc, and 50 μg mL−1 kanamycin (Sigma). The paclobutrazol sensitivity of seeds was determined as described previously (Jacobsen and Olszewski, 1993).
SPY Localization by Immunoblot Analysis
Antibodies against two peptides, DTKQKQVEELVRLPDC (anti-DTKQ) and LQKEVHDDPLISKDLGP (anti-LQKE),located in the C-terminal half of SPY, were prepared and affinity purified by Quality Controlled Biochemicals, Inc. (Hopkinton, MA).
To detect SPY in Arabidopsis, 0.1 g of seeds was imbibed in water in a petri dish and placed under constant light at 4°C. After 3 d, the plate was placed at room temperature under constant light. After 2 d, excess water was drained from the seedlings and they were transferred to a chilled mortar and ground to a fine powder in liquid nitrogen. The powder was mixed with 2 mL of ice-cold extract buffer (10 mm Tris, pH 7.2; 5 mm MgCl2; and 5 mm 2-mercap-toethanol) with 1 mm phenylmethylsulfonyl fluoride. The paste was transferred to tubes and centrifuged at 16,000g for 15 min to pellet insoluble material. The supernatant was transferred to a fresh tube and ammonium sulfate was added to 50% (w/v) saturation. Proteins were allowed to precipitate overnight at 4°C. Precipitated proteins were collected by centrifugation at 16,000g for 15 min. The supernatant was discarded and the pellet was resuspended in extraction buffer, desalted by gel filtration chromatography, and boiled in SDS-PAGE sample buffer.
For nuclear isolation from cauliflower (Brassica oleracea var. botrytis), 150 g of inflorescence meristem was harvested and nuclei were isolated as described by Olszewski et al. (1982), except that ethidium bromide was omitted from all buffers and the tissue was not washed with diethylether. The first supernatant (crude cytosolic fraction) and the purified nuclei obtained after washing away residual Percoll (Amersham-Pharmacia Biotech, Uppsala) were analyzed further. For immunoblot analysis, protein concentrations were approximated by the Bradford assay (Bio-Rad Laboratories, Hercules, CA) and by visual comparison with protein markers of known concentration on Coomassie Blue-stained gels. Purified nuclear and crude cytosolic proteins (35 μg) were separated by SDS-PAGE and blotted onto Immobilon-P transfer membrane (Millipore, Bedford, MA). Three identical blots were made containing both fractions and these were probed with SPY antipeptide antibodies, monoclonal anti-β-tubulin antibodies (Sigma), or antihistone H1 antibodies (gift of Dr. Steve Gantt, University of Minnesota, St. Paul). In brief, blots were blocked for at least 1 h at room temperature with constant shaking in blocking solution (5% [w/v] nonfat dry milk in Tris-buffered saline [TBS] + 0.75% [w/v] Tween 20). Blots were then incubated at room temperature with constant shaking in blocking solution with the primary antibody for 1 h for histone H1 and β-tubulin detection or overnight for SPY detection. The blots were washed two times for 5 min each with TBS + 3% (w/v) Tween 20 and then two times for 5 min each with TBS. Blots were then incubated for 30 min at room temperature with constant shaking in blocking solution with peroxidase-conjugated protein A (Sigma) for SPY and histone H1 detection or peroxidase-conjugated goat anti-mouse IgG (Boehringer Mannheim/Roche, Indianapolis) for β-tubulin detection. Blots were washed as described above and signals were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Chemical, Rockford, IL) as per the manufacturer's instructions.
ACKNOWLEDGMENTS
We thank Liz Cebula for help with generating and analyzing transgenic lines, David Marks for help with photography, Steve Jacobsen for sharing in situ hybridization data, and Lynn Hartweck for comments on the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–9604126 and MCB–9983583 to N.E.O.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.020002.
LITERATURE CITED
- Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A. Rice gibberellin-insensitive dwarf mutant gene Dwarf1 encodes the alpha-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Chiu W-l, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, Garcia-Lepe R, Lewis MJ, Hedden P. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E. The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell. 1999;11:1019–1032. doi: 10.1105/tpc.11.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T, Asahi T, Iwasaki Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci USA. 1999;96:7575–7580. doi: 10.1073/pnas.96.13.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 1999;17:1–9. doi: 10.1046/j.1365-313x.1999.00346.x. [DOI] [PubMed] [Google Scholar]
- Harberd NP, King KE, Carol P, Cowling RJ, Peng J, Richards DE. Gibberellin: inhibitor of an inhibitor of… ? Bioessays. 1998;20:1001–1008. doi: 10.1002/(SICI)1521-1878(199812)20:12<1001::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Hull GA, Devic M. The beta-glucuronidase (GUS) reporter gene system. Gene fusions: spectrophotometric, fluorometric, and histochemical detection. Methods Mol Biol. 1995;49:125–141. doi: 10.1385/0-89603-321-X:125. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE, Meyerowitz EM. SPINDLY's role in the gibberellin response pathway. Symp Soc Exp Biol. 1997;51:73–78. [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Smith SJ, Desikan R, Plakidou-Dymock S, Lovegrove A, Hooley R. Heterotrimeric G proteins are implicated in gibberellin induction of α-amylase gene expression in wild oat aleurone. Plant Cell. 1998;10:245–254. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A transformation competent Arabidopsis genomic library in Agrobacterium. Bio/Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Lovegrove A, Hooley R. Gibberellin and abscisic acid signaling in aleurone. Trends Plant Sci. 2000;5:102–110. doi: 10.1016/s1360-1385(00)01571-5. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- Mullen JL, Turk E, Johnson K, Wolverton C, Ishikawa H, Simmons C, Soll D, Evans ML. Root-growth behavior of the Arabidopsis mutant rgr1. Roles of gravitropism and circumnutation in the waving/coiling phenomenon. Plant Physiol. 1998;118:1139–1145. doi: 10.1104/pp.118.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kusano T, Katsumi M, Sano H. Rice gibberellin-insensitive gene homolog, OsGAI, encodes a nuclear-localized protein capable of gene activation at transcriptional level. Gene. 2000;245:21–29. doi: 10.1016/s0378-1119(00)00018-4. [DOI] [PubMed] [Google Scholar]
- Olszewski N, Hagen G, Guilfoyle TJ. A transcriptionally active, covalently closed minichromosome of cauliflower mosaic virus DNA isolated from infected turnip leaves. Cell. 1982;29:395–402. doi: 10.1016/0092-8674(82)90156-8. [DOI] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F et al. “Green revolution” genes encode mutant gibberellin response modulators. Nature. 1999a;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Peng J, Richards DE, Moritz T, Cano-Delgado A, Harberd NP. Extragenic suppressors of the Arabidopsis gai mutation alter the dose-response relationship of diverse gibberellin responses. Plant Physiol. 1999b;119:1199–1208. doi: 10.1104/pp.119.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Richards DE, King KE, Ait-Ali T, Harberd NP. How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:67–88. doi: 10.1146/annurev.arplant.52.1.67. [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE. Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell. 1998;10:995–1007. doi: 10.1105/tpc.10.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos MD, Hanover JA. Structure of O-linked GlcNAc transferase: mediator of glycan-dependent signaling. Biochem Biophys Res Commun. 2000;271:275–280. doi: 10.1006/bbrc.2000.2600. [DOI] [PubMed] [Google Scholar]
- Rutherford R, Masson PH. Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiol. 1996;111:987–998. doi: 10.1104/pp.111.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell. 1997;9:355–365. doi: 10.1105/tpc.9.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Chang C, Krol E, Sun T-p. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997b;12:9–19. doi: 10.1046/j.1365-313x.1997.12010009.x. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-p. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-s, Dill A, Kawaide H, Kamiya Y, Sun T-p. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1565. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martinez ES, Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997a;146:1087–1090. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow DM, Hart GW. Nuclear and cytoplasmic glycosylation. Int Rev Cytol. 1998;181:43–74. doi: 10.1016/s0074-7696(08)60416-7. [DOI] [PubMed] [Google Scholar]
- Steber CM, Cooney SE, McCourt P. Isolation of the GA response mutant sly1 a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics. 1998;149:509–521. doi: 10.1093/genetics/149.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T-p. Gibberellin signal transduction. Curr Opin Plant Biol. 2000;3:374–380. doi: 10.1016/s1369-5266(00)00099-6. [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng T-s, Olszewski NE. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol. 2001;126:1174–1185. doi: 10.1104/pp.126.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, Krepel L, Hart G, Olszewski N. Genetic and biochemical analysis of Arabidopsis SPY. In: Altman A, Ziv M, Izhar S, editors. Plant Biotechnology and In Vitro Biology in the 21st Century. New York: Kluwer Academic Publishers; 1999b. pp. 445–448. [Google Scholar]
- Thornton T, Swain SM, Olszewski N. Gibberellin signal transduction presents: the SPY who O-GlcNAc'd me. Trends Plant Sci. 1999a;4:424–428. doi: 10.1016/s1360-1385(99)01485-5. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, Matsuoka M. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci USA. 2000;97:11638–11643. doi: 10.1073/pnas.97.21.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-l, Li L, Gage DA, Zeevaart JA. Feedback regulation of GA5 expression and metabolic engineering of gibberellin levels in Arabidopsis. Plant Cell. 1999;11:927–936. doi: 10.1105/tpc.11.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley JR, Ross JJ, Sherriff LJ, Reid JB. Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 1999;125:627–633. doi: 10.1104/pp.125.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M. Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 1999;19:249–257. doi: 10.1046/j.1365-313x.1999.00520.x. [DOI] [PubMed] [Google Scholar]