Abstract
Photosystem I (PSI) of higher plants contains 18 subunits. Using Arabidopsis En insertion lines, we have isolated knockout alleles of the genes psaG, psaH2, and psaK, which code for PSI-G, -H, and -K. In the mutants psak-1 and psag-1.4, complete loss of PSI-K and -G, respectively, was confirmed, whereas the residual H level in psah2-1.4 is due to a second gene encoding PSI-H, psaH1. Double mutants, lacking PSI-G, and also -K, or a fraction of -H, together with the three single mutants were characterized for their growth phenotypes and PSI polypeptide composition. In general, the loss of each subunit has secondary, in some cases additive, effects on the abundance of other PSI polypeptides, such as D, E, H, L, N, and the light-harvesting complex I proteins Lhca2 and 3. In the G-less mutant psag-1.4, the variation in PSI composition suggests that PSI-G stabilizes the PSI-core. Levels of light-harvesting complex I proteins in plants, which lack simultaneously PSI-G and -K, indicate that PSI subunits other than G and K can also bind Lhca2 and 3. In the same single and double mutants, psag-1.4, psak-1, psah2-1.4, psag-1.4/psah2-1.4, and psag-1.4/psak-1 photosynthetic electron flow and excitation energy quenching were analyzed to address the roles of the various subunits in P700 reduction (mediated by PSI-F and -N) and oxidation (PSI-E), and state transitions (PSI-H). Based on the results, we also suggest for PSI-K a role in state transitions.
PSI mediates light-driven electron transport from plastocyanin to ferredoxin across the thylakoid membrane. The PSI complexes in plants and cyanobacteria are basically similar, but there are some notable differences (Scheller et al., 1997): The plant PSI is slightly larger (Kitmitto et al., 1997), trimer formation has been observed only in cyanobacteria (Chitnis and Chitnis, 1993), and the plant PSI core is associated with the light-harvesting complex I (LHCI), which is made up of the four different chlorophyll (Chl)/carotenoid-binding polypeptides Lhca1-4 (Jansson, 1999). In addition, among the 14 subunits that form the core of PSI in flowering plants (PSI-A through -O; Scheller et al., 1997, 2001; H.V. Scheller, personal communication), four subunits (PSI-G, -H, -N, and -O; Okkels et al., 1989, 1992; Knoetzel and Simpson, 1993; H.V. Scheller, personal communication) are not present in cyanobacteria. Conversely, no homolog of the cyanobacterial PSI-M has been found yet in higher plants.
Most essential for PSI function are the three subunits PSI-A, -B, and -C, which bind the electron acceptors. These acceptor-binding proteins and PSI-I and -J are encoded by the chloroplast genome, whereas all other subunits are nucleus encoded; in Arabidopsis, there are two functional gene copies each for PSI-D, -E, and -H (Naver et al., 1999; Varotto et al., 2000; Pesaresi et al., 2001).
Insights into the structure and function of PSI in flowering plants have been obtained by electron microscopy, analyses of pigment stoichiometry, chemical cross-linking studies, and, more recently, by the analysis of Arabidopsis lines deficient for specific subunits of PSI (for review, see Pesaresi et al., 2001; Scheller et al., 2001). Such analyses have revealed that PSI-K plays a role in organizing LHCI (Jensen et al., 2000), PSI-N is necessary for the interaction of plastocyanin with PSI (Haldrup et al., 1999), and PSI-H putatively is the attachment site for LHCII during state transitions (Lunde et al., 2000). Loss of PSI-E or -F has far more dramatic consequences on the photoautotrophic growth of Arabidopsis than corresponding mutations in cyanobacteria or algae (Chitnis et al., 1989, 1991; Zhao et al., 1993; Xu et al., 1994; Farah et al., 1995): In Arabidopsis, the entire stromal side of PSI is affected by disruption of the psaE1 gene (Varotto et al., 2000; unpublished data in our lab), whereas deficiency of PSI-F results in significantly decreased levels of most PSI polypeptides (Haldrup et al., 2000), making it difficult to distinguish between primary and secondary effects of the loss of these subunits in Arabidopsis.
Here, we report for the first time, to our knowledge, the generation and characterization of a loss-of-function mutant for PSI-G in Arabidopsis (see “Note Added in Proof”). Like the loss of subunit H or K, absence of PSI-G does not interfere with photoautotrophy. To determine whether this lack of severe phenotypic effects is due to redundancy, arising because one or more of the remaining PSI subunits can functionally substitute for the product of the defective gene, PSI-G mutants that were partially deficient in -H or completely lacked -K were tested for photosynthetic electron flow and the organization of PSI was characterized. The consequences of the simultaneous loss of PSI-G and -K were of particular interest because these two proteins are evolutionarily related and have been proposed to play similar roles in the organization of LHCI (Kjaerulff et al., 1993; Jansson et al., 1996; Jensen et al., 2000).
RESULTS
Generation of Single and Double Mutant Lines for PSI-G, -H, and -K
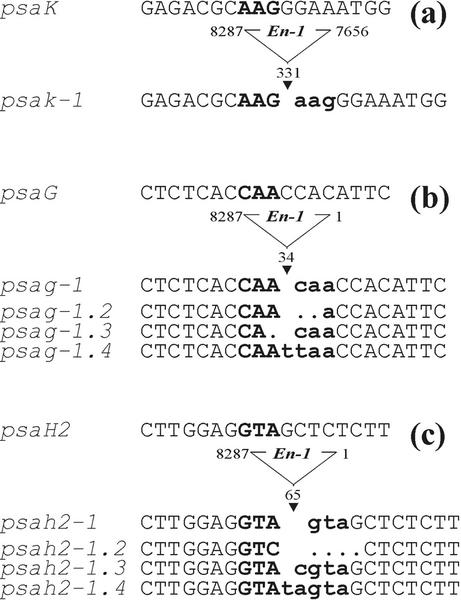
The mutants psag-1 (psag::En1), psah2-1 (psah2::En1), and psak-1 (psak::ΔEn1), each of which has an En transposon inserted in the coding region of the corresponding gene, were identified by a reverse genetic screening approach as described by Wisman et al. (1998). The stable psak-1 allele contains an immobilized fragment of about 600 bp from the right end of the En transposon within the second exon (Fig. 1a). For psag-1 and psah2-1, empty donor sites were amplified by PCR from the genomic DNA of single plants and subjected to high-resolution PAGE to identify germinal footprints leading to frameshift mutations (Fig. 1, b and c). Alleles with frameshifts ranging from +1 to +5 bp were identified for the two loci. For all further molecular and physiological analyses, psak-1, and the (+4) frameshift mutant psag-1.4 and the (+5) frameshift mutant psah2-1.4, were used. The double mutants psag-1.4/psah2-1.4, and psag-1.4/psak-1 were obtained by crossing the corresponding single mutants and genotyping their F2 progenies to identify homozygous double mutants.
Figure 1.
Mutations in the loci psaG, psaH2, and psaK. En insertions are located in the second exon of psaK (a), and the first exons of psaG (b) and psaH2 (c). DNA sequences of the transposon donor sites from independent germinal revertants of psag-1 and psah2-1 were obtained by PCR amplification. The footprints left at each locus after En excision are indicated by bold lowercase letters, whereas bold uppercase letters indicate the target site in the wild type (WT). The psak-1 allele contains a stable insertion of a deletion derivative of En in the second exon.
Photoautotrophic Growth of Single and Double Mutants
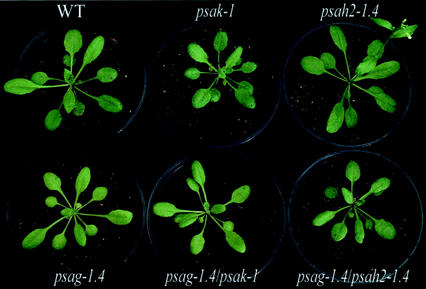
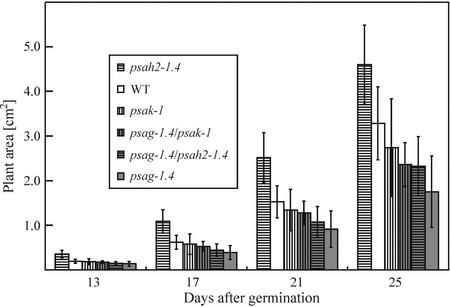
All single and double mutants could grow photoautotrophically in a growth chamber. Germination of seeds was not affected in the mutants and visual inspection did not reveal any drastic change in phenotype compared with WT plants, except for a slightly lighter pigmentation in both psag-1.4 and psag-1.4/psak-1 (Fig. 2). In two genotypes, flowering time was altered: Under long-day conditions, psag-1.4 plants flowered 8 d later, and psah2-1.4 plants 3 d earlier than the WT (Fig. 2). Variation in flowering time was increased in psag-1.4 and psag-1.4/psak-1 plants with respect to the other genotypes: 90% of individuals of the former two genotypes flowered within 7 d, whereas the latter genotypes required only a period of 5 d. Plant size was quantified systematically by noninvasive image analysis (Leister et al., 1999) between 1 and 4 weeks after germination, and differences in growth behavior were recorded (Fig. 3): psak-1 and psag-1.4 plants showed a decrease in mean size, whereas psah2-1.4 plants were in average larger than the WT. The double mutants were intermediate in mean size when compared with the corresponding single mutants.
Figure 2.
Phenotypes of WT plants, single (psag-1.4, psak-1, and psah2-1.4), and double (psag-1.4/psah2-1.4 and psag-1.4/psak-1) mutants for PSI subunits. Arabidopsis plants (5–6 weeks old) were grown in a greenhouse under long-day conditions.
Figure 3.
Growth kinetics of single and double mutants compared with WT plants. Areas of 54 plants for each genotype were measured in the period from 13 to 25 d after germination. Mean values are shown and error bars indicate sds.
PSI Polypeptide and Thylakoid Pigment Composition in WT and Mutant Plants
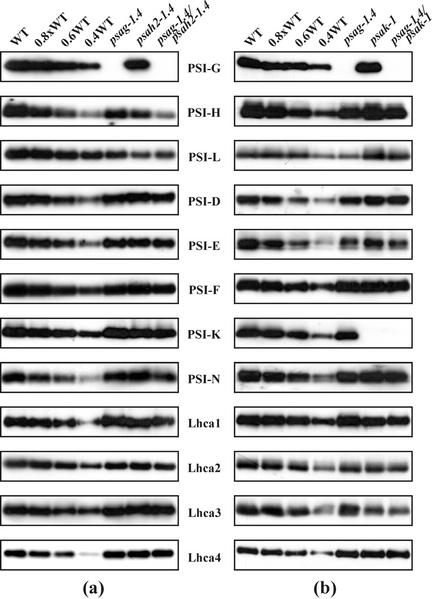
Western analyses demonstrated that the psag-1.4 mutation leads to complete loss of PSI-G in thylakoids, whereas about 70% of the WT level of PSI-H is still present in psah2-1.4 plants (Fig. 4a; Table I) due to the activity of a second gene coding for PSI-H, psaH1 (Naver et al., 1999). The absence of PSI-G also affected the abundance of almost all other PSI polypeptides, in particular PSI-L (present at about 40% of WT levels). The abundance of the LHCI proteins was slightly affected: The abundance of Lhca2 was about 80%, and of Lhca4 about 110%, that of WT.
Figure 4.
Immunoblot analysis of single and double mutants and WT plants. Aliquots (5 μg) of thylakoid proteins from WT and single- or double-mutant plants were loaded in lanes WT and psag-1.4, psah2-1.4, psag-1.4/psah2-1.4 (a), or psag-1.4, psak-1, psag-1.4/psak-1 psae1-1 (b), and decreasing amounts of WT thylakoid membrane proteins were added to lanes 0.8×, 0.6×, and 0.4× WT. Replicate filters were immunolabeled with antibodies raised against PSI-G, -H, -L, -D, -E, -F, -K, -N, and Lhca1-4. At least three independent experiments were performed of which representative results are shown. Mean values of PSI polypeptide levels in the mutant genotypes with respect to WT values are listed in Table I.
Table I.
Levels of PSI polypeptides in psag1-4, psah2-1.4, psak-1, psag1-4/psah2-1.4, and psag1-4/psak-1.4
| Mutant | PSI Polypeptide Level
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | E | F | G | H | K | L | N | Lhca1 | Lhca2 | Lhca3 | Lhca4 | |
| % of WT | ||||||||||||
| psag-1.4 | 80 | 80 | 80 | 0 | 80 | 100 | 40 | 100 | 100 | 80 | 100 | 110 |
| psah2-1.4 | 100 | 80 | 90 | 100 | 70 | 90 | 30 | 100 | 100 | 90 | 100 | 110 |
| psak-1 | 100 | 90 | 100 | 100 | 90 | 0 | 90 | 100 | 80 | 60 | 50 | 110 |
| psag-1.4/psah2-1.4 | 80 | 80 | 80 | 0 | 50 | 80 | 30 | 80 | 80 | 60 | 100 | 110 |
| psag-1.4/psak-1 | 90 | 70 | 80 | 0 | 80 | 0 | 50 | 100 | 70 | 50 | 40 | 110 |
Signals obtained in three independent experiments (see Fig. 4) were quantified using the Lumi Analyst 3.0 (Boehringer Mannheim/Roche, Basel). Values were then normalized to the WT before calculating mean values. The se was 10% or less.
The partial knockout of PSI-H led to a slight decrease in the level of PSI-E, -F, and -K. In addition, we detected only about 30% of the normal amount of PSI-L in psah2-1.4. This secondary loss of PSI-L is more marked than that seen in PSI-H-less cosuppression lines, which have 60% of WT PSI-L levels (Lunde et al., 2000). In the double mutant psag-1.4/psah2-1.4, an additive effect of the two mutations on the amount of the H subunit was observed (50% of WT in psag-1.4/psah2-1.4, 70% in psah2-1.4, and 80% in psag-1.4). Further additive effects of the two mutations were observed for PSI-N and Lhca1.
In thylakoids of psak-1 plants, the K subunit is completely missing (Fig. 4b; Table I). The levels of the PSI core polypeptides are only slightly affected by the absence of PSI-K: Only the amounts of PSI-E -H, and -L are slightly decreased (by about 10%). In contrast, the abundance of Lhca2 and 3 is decreased by about 40% and 50%, respectively. In psag-1.4/psak-1 thylakoids, an additive effect of the two mutations on the abundance of Lhca2 and 3 (50% and 40% of WT levels, respectively) was observed.
Additional consequences of the mutations were studied by examining leaf pigment composition by HPLC (Table II). In the PSI-G knockout lines psag-1.4, psag-1.4/psah2-1.4, and psag-1.4/psak-1, the level of the xanthophyll cycle pigments (VAZ-pool) was higher than in WT plants, a finding that could be related to the altered antenna composition in the psag-1.4 background. In psag-1.4, the total Chl content (Chl a + b) was decreased by about 25%, which could, in part, be attributable to a delay in plant development, and therefore reduced chloroplast content in this genotype. Interestingly, the drop in total Chl level was not detected in psag-1.4/psah2-1.4, or psag-1.4/psak-1. The two genotypes lacking PSI-K, psak-1, and psak-1/psag-1.4 showed an increase in the Chl a/b ratio, which is consistent with the increased PSI to PSII ratio monitored in PSI-K cosuppression lines (Jensen et al., 2000). In psag-1.4 plants, the Chl a/b ratio was slightly reduced, indicating either an increased peripheral antenna or more likely, in light of the immunoblot analyses described above, a somewhat lower PSI/PSII ratio.
Table II.
Pigment composition of WT and mutant plants
| Leaf Pigments | WT | psag-1.4 | psah2-1.4 | psak-1 | psag-1.4/psah2-1.4 | psag-1.4/psak-1 |
|---|---|---|---|---|---|---|
| Nx | 41 ± 3 | 43 ± 2 | 43 ± 2 | 40 ± 2 | 42 ± 3 | 40 ± 0 |
| VAZ | 42 ± 2 | 49 ± 2 | 43 ± 1 | 40 ± 0 | 46 ± 1 | 49 ± 1 |
| Lu | 144 ± 5 | 155 ± 2 | 137 ± 6 | 145 ± 5 | 139 ± 7 | 141 ± 7 |
| β-Car | 61 ± 2 | 56 ± 1 | 58 ± 1 | 61 ± 0 | 56 ± 1 | 57 ± 1 |
| Chl a+b | 1,169 ± 18 | 891 ± 34 | 1,139 ± 102 | 1,220 ± 49 | 1,278 ± 131 | 1,110 ± 21 |
| Chl a/b | 3.18 ± 0.02 | 3.11 ± 0.04 | 3.21 ± 0.05 | 3.61 ± 0.05 | 3.19 ± 0.03 | 3.61 ± 0.02 |
The pigment content of WT (n = 5) and mutant (n = 5 each) plants was determined by HPLC. The carotenoid content is given in mmol per mol Chl (a + b), and the Chl content is expressed as nmol Chl (a + b) per g fresh wt. Mean values (±sd) are shown. Nx, Neoxanthin; VAZ, sum of xanthophyll's cycle pigments (violaxanthin + antheraxanthin + zeaxanthin); Lu, lutein; β-Car, β-carotene.
Alteration of the Redox States of PSI and PSII in Single and Double Mutants
Photosynthetic electron flow and the levels of state transitions were characterized by measuring parameters of Chl fluorescence and P700+ absorbance in the mutants (Table III). In all mutants, the maximum quantum yield of PSII (Fv/Fm) was not markedly altered. Moreover, no major change was detected in the redox state of the plastoquinone pool.
Table III.
Spectroscopic analyses of WT (n = 5) and mutant (n = 5 each) plants
| Parameter | WT | psag-1.4 | psah2-1.4 | psak-1 | psag-1.4/psah2-1 | psag-1.4/psak-1 |
|---|---|---|---|---|---|---|
| FV/FM | 0.83 ± 0.005 | 0.81 ± 0.005 | 0.82 ± 0.009 | 0.84 ± 0.002 | 0.82 ± 0.001 | 0.84 ± 0.004 |
| φ∥ | 0.77 ± 0.008 | 0.77 ± 0.006 | 0.76 ± 0.016 | 0.77 ± 0.010 | 0.76 ± 0.009 | 0.78 ± 0.007 |
| 1-qP | 0.024 ± 0.014 | 0.029 ± 0.005 | 0.027 ± 0.014 | 0.026 ± 0.011 | 0.029 ± 0.011 | 0.022 ± 0.011 |
| Fr (in %WT) | 0.82 ± 0.017 (100) | 0.57 ± 0.028 (69.5) | 0.77 ± 0.030 (93.9) | 0.54 ± 0.085 (65.9) | 0.43 ± 0.047 (52.4) | 0.31 ± 0.060 (37.8) |
| t1.2red | 54 ± 1 ms | 59 ± 1 ms | 59 ± 1 ms | 55 ± 2 ms | 60 ± 1 ms | 52 ± 1 ms |
| t1.2ox | 0.39 ± 0.01 s | 0.46 ± 0.02 s | 0.50 ± 0.02 s | 0.46 ± 0.02 s | 0.51 ± 0.02 s | 0.62 ± 0.01 s |
Mean values (±sd) are shown.
The rate of reduction and reoxidation of P700 was determined by measuring the parameters t1/2red and t1/2ox. An alteration in the reduction rate of P700 was recorded in psag-1.4/psah2-1.4 plants, where t1/2red was increased by about 10%. This effect could be due to the slightly decreased levels of PSI-N, which is necessary for the interaction of plastocyanin with PSI (Haldrup et al., 1999). Of the five genotypes tested, in fact, abundance of PSI-N was reduced in psag-1.4/psah2-1.4 only (Table I). In all mutants, t1/2ox was increased, particularly in psag-1.4/psak-1. The pronounced delay in P700 reoxidation seen in the last genotype is probably due to the secondary loss of PSI-E (70% of WT levels), which is necessary for efficient electron transfer from PSI to ferredoxin (Varotto et al., 2000). This conclusion is supported by the concomitance of reduction in PSI-E amounts and increase in t1/2ox in all five genotypes. However, it cannot be excluded that the decreased functional PSI antenna size in psag-1.4/psak-1 also contributes to the delay in P700 oxidation.
A Role of PSI-K in State Transitions
State transitions were followed by measuring PSII Chl fluorescence signals in states 1 and 2. The parameter relative fluorescence change (Fr; Lunde et al., 2000) is a measure for the redistribution of excitation energy between the photosystems, and was decreased in all mutant genotypes (Table III). The Fr level in both psag-1.4 and psak-1 indicates a decrease in state transitions by about 30%, whereas the partial knockout of PSI-H (psah2-1.4) has only a slight effect on this parameter. In the double mutants psag-1.4/psah2-1 and psag-1.4/psak-1, Fr was drastically reduced (to about 50% and 40% of WT levels, respectively).
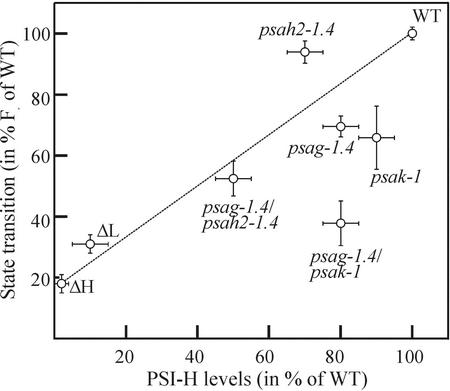
When the levels of state transitions, quantified by measuring Fr, and the abundance of PSI-H in the five mutant genotypes and the cosuppression lines ΔL and ΔH (Lunde et al., 2000) are displayed in form of a scatter plot (Fig. 5), major deviations from a linear correlation between the two parameters became evident. In particular, psak-1 and psag-1.4/psak-1 exhibited a decrease in state transitions that cannot be attributed solely to the decrease in their PSI-H content. This finding suggests that the loss of PSI-K also affects state transitions.
Figure 5.
Scatter plot of the levels of state transitions and PSI-H in mutant and WT plants. Levels of state transitions (Fr) were quantified as described in “Materials and Methods” and are given as percent of the WT value (see also Table II). The abundances of PSI-H in the different genotypes were quantified from Figure 4. Data for the cosuppression lines for PSI-L (ΔL) and -H (ΔH) were extracted from Lunde et al. (2000). Error bars indicate sds for the state transition measurements, whereas for the PSI-H levels the error bars reflect an estimated inaccuracy of ±10% (for genotypes with a PSI-H content ranging from 40%–100%), ±5% (for ΔL), and ±2% (for ΔH) of WT levels.
DISCUSSION
Mutants or cosuppression lines have previously been isolated for all nucleus-encoded subunits of PSI, with the exception of PSI-D, -G, and the recently discovered -O (H.V. Scheller, personal communication). The isolation of a PSI-G knockout line, and the construction of double mutants for PSI-G/-K and -G/-H, allowed us to carry out a functional characterization of PSI-G to scrutinize the current working model of PSI-LHCI structure and to define structural requirements for state transitions in PSI.
Lack of PSI-G is associated with a decrease in the abundance of most PSI core proteins, but does not markedly affect the level of Lhca proteins. This finding is compatible with a role for PSI-G in stabilizing the PSI core. The evolutionarily related PSI-K subunit, on the other hand, does not have such a stabilizing function (Jensen et al., 2000; this study). According to the current working model for the structure and topology of PSI (Scheller et al., 2001), PSI-G is located peripherally, and forms contacts with an Lhca2 homodimer (Jansson et al., 1996). PSI-K is at the opposite pole of PSI, and interacts with an Lhca3 homodimer, whereas the two Lhca1/Lhca4 heterodimers are associated with the PSI-F/J region (Haldrup et al., 2000; Boekema et al., 2001; Scheller et al., 2001). The only slight decrease in amounts of Lhca2 seen in psag-1.4 does not support the idea that Lhca2 is exclusively bound to PSI-G. However, the marked drop in levels of Lhca3 in plants lacking PSI-K (Jensen et al., 2000; this study), imply that, in the WT, homodimers of Lhca3 are bound to PSI-K. Three lines of evidence strongly suggest that Lhca2 and Lhca3 homodimers are in physical contact with each other: (a) Lhca2 and Lhca3 can be chemically cross-linked (Jansson et al., 1996), (b) PSI-K-less lines exhibit a secondary loss of Lhca2 (Jensen et al., 2000; this study), and (c) antisense inhibition of either Lhca2 or Lhca3 synthesis results in a concomitant decrease in the levels of both proteins (Ganeteg et al., 2001). Cross-linking experiments provide no support for the idea that Lhca2 and Lhca3 form heterodimers (Jansson et al., 1996), which would otherwise explain this interdependence. Furthermore, WT levels of Lhca3 are present in psag1.4 plants, although the Lhca2 concentration is decreased. Taken together, these data imply that Lhca2/Lhca3 heterodimers may exist under normal conditions, but do not accumulate in significant amounts.
Overall, the changes in LHCI observed in PSI mutants indicate a complex quantitative interdependence between the various Lhca proteins, and also suggest that some degree of flexibility in its composition is compatible with function. One example of such changes is provided by psag-1.4, where, in addition to the decrease in Lhca2, the abundance of Lhca4 is slightly increased with respect to the WT. Furthermore, plants containing only 5% of the normal amount of PSI-F (Haldrup et al., 2000) exhibit multiple changes in LHCI composition (Lhca1 drops to 70%, Lhca2 drops to 40%, and Lhca3 drops to 30% of WT, whereas Lhca4 reaches 120% of WT levels). In the double mutant psag-1.4/psak-1, which according to the current working model should lack the attachment sites for both Lhca2 and Lhca3 homodimers, about one-half the normal level of Lhca2 and Lhca3 is still detectable. This indicates that both Lhca2 and Lhca3 can bind to other PSI proteins, in addition to PSI-G and -K, and thus supports the idea that Lhca proteins can exist in alternative dimeric forms and stoichiometries, depending on environmental conditions (Bailey et al., 2001; Ganeteg et al., 2001), and thus that LHCI can be organized in several different ways. Taking into account the existence of barley (Hordeum vulgare) mutants, which accumulate LHCI but not PSI core to WT levels (Hoyer-Hansen et al., 1988; Nielsen et al., 1996), additional experiments are required to prove the existence and extent of LHCI reorganization. In this sense, the psag-1.4, psak-1, and psag-1.4/psak-1 mutants represent a useful tool for further studies.
In the absence of PSI-H, state transitions are almost completely suppressed (Lunde et al., 2000). Although all mutant genotypes analyzed in this study show a reduction in both Fr (as a measure of state transitions) and PSI-H content, no strict correlation between the two parameters was evident. Moreover, both Arabidopsis genotypes lacking PSI-K displayed an additional drop in the level of state transitions, implying that in such thylakoids LHCI is present in a novel form, which affects the interaction between PSI and LHCII (Jensen et al., 2000). Interestingly, in the mutants psag-1.4 and psag-1.4/psah2-1.4, which have reduced amounts of both PSI-H and Lhca2, the decrease in state transitions seems proportional to the degree of loss of PSI-H (see Fig. 5). One may conclude from this that Lhca3, rather than Lhca2, mediates the role of PSI-K in state transitions. Alternatively, it cannot be excluded that plants devoid of PSI-K bind LHCII in a different and more stable way, giving rise to a PSI-LHCII complex even under state 1-favoring conditions. However, given the uncertainties regarding the exact topology and structure of LHCI (Scheller et al., 2001), the molecular basis of the involvement of PSI-K and LHCI in state transitions remains unclear.
Knockout of PSI-G or -K, as well as the simultaneous loss of -G and -K, or of -G and -H, affects only marginally, if at all, the photoautotrophic growth of Arabidopsis plants, at least when this is evaluated on the basis of leaf area measurements. Plants that are partially deficient in PSI-H apparently grow even better than the WT under optimal conditions. In addition, in none of the mutants tested in this study was overall photosynthetic electron flow affected severely, as indicated by the fact that parameters such as maximum and effective quantum yield, and photochemical quenching, were unchanged. Several other mutant studies (Haldrup et al., 1999; Naver et al., 1999; Jensen et al., 2000; Li et al., 2000; Graβes et al., 2002) have also shown that under optimal conditions, Arabidopsis can tolerate the loss of single polypeptides of the photosynthetic apparatus and their associated functions; for example, energy dissipation by ΔpH-dependent quenching (Li et al., 2000; Graβes et al., 2002), or photosynthetic state transitions (Lunde et al., 2000; unpublished data in our lab) without obvious consequences for photoautotrophic growth. However, even if growth under growth chamber conditions appears to be unaffected in such mutants, there is increasing evidence that fitness in natural environments, particularly under rapidly changing illumination conditions, is reduced by such mutations (Graβes et al., 2002; S. Jansson, personal communication).
The study of subunit function in the PSI of higher plants has reached an advanced state in Arabidopsis. For all of the nucleus-encoded PSI subunits, except PSI-D and the recently discovered PSI-O (H.V. Scheller, personal communication), mutants and/or cosuppression lines are now available, allowing an assessment of the contribution of individual polypeptides to PSI function and composition. Previous analyses (Haldrup et al., 2000; Varotto et al., 2000) have shown, and this study confirms, that in the absence of specific PSI subunits, the abundance of several other PSI polypeptides may be altered, which hampers the unambiguous assignment of specific functions to individual polypeptides of PSI. The generation of mutants lacking two or more PSI subunits, and their systematic analysis with respect to PSI composition, electron flow through PSI, and overall photosynthetic electron flow, will allow us to supplement the existing data collection, making possible a statistically reliable analysis of structure-function relationships between the protein components of PSI.
MATERIALS AND METHODS
Plant Propagation and Growth Measurement
The En-mutagenized Arabidopsis (ecotype Columbia 0) population has been described before (Wisman et al., 1998). Plants were grown on Minitray soil (Gebr. Patzer GmbH & Co. KG, Sinntal-Jossa, Germany) in plastic trays in an air-conditioned greenhouse (day length of 14 h at 20°C and 80 μmol photons m−2 s−1; 10-h night at 16°C). Methods for the measurement of growth have been described previously (Leister et al., 1999).
Isolation and Stabilization of psaG::En1, psaH2::En1, and psaK::ΔEn1 Alleles
Transposon insertions within psaG, psaH2, and psaK were identified by screening the En-tagged population using gene-specific primers in combination with En-specific primers, followed by hybridization with a gene-specific probe, as described by Varotto et al. (2000). Gene-specific primers were: G-1s (5′-ATGGCCACAAGCGCATCAGCTTTGCTC-3′), G-450as (5′-GGAAGTAGCCAAGATGTAGTAAGCAACG-3′), H2-234s (5′-AGCTTGCCGCGAGGAC CGAGCTTAGG-3′), H2-603as (5′-TGTGGTGGCTAAGTATGGAGACAAAAGT-3′), K-6s (5′-AAGAAAATGGCTAGCACTATGATGACTA-3′), and K-690as (5′-TTCAAATAGCA CCAATGTTTTTAAGGCC-3′). Positive lines were confirmed by sequencing PCR-amplified insertion sites.
The stable frameshift mutants psag1-1.4 and psah2-1.4 were identified among the progenies of corresponding En insertion lines by separating PCR-amplified empty donor sites on 6% (w/v) denaturing polyacrylamide gels, followed by sequencing of candidate bands that differed in size from those derived from the WT allele. For the identification of homozygous frameshift mutants, lines putatively containing footprints were selfed and the next generation was analyzed as described above. Primers used for the amplification of empty donor sites were: G-1s, G-144as (5′-GGTTGATAGTTTGGGTAGGG-3′), H2-29s (5′-TACAACCTTCTGCCGC CGTG-3′), and H2-253as (5′-CTCCATACTTAGCCACCACA-3′). For psak-1, the entire insertion site, including the 600-bp deletion derivative of En, was amplified by PCR with primers K-291s (5′-TTTGTCATCCCAGGCAAGTG-3′) and K-490as (5′-AACATCAGGGTCGTCGACGT-3′), and sequenced.
Sequence Analysis
Sequence data were analyzed with the Wisconsin Package version 10.0 (Genetics Computer Group, Madison, WI; Devereux et al., 1984) and sequences were aligned using the CLUSTAL W program (version 1.7; Thompson et al., 1994).
Immunoblot Analysis of Proteins
Thylakoid proteins from 4-week-old plants were isolated as described (Bassi et al., 1985). For denaturing PAGE analysis, protein amounts equivalent to 5 μg of Chl were loaded for each genotype. Decreasing amounts of WT proteins (4, 3, and 2 μg of Chl) were loaded in parallel lanes (0.8× WT, 0.6× WT, and 0.4× WT). For immunoblot analyses, proteins were transferred to Immobilon-P membranes (Millipore, Bedford, MA) and incubated with antibodies specific for individual subunits of PSI (PSI-G, -H, -L, -D, -E, -F, -K, and -N) and LHCI (Lhca1, 2, 3, and 4). Signals were detected using the Enhanced Chemiluminescence Western-Blotting Kit (Amersham, Buckinghamshire, UK) and quantified using the Lumi Analyst 3.0 (Boehringer Mannheim/Roche).
Pigment Analysis
Pigments were analyzed by reversed-phase HPLC as described (Färber et al., 1997). The following conversion factors (peak area per picomole of the respective pigment) were used: 3,097 (neoxanthin), 3,656 (violaxanthin), 2,902 (antheraxanthin and lutein), 3,006 (zeaxanthin), 1,300 (Chl b), 1,350 (Chl a), and 3,598 (β-carotene). For pigment extraction, leaf discs were frozen in liquid nitrogen and disrupted in a mortar in the presence of acetone. After a short centrifugation, pigment extracts were filtered through a 0.2-μm membrane filter and either used directly for HPLC analysis or stored for up to 2 d at −20°C.
Chl Fluorescence and P700 Absorption Measurements
In vivo Chl a fluorescence of single leaves was measured using the PAM 101/103 device (Walz, Effeltrich, Germany) as described by Varotto et al. (2000). Pulses (800 ms) of white light (6,000 μmol photons m−2 s−1; Walz FL-103/E.220) were used to determine the maximum fluorescence (Fm) and the ratio (Fm − F0)/ Fm = Fv/Fm. A 20-min illumination with actinic light (Walz FL-101/E) at 80 μmol photons m−2 s−1 was used to drive electron transport between the two photosystems before measuring the effective quantum yield of PSII (ΦII) and photochemical quenching [qP = (Fm′ − FS)/(Fm′ − F0′); Schreiber, 1986].
A dual-wavelength pulse-modulation system (Walz ED-P700DW-E) was used to record changes in the absorbance of P700+ (Klughammer and Schreiber, 1994). Leaves were illuminated with background far-red light (15 W m−2, equaling 90 μmol photons m−2 s−1; Walz 102-FR), and immediately after full oxidation of P700, a saturating xenon light pulse (50 ms) was applied (Walz XMT-103) to reduce P700+. t1/2red and t1/2ox were calculated from the recorded kinetics of P700 reduction by the xenon light and reoxidation by the far-red light.
To measure state transitions, the parameter Fr was monitored in five WT and five mutant plants each as described before (Lunde et al., 2000). The maximum fluorescence (Fm) was measured as described above, and leaves were illuminated subsequently for 15 min with blue light (100 μmol photons m−2 s−1) from a KL-1500 lamp (Schott, Mainz, Germany) equipped with a Walz BG39 filter. Far-red light (Walz 102-FR; peak emission 730 nm, 90 μmol photons m−2 s−1) was turned on, and after 15 min the maximum fluorescence yield in state 1 (Fm1) was determined. The far-red light was switched off and the fluorescence recorded for 15 min, after which the maximum fluorescence yield in state 2 (Fm2) was measured. The relative change in fluorescence was calculated as: Fr = [(FI′ − FI) − (FII′ − FII)]/FI′ − FI (Lunde et al., 2000), where FI and FII designate fluorescence in the presence of PSI light in states 1 and 2, respectively, whereas FI′ and FII′ designate fluorescence in the absence of PSI light in states 1 and 2, respectively.
Note Added in Proof
When this paper was reviewed, we learned that independently of our work an analysis of PSI-G antisense line has been published (P.E. Jensen, L. Rosgaard, J. Knoetzel, H.V. Scheller [2002] J Biol Chem 277: 2798–2803).
ACKNOWLEDGMENTS
We thank Stefan Jansson for providing us with LHCI antibodies, and Poul Erik Jensen and Henrik Vibe Scheller for their generosity in sharing their PSI antibodies with us. Grateful acknowledgments are extended to Paul Hardy, Koen Dekker, and Heinz Saedler.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. Ja 665/2–1 to P.J. and Le 1265/1–1, 2 to D.L.) and by the Bundesministerium für Bildung und Forschung (to D.L.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002089.
LITERATURE CITED
- Bailey S, Walters RG, Jansson S, Horton P. Acclimation of Arabidopsis thaliana to the light environment: the existence of separate low light and high light responses. Planta. 2001;213:794–801. doi: 10.1007/s004250100556. [DOI] [PubMed] [Google Scholar]
- Bassi R, dal Belin Peruffo A, Barbato R, Ghisi R. Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur J Biochem. 1985;146:589–595. doi: 10.1111/j.1432-1033.1985.tb08692.x. [DOI] [PubMed] [Google Scholar]
- Boekema EJ, Jensen PE, Schlodder E, van Breemen JF, van Roon H, Scheller HV, Dekker JP. Green plant photosystem I binds light-harvesting complex I on one side of the complex. Biochemistry. 2001;40:1029–1036. doi: 10.1021/bi0015358. [DOI] [PubMed] [Google Scholar]
- Chitnis PR, Purvis D, Nelson N. Molecular cloning and targeted mutagenesis of the gene psaF encoding subunit III of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1991;266:20146–20151. [PubMed] [Google Scholar]
- Chitnis PR, Reilly PA, Miedel MC, Nelson N. Structure and targeted mutagenesis of the gene encoding 8-kDa subunit of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 1989;264:18374–18380. [PubMed] [Google Scholar]
- Chitnis VP, Chitnis PR. PsaL subunit is required for the formation of photosystem I trimers in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1993;336:330–334. doi: 10.1016/0014-5793(93)80831-e. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farah J, Rappaport F, Choquet Y, Joliot P, Rochaix JD. Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunit. EMBO J. 1995;14:4976–4984. doi: 10.1002/j.1460-2075.1995.tb00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Färber A, Young AJ, Ruban AV, Horton P, Jahns P. Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants: the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching. Plant Physiol. 1997;115:1609–1618. doi: 10.1104/pp.115.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg U, Strand A, Gustafsson P, Jansson S. The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 2001;127:150–158. doi: 10.1104/pp.127.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graβes T, Pesaresi P, Schiavon F, Varotto C, Salamini F, Jahns P, Leister D. The role of ΔpH-dependent dissipation of excitation energy in protecting photosystem II against light-induced damage in Arabidopsis thaliana. Plant Physiol Biochem. 2002;40:41–49. [Google Scholar]
- Haldrup A, Naver H, Scheller HV. The interaction between plastocyanin and photosystem I is inefficient in transgenic Arabidopsis plants lacking the PSI-N subunit of photosystem I. Plant J. 1999;17:689–698. doi: 10.1046/j.1365-313x.1999.00419.x. [DOI] [PubMed] [Google Scholar]
- Haldrup A, Simpson DJ, Scheller HV. Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J Biol Chem. 2000;275:31211–31218. doi: 10.1074/jbc.M002933200. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen G, Bassi R, Honberg LS, Simpson DJ. Immunological characterization of chlorophyll a/b-binding proteins of barley thylakoids. Planta. 1988;173:12–21. doi: 10.1007/BF00394481. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jansson S, Andersen B, Scheller HV. Nearest-neighbor analysis of higher-plant photosystem I holocomplex. Plant Physiol. 1996;112:409–420. doi: 10.1104/pp.112.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gilpin M, Knoetzel J, Scheller HV. The PSI-K subunit of photosystem I is involved in the interaction between light-harvesting complex I and the photosystem I reaction center core. J Biol Chem. 2000;275:24701–24708. doi: 10.1074/jbc.M000550200. [DOI] [PubMed] [Google Scholar]
- Kitmitto A, Holzenburg A, Ford RC. Two-dimensional crystals of photosystem I in higher plant grana margins. J Biol Chem. 1997;272:19497–19501. doi: 10.1074/jbc.272.31.19497. [DOI] [PubMed] [Google Scholar]
- Kjaerulff S, Andersen B, Nielsen VS, Moller BL, Okkels JS. The PSI-K subunit of photosystem I from barley (Hordeum vulgare L.). Evidence for a gene duplication of an ancestral PSI-G/K gene. J Biol Chem. 1993;268:18912–18916. [PubMed] [Google Scholar]
- Klughammer C, Schreiber U. An improved method, using saturating light pulses, for the determination of photosystem I quantum yield via P700+-absorbance changes at 830 nm. Planta. 1994;192:261–268. [Google Scholar]
- Knoetzel J, Simpson DJ. The primary structure of a cDNA for PsaN, encoding an extrinsic lumenal polypeptide of barley photosystem I. Plant Mol Biol. 1993;22:337–345. doi: 10.1007/BF00014940. [DOI] [PubMed] [Google Scholar]
- Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F. Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol Biochem. 1999;37:671–678. [Google Scholar]
- Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- Lunde C, Jensen PE, Haldrup A, Knoetzel J, Scheller HV. The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature. 2000;408:613–615. doi: 10.1038/35046121. [DOI] [PubMed] [Google Scholar]
- Naver H, Haldrup A, Scheller HV. Cosuppression of photosystem I subunit PSI-H in Arabidopsis thaliana. Efficient electron transfer and stability of photosystem I is dependent upon the PSI-H subunit. J Biol Chem. 1999;274:10784–10789. doi: 10.1074/jbc.274.16.10784. [DOI] [PubMed] [Google Scholar]
- Nielsen VS, Scheller HV, Moller BL. The photosystem I mutant viridis-zb63 of barley (Hordeum vulgare) contains low amounts of active but unstable photosystem I. Physiol Plant. 1996;98:637–644. [Google Scholar]
- Okkels JS, Nielsen VS, Scheller HV, Moller BL. A cDNA clone from barley encoding the precursor from the photosystem I polypeptide PSI-G: sequence similarity to PSI-K. Plant Mol Biol. 1992;18:989–994. doi: 10.1007/BF00019215. [DOI] [PubMed] [Google Scholar]
- Okkels JS, Scheller HV, Jepsen LB, Moller BL. A cDNA clone encoding the precursor for a 10.2 kDa photosystem I polypeptide of barley. FEBS Lett. 1989;250:575–579. doi: 10.1016/0014-5793(89)80799-9. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Richly E, Kurth J, Salamini F, Leister D. Functional genomics of Arabidopsis photosynthesis. Plant Physiol Biochem. 2001;39:285–294. [Google Scholar]
- Scheller HV, Jensen PE, Haldrup A, Lunde C, Knoetzel J. Role of subunits in eukaryotic PSI. Biochim Biophys Acta. 2001;1507:41–60. doi: 10.1016/s0005-2728(01)00196-7. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Naver H, Moller BL. Molecular aspects of photosystem I. Physiol Plant. 1997;100:842–851. [Google Scholar]
- Schreiber U. Detection of rapid induction kinetics with a new type of high-frequency modulated chlorophyll fluorometer. Photosynth Res. 1986;9:261–272. doi: 10.1007/BF00029749. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Pesaresi P, Meurer J, Oelmuller R, Steiner-Lange S, Salamini F, Leister D. Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 2000;22:115–124. doi: 10.1046/j.1365-313x.2000.00717.x. [DOI] [PubMed] [Google Scholar]
- Wisman E, Hartmann U, Sagasser M, Baumann E, Palme K, Hahlbrock K, Saedler H, Weisshaar B. Knock-out mutants from an En-1 mutagenized Arabidopsis thaliana population generate phenylpropanoid biosynthesis phenotypes. Proc Natl Acad Sci USA. 1998;95:12432–12437. doi: 10.1073/pnas.95.21.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Yu L, Chitnis VP, Chitnis PR. Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunits. J Biol Chem. 1994;269:3205–3211. [PubMed] [Google Scholar]
- Zhao J, Snyder WB, Mühlenhoff U, Rhiel E, Warren PV, Golbeck JH, Bryant DA. Cloning and characterization of the psaE gene of the cyanobacterium Synechococcus sp. PCC 7002: characterization of a psaE mutant and overproduction of the protein in Escherichia coli. Mol Microbiol. 1993;9:183–194. doi: 10.1111/j.1365-2958.1993.tb01680.x. [DOI] [PubMed] [Google Scholar]