Abstract
The RHD3 (ROOT HAIR DEFECTIVE3) gene encodes a putative GTP-binding protein required for appropriate cell enlargement in Arabidopsis. To obtain insight into the mechanisms of RHD3 regulation, we conducted a molecular genetic dissection of RHD3 gene expression and function. Gene fusion and complementation studies show that the RHD3 gene is highly expressed throughout Arabidopsis development and is controlled by two major regulatory regions. One regulatory region is located between −1,500 and −600 bp upstream of the RHD3 gene and is required for vascular tissue expression. The other region is intragenically located and includes the 558-bp first intron, which is responsible for high-level expression of RHD3 throughout the plant. The presence and location of this intron is essential for gene function because constructs lacking this intron or constructs with the intron in an abnormal position are unable to functionally complement the rhd3 mutations. We also analyzed the role of other RHD genes and the plant hormones auxin and ethylene in RHD3 regulation, and we determined that these act downstream or independently from the RHD3 pathway. This study shows that multiple levels of regulation are employed to ensure the appropriate expression of RHD3 throughout Arabidopsis development.
The proper development of plant shape is at least partly dependent on regulated cell expansion (Steeves and Sussex, 1989). Although plant hormones, cytoskeletal components, and wall-loosening proteins are known to influence this process (Evans, 1984; Abeles et al., 1992; Cosgrove, 1997), the molecular mechanisms that regulate their function and the underlying genetic control of cell morphogenesis remain largely unknown.
The Arabidopsis root epidermis has been exploited for studying the molecular and cellular basis of plant cell morphogenesis because of its simplicity and accessibility (Schiefelbein et al., 1997; Paquette and Benfey, 2001). During the development of the root epidermis, the individual cells undergo a large increase in cell size and they become highly elongated, indicating that the orientation and extent of cell expansion is regulated during cell differentiation. Epidermal cells are organized into columns (files) along the length of the root, with newly formed cells located near the apex in the meristematic region and the “older” cells located farther from the apex. Thus, each cell in a file is more developmentally advanced than the one below it, which provides an opportunity to easily study all developmental stages along the root (Dolan et al., 1993; Schiefelbein et al., 1997).
In previous studies, several genetic loci involved in Arabidopsis root epidermal cell morphogenesis have been identified. For example, the RHD (ROOT HAIR DEFECTIVE) 1-4 genes (Schiefelbein and Somerville, 1990) and the COW1 gene (Grierson et al., 1997) are required for normal root hair growth, and the RHD6 gene is required for root hair initiation (Masucci and Schiefelbein, 1994). The RHD3 gene is unique because it is required for regulated cell expansion throughout root epidermis development because root epidermal cells of rhd3 mutants are reduced in size and abnormal in shape. In addition, the RHD3 gene appears to act as a general regulator of cell enlargement in Arabidopsis because rhd3 mutations cause a reduction in cell size throughout the plant and because RHD3 transcripts accumulate in all major plant organs (Wang et al., 1997; Galway et al., 1997). The RHD3 gene has been shown to encode a novel protein with putative GTP-binding sites, and RHD3-like genes exist in diverse organisms, suggesting a common cellular function in eukaryotes (Wang et al., 1997).
To gain insight into the molecular regulation of RHD3, we analyzed RHD3 expression by in situ hybridization, RHD3 promoter-GUS reporter gene fusions, and molecular complementation experiments. These studies document the critical role of a 5′- non-transcribed region and the large first intron in regulating RHD3 gene expression and function. In addition, we analyzed the potential role of other RHD genes and plant hormones in regulating the RHD3 gene.
RESULTS
RHD3 Is Expressed throughout the Arabidopsis Plant
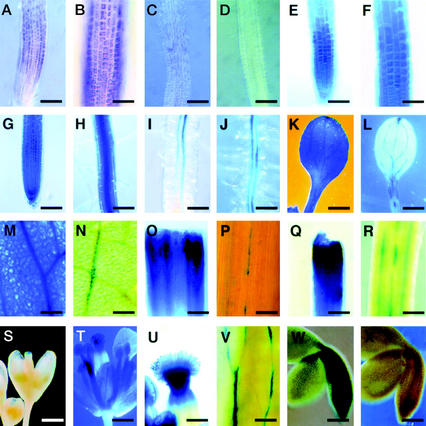
To study the expression pattern of the RHD3 gene, we first employed whole-mount in situ RNA hybridization experiments with RHD3 probes on Arabidopsis seedling roots. RHD3 transcripts were detected in the elongation zone of wild-type seedling roots (Fig. 1, A and B). This result is consistent with the postulated RHD3 role in regulating cell enlargement. The rhd3-3 mutant, which bears a nonsense mutation, exhibits a greatly reduced transcript level in a northern-blot analysis (Wang et al., 1997) and very weak in situ RNA hybridization signal (Fig. 1C). No signal was detected when the hybridization probe was excluded (Fig. 1D).
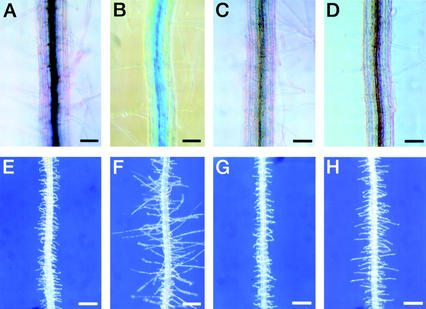
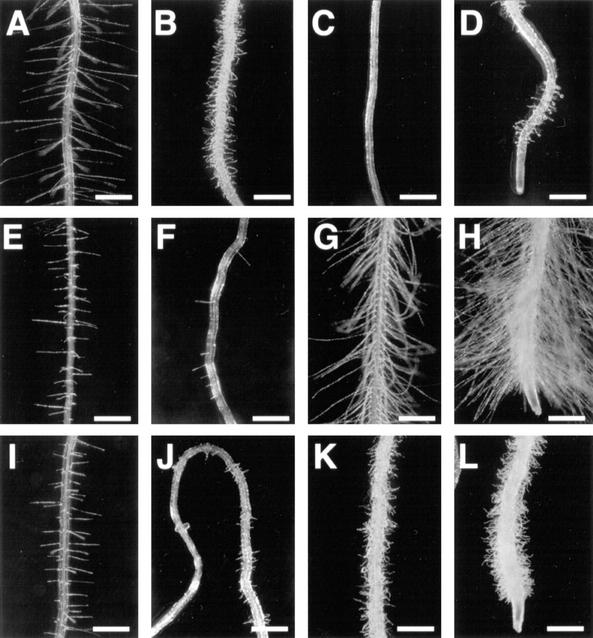
Figure 1.
Expression of the Arabidopsis RHD3 gene. A through D, In situ RNA hybridization; E through X, RHD3 promoter::GUS reporter gene expression. A and B, Four-day-old wild-type root hybridized with RHD3 antisense RNA probe shows RHD3 transcript accumulation in the root elongation zone. Bar in A = 100 μm. Bar in B = 50 μm. C, Four-day-old rhd3-3 mutant root hybridized with RHD3 antisense RNA probe shows reduced RHD3 transcript accumulation. Bar = 100 μm. D, Four-day-old wild-type root incubated without RNA probe shows no hybridization signal. Bar = 100 μm. E and F, Four-day-old root from transgenic line harboring the X1 RHD3::GUS construct incubated for 30 min in X-Gluc substrate shows expression in the elongation zone. Bar in E = 100 μm. Bar in F = 50 μm. G and H, Four-day-old root apex (G) and mature root segment (H) from transgenic line harboring the X1 RHD3::GUS construct incubated for 4 h in X-Gluc substrate shows expression throughout the root. Bars = 100 μm. I and J, Four-day-old root elongation zone (I) and mature root segment (J) from transgenic line harboring the H1 RHD3::GUS construct incubated for 16 h in X-Gluc substrate shows preferential expression in the vascular tissue. Bar in I = 50 μm. Bar in J = 100 μm. K and L, Six-day-old seedling cotyledons from transgenic lines harboring the X1 (K) or H1 (L) RHD3::GUS constructs incubated for 16 h in X-Gluc substrate shows expression throughout the cotyledon (K) or in vascular tissue (L). Bars = 500 μm. M and N, Three-week-old rosette leaves from transgenic lines harboring the X1 (M) or H1 (N) RHD3::GUS constructs incubated for 16 h in X-Gluc substrate shows expression throughout the leaf (M) or in vascular tissue (N). Bars = 100 μm. O and P, Five-week-old stems from transgenic lines harboring the X1 (O) or H1 (P) RHD3::GUS constructs incubated for 16 h in X-Gluc substrate shows expression throughout the stem (O) or in vascular tissue (P). Bar in O = 200 μm. Bar in P = 100 μm. Q and R, Five-week-old pedicels from transgenic lines harboring the X1 (Q) or H1 (R) RHD3::GUS constructs incubated for 16 h in X-Gluc substrate shows expression throughout the pedicel (Q) or in particular cell files (R). Bar in Q = 200 μm. Bar in R = 100 μm. S through U, Five-week-old transgenic lines harboring the X1 RHD3::GUS constructs showing an immature flower (S), mature flower (T), or stigmatic tissue from mature flower (U) incubated for 16 h in X-Gluc substrate. Bars in S and T = 800 μm. Bar in U = 200 μm. V, Five-week-old sepal tissue from flowers of transgenic line harboring the H1 RHD3::GUS construct incubated for 16 h in X-Gluc substrate shows expression in vascular tissue. Bars = 100 μm. W and X, Embryos at curled cotyledon stage from transgenic lines harboring the X1 (W) or H1 (X) RHD3::GUS constructs incubated for 16 h in X-Gluc substrate shows expression throughout the embryo (W) or absent (X). Bars = 100 μm.
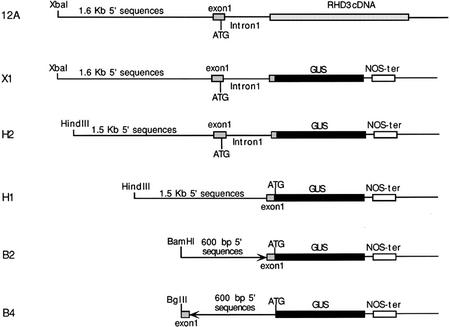
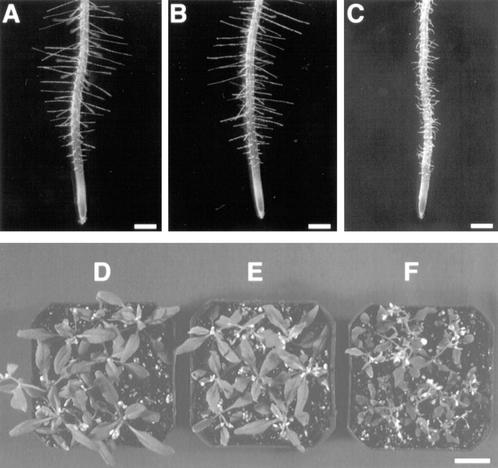
To examine RHD3 expression in greater detail, we constructed and analyzed a RHD3 promoter-β-glucuronidase (GUS) reporter gene fusion. The construction of this fusion was based on results from studies to define the DNA sequences necessary for RHD3 gene function. In previously published work, a construct shown in Figure 2 (designated 12A) containing 1.6 kb of 5′-non-coding sequence, the first exon, and the first intron of the RHD3 gene fused to the rest of RHD3 cDNA was found to rescue the rhd3 mutant root hair phenotype in transformed green callus tissue (Wang et al., 1997). In the present study, we extended these findings by generating stable transgenic plants that contain the 12A construct in the rhd3-2 (Columbia background) or rhd3-3 allele (Nossen background). Like the transformed callus tissue, these transgenic plants produce root hairs with normal length and morphology (Fig. 3, A–C). In addition, these plants possess leaves that are comparable in size to the wild type (Fig. 3, D–F). Furthermore, their root epidermal cell length and root growth rate were indistinguishable from the wild type (Table I). Together, these results show that the DNA sequences fused to the RHD3 cDNA in the 12A construct are sufficient to rescue the rhd3 mutant defects.
Figure 2.
Structure of RHD3 gene fusions. The 12A construct represents an in-frame fusion of the 5′ end of the RHD3 gene (including first intron) to the RHD3 cDNA. The other constructs shown are RHD3::GUS reporter gene fusions that include (X1 and H2) or do not include (H1, B2, and B4) the first RHD3 intron. Details of the DNA constructions are given in “Materials and Methods.”
Figure 3.
Complementation of the rhd3 mutant phenotype by the 12A RHD3 gene::cDNA construct. A through C, Four-day-old seedling roots. Bars = 300 μm. D through F, Rosettes of 4-week-old plants. Bar = 2 cm. A and D, Wild type (Columbia ecotype). B and E, Transgenic rhd3-2 harboring the 12A construct. C and F, rhd3-2 mutant.
Table I.
Effect of RHD3 gene constructs on root growth and cell elongation
| Genotype | Root Growth Rate
|
Epidermis Cell Length
|
||
|---|---|---|---|---|
| mm d−1a | % Wild type | μm cell−1a | % Wild type | |
| Columbiab | 5.3 ± 0.8 | 100 | 207 ± 36 | 100 |
| rhd3-2 (Col) | 2.6 ± 0.3 | 48 | 92 ± 13 | 45 |
| rhd3-2, 12A | 5.2 ± 0.8 | 98 | 222 ± 36 | 107 |
| rhd3-2, 12A-I | NDc | – | 94 ± 15 | 45 |
| Nossenb | 5.7 ± 0.5 | 100 | 166 ± 29 | 100 |
| rhd3-3 (No) | 2.8 ± 0.4 | 48 | 85 ± 12 | 51 |
| rhd3-3, 12A | 5.6 ± 0.5 | 98 | 184 ± 33 | 111 |
| rhd3-3, 12A-I | ND | – | 89 ± 11 | 54 |
Values given represent the mean ± sd.
Wild type.
ND, Not determined.
Based on these results, we used the same genomic RHD3 DNA sequences (1.6 kb of 5′-non-coding sequence, the first exon, and the first intron) to generate a translational fusion to the GUS reporter gene (designated construct X1; Fig. 2). Twelve independent transgenic lines bearing this construct were obtained and analyzed, and the pattern of GUS accumulation was similar in all lines. These RHD3 regulatory sequences direct high levels of GUS expression throughout the root, including the root elongation zone (Fig. 1, E–H). When the transgenic X1 plants were incubated in GUS substrates for an extended period of time (8–16 h), stained cells were detected in all plant tissues. For example, GUS expression was observed in the cotyledons (Fig. 1K) and hypocotyl (data not shown) of seedlings, the leaf (Fig. 1M), stem (Fig. 1O), and pedicel (Fig. 1Q) of mature plants, and the stigmatic papillae cells and anther of flowers (Fig. 1, S–U). In addition, GUS expression was detected throughout the developing embryo of these RHD3::GUS lines (Fig. 1W). These results suggest that the RHD3 gene is expressed throughout the Arabidopsis plant and therefore may participate in a fundamental cellular process.
Identification of RHD3 Regulatory Regions
To study the regulation of RHD3, a series of deletion constructs of the X1 RHD3::GUS fusion were generated (Fig. 2). The H2 construct is essentially the same as X1 except that it lacks 100 bp at the 5′ end (Fig. 2). Transgenic H2 plants exhibit the same GUS expression pattern as construct X1 (data not shown), suggesting that the distal 100-bp upstream sequence is not required for regulating RHD3 expression.
The RHD3::GUS construct H1 is similar to H2 except that it is a transcriptional fusion and lacks the entire 558-bp first intron of RHD3 as well as 73 bp of the first exon that flanks this first intron (Fig. 2). GUS expression in H1 transgenic plants was found to be restricted to the vascular tissues of the roots (Fig. 1, I and J), and discontinuous weak expression in the vascular tissue of the cotyledon (Fig. 1L), leaf (Fig. 1N), stem (Fig. 1P), pedicel (Fig. 1R), and the flower sepals (Fig. 1V), and no detectable expression in the developing embryo (Fig. 1X). In addition, quantitative GUS assays showed that the average GUS activity in the seedlings roots from the H1 lines is less than 1% of the activity present in the H2 lines (29,300 versus 210 pmol methylumbelliferone min−1 mg protein−1). These results indicate that the DNA segment containing the proximal 73-bp first exon sequence and the first intron is required for the appropriate qualitative and quantitative expression of the RHD3 gene.
Two additional constructs, B2 and B4, were generated that contain approximately 600 bp of the 5′ non-transcribed sequence plus 90 bp of the distal first exon sequence transcriptionally fused to the GUS reporter gene (Fig. 2). The B2 construct has this RHD3 DNA segment in the correct orientation, and B4 has this in the reverse orientation (as a negative control), with respect to the GUS gene-coding region (Fig. 2). No GUS staining was observed in transgenic plants harboring either of these constructs (data not shown), suggesting that these sequences are not sufficient to drive gene expression at a detectable level. Comparing the expression patterns of constructs B2 and H1 leads to the conclusion that sequences located between −600 bp and −1,500 bp are responsible for vascular tissue expression of RHD3.
The First Intron Is Required to Confer Appropriate RHD3 Expression
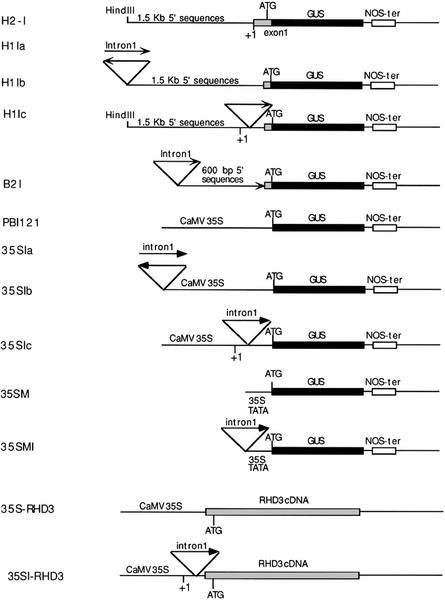
The dramatically different expression patterns conferred by constructs H2 and H1 indicate that the 631-bp intragenic sequence plays an important role in regulating RHD3 expression. To determine whether it is the 73-bp proximal first exon segment or the 558-bp intron sequence that possesses a regulatory function, a construct (designated H2-I) was generated that is identical to H2 except that it lacks the first intron (Fig. 4). The GUS gene expression pattern in H2-I transgenic plants is similar to H1 (Fig. 5A; data not shown), indicating that the 73-bp proximal first exon sequence does not have a major effect on the reporter gene expression; thus, the RHD3 expression pattern is dependent on the first intron sequence.
Figure 4.
Structure of RHD3 gene fusions containing or lacking the first intron. The H2-I construct contains the same upstream regulatory sequences as H2 except that the first intron is absent. For other constructs, a PCR fragment containing the first intron was added to the upstream or in the transcribed regions of various GUS reporter or RHD3 cDNA gene fusions. The orientation of the intron sequences is indicated by the arrows. The +1 symbol indicates the transcription initiation site. Details of the cloning procedures are described in “Materials and Methods.”
Figure 5.
Effects of the RHD3 first intron on gene expression. A through D, Five-day-old seedling roots harboring the H2-I (A), pBI121 (B) 35SIc (C), or H1Ic (D) construct and incubated with X-gluc for 16 h. Bars = 100 μm. E through H, Root phenotype of 5-day-old rhd3-3 seedlings harboring the 35S::RHD3 (E and F) or the 35SI::RHD3 (G and H) constructs. Bars = 200 μm. E and G, Mutant phenotypes. F and H, Partially rescued phenotypes.
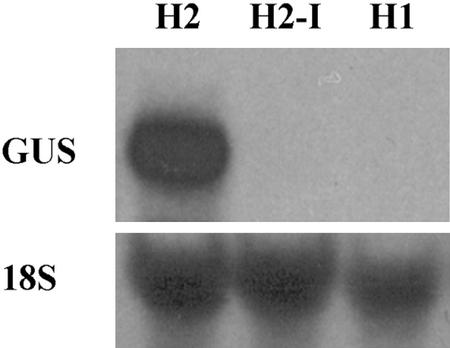
The effect of the intron sequence was explored by conducting RNA-blot (northern) analysis with the H2, H2-I, and H1 lines. Using a GUS-specific probe, we discovered an abundant RNA of the appropriate size in seedling RNA from the H2, but no hybridizing band was detected in the H2-I or H1 lines (Fig. 6). This indicates that the RHD3 intron is required for proper gene transcription or RNA stability.
Figure 6.
RNA-blot analysis of GUS reporter expression. RNA was isolated from 7-d-old seedlings from the H1, H2, and H2-I lines, and the blot was probed with a GUS-specific DNA fragment. Approximately 20 μg of total RNA was loaded per lane. The blot was subsequently hybridized with an 18S rRNA probe as a loading control.
To test the possibility that the first intron may possess a transcriptional enhancer-like element, a GUS reporter fusion was constructed with the first intron sequence placed upstream of the 1.5-kb RHD3 native promoter (construct H1Ia; Fig. 4) and upstream of the 0.6-kb promoter fragment in the B2 construct (construct B2I; Fig. 4). In addition, the RHD3 first intron was placed upstream of the heterologous cauliflower mosaic virus 35S minimal promoter (containing 46-bp promoter sequences and unable to induce detectable expression by itself) and the cauliflower mosaic virus 35S constitutive promoter fused to the GUS reporter (constructs 35SMI and 35Sia, respectively; Fig. 4). Furthermore, GUS fusions were made with the intron placed in an inverse orientation upstream of the 1.5-kb RHD3 native promoter and the 35S constitutive promoter (constructs H1Ib and 35Sib, respectively; Fig. 4). If the RHD3 intron sequence acts as an enhancer-like element to influence gene transcription, these constructs would be expected to confer a high level of GUS expression. However, transgenic lines bearing these constructs failed to exhibit a detectable difference in GUS expression (assessed by histochemical staining) compared with lines with the intron-less control constructs (data not shown).
One possible explanation for the results above is that the first RHD3 intron regulates expression only when it is located within the transcribed portion of a gene. To test this, GUS constructs were generated with the first intron (in its correct orientation) within the transcribed non-coding region downstream of the 1.5-kb native RHD3 promoter and the 35S constitutive promoter (constructs H1Ic and 35Sic, respectively; Fig. 4). Surprisingly, transgenic plants bearing these constructs actually exhibited a reduction in GUS expression, compared with plants containing the intron-less control constructs (Fig. 5, B–D). Reverse transcriptase-PCR experiments indicated that the intron was spliced properly from the H1Ic transcript (data not shown). Together, comparing the effects of the H1Ic and H2 constructs, the first RHD3 intron apparently affects gene expression only when located at its normal position.
RHD3 Gene Function Is Dependent on the Presence of the First Intron
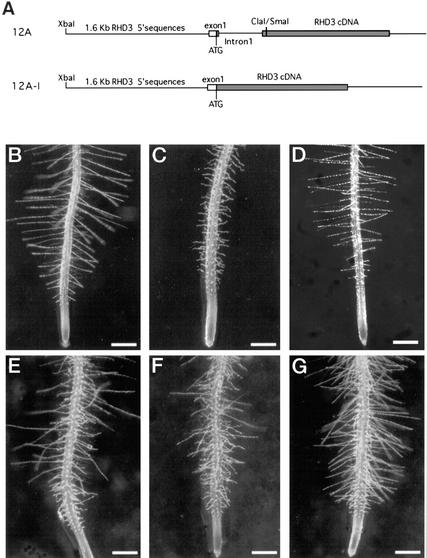
Although the above results show that the first intron is required for appropriate RHD3 gene expression, the significance of this intron for RHD3 function was not clear. As described, the 12A RHD3 construct, which contains the RHD3 promoter and first intron fused to the RHD3 cDNA, can fully complement the rhd3 mutant phenotype (Fig. 3; Table I). A construct was generated (designated 12A-I) that contains the same upstream regulatory sequences as the 12A construct fused to the RHD3 cDNA, except that the first intron was excluded (Fig. 7A). This construct was introduced into the rhd3-2 and rhd3-3 mutant lines, and the resulting transgenic plants exhibited either the rhd3 mutant phenotype or a phenotype intermediate between the rhd3 mutant and wild type (Fig. 7, B–G; Table I). The inability of this construct to complement the rhd3 mutant shows that the first intron is required for RHD3 gene function.
Figure 7.
The first intron of the RHD3 gene is required for complete complementation of the rhd3 mutant phenotype. A, RHD3 construct maps showing the 12A construct (including the first RHD3 intron) and the 12A-I construct (lacking the first intron). B through G, Phenotype of 4-day-old seedling roots and root hairs. Bars = 300 μm. B, Wild type (no ecotype). C, rhd3-3 mutant. D, Transgenic rhd3-3 seedling harboring the 12A construct. E through G, Independent transgenic rhd3-3 seedlings harboring the 12A-I construct showing the variation in the partial complementation phenotypes.
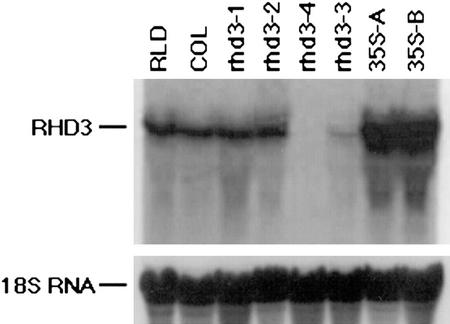
To test whether increased transcription of the RHD3 gene could substitute for the presence of the first intron, a construct was generated (35S::RHD3, Fig. 4) in which the RHD3 cDNA was placed under control of the constitutive 35S promoter. After transformation of rhd3-3 mutant plants, this transgene did not rescue the rhd3 mutant phenotype (Fig. 5E), although a partially complemented phenotype was observed in three of 14 lines (Fig. 5F). The results from a northern-blot analysis showed that the failed complementation was not due to transgene inactivation (e.g. cosuppression); in fact, the overall level of RHD3 RNA is much greater in the 35S-RHD3 lines than in wild-type plants (Fig. 8). This indicates that the high level of gene transcription directed by the 35S promoter is not sufficient to ensure RHD3 gene function.
Figure 8.
RNA-blot analysis of RHD3 expression. RNA was isolated from 7-d-old seedlings from two wild-type lines (Rschew [RLD] and Columbia), four rhd3 mutants (rhd3-1, rhd3-2 rhd3-4, and rhd3-3), and two independent transgenic lines bearing the 35S-RHD3 cDNA construct in the rhd3-3 mutant background (35S-A and 35S-B). Approximately 20 μg of total RNA was loaded per lane; the 18S rRNA probe used as a loading control is shown.
In a related experiment, we sought to determine whether the 35S promoter might complement the rhd3 mutant if the first intron is included in the transcribed (non-translated) region in the proper orientation (construct 35SI::RHD3, Fig. 4). However, when introduced into rhd3 mutant plants, this transgene did not fully complement the mutant phenotype (Fig. 5, G and H), although reverse transcriptase-PCR analysis indicated that the intron was spliced properly from the 35SI::RHD3 transcript (data not shown). This result is consistent with the reporter gene expression studies with the 35SIc construct (Fig. 5C). Taken together, these data indicate that the first intron is necessary for appropriate RHD3 expression and function, and it does not act properly when placed in abnormal locations.
Root Hair Development Genes and Plant Hormones Do Not Affect RHD3 Gene Expression
To attempt to define factors that regulate the activity of the RHD3 promoter and/or first intron, we examined the possibility that RHD3 expression may be regulated by its own protein or by other RHD gene products. Thus, the full-length RHD3 promoter::GUS reporter gene constructs (X1 and H2) were introduced into the rhd3 mutant or into the rhd1, rhd2, rhd4, and rhd6 mutants. GUS activity in each of these lines was found to be indistinguishable from the wild-type RHD3::GUS plants, based on histochemical analysis in roots (data not shown). This indicates that these RHD genes do not regulate the activity of the RHD3 promoter or first intron. In addition, double-mutant combinations between rhd3 and rhd1 (Schiefelbein and Somerville, 1990), rhd3 and rhd4 (Schiefelbein and Somerville, 1990), and rhd3 and rhd6 (Fig. 9, A–D) display an additive phenotype, which further supports the notion that RHD3 acts in a pathway separate from these genes during root hair development.
Figure 9.
Root phenotypes of 5-d-old seedlings from rhd3 mutant and double mutants. A, Wild type (Columbia ecotype). B, rhd3-1 mutant. C, rhd6-1 mutant. D, rhd3-1 rhd6-1 double mutant. E, aux1-7 mutant. F, axr2-1 mutant. G, eto1-1 mutant. H, ctr1-2 mutant. I, ein2-1 mutant. J, rhd3-1 axr2-1 double mutant. K, rhd3-1 eto1-1 double mutant. L, rhd3-1 ctr1-2 double mutant. Bars = 400 μm.
To determine whether the plant hormones auxin and ethylene may affect RHD3 expression, three sets of experiments were conducted. First, transgenic seedlings bearing the full-length RHD3::GUS constructs (X1 and H2) were transferred from standard growth media to media supplemented with 30 and 300 nm indole-3-acetic acid (an auxin), 5 and 50 μm 1-aminocyclopropane-1-carboxylic acid (a precursor to ethylene biosynthesis), or 5 μm aminoethoxyvinyl-Gly (an ethylene biosynthesis inhibitor), and then tested for GUS activity by histochemical staining. Although these hormone and inhibitor treatments significantly altered root morphogenesis, they did not cause a detectable change in the RHD3::GUS expression (data not shown).
In a second test of the effect of hormones on RHD3 gene expression, RHD3::GUS activity was examined in various hormone-related mutant backgrounds. These mutants included the aux1 (Pickett et al., 1990), axr2 (Wilson et al., 1990), eto1 (Guzman and Ecker, 1990), ein2 (Guzman and Ecker, 1990), and ctr1 (Kieber et al., 1993). As shown in Figure 9, E through I, each of these mutations affects the development of the root and/or root hairs of Arabidopsis. Despite these effects, none of these mutations caused an observable effect on the expression of the RHD3::GUS X1 or H2 constructs (data not shown), indicating that these hormone-related genes do not directly regulate the RHD3 promoter or first intron activity.
In a third test of the relationship between RHD3 and plant hormones, double mutants were constructed and analyzed that contained the rhd3 and either the eto1, ctr1, or axr2 mutations. Because each of these mutants exhibit a cell size defect in the root epidermis (Table II), we were able to use this character to determine the genetic relationships between the rhd3 and these mutations. As shown in Table II and Figure 9, J through L, the rhd3 ctr1, rhd3 eto1, and rhd3 axr2 double mutants each display a phenotype indicative of an additive interaction of the single mutations, consistent with the possibility that the genes act in separate pathways.
Table II.
Effect of mutations on root epidermis cell length
| Genotype | Cell Lengtha |
|---|---|
| μm cell−1 | |
| Columbia | 207 ± 36 |
| rhd3-1 | 86 ± 19 |
| eto1–1 | 59 ± 7 |
| rhd3-1 eto1–1 | 32 ± 4 |
| ctr1–2 | 37 ± 6 |
| rhd3-1 ctr1–2 | 26 ± 5 |
| axr2 | 169 ± 23 |
| rhd3-1 axr2 | 64 ± 8 |
Mean ± sd for root hair-bearing epidermal cells.
DISCUSSION
We have conducted a detailed study of the expression and regulation of the Arabidopsis RHD3 gene, which encodes a putative GTP-binding protein required for normal cell enlargement throughout plant development. One of the major findings is the critical role of the first intron of RHD3 for appropriate gene expression and function. This intron is notable because of its relatively large size (558 bp) and its location adjacent to the translation start codon (in codon no. 2). These features initially suggested a possible regulatory function for the first intron sequences. Comparison of reporter gene expression patterns between constructs containing this region (H2) and lacking this region (H1) shows a qualitative and quantitative difference in the expression pattern. In the absence of the intron, gene expression is reduced by two orders of magnitude (as determined both by RNA-blot hybridization and reporter enzyme activity) and is restricted to vascular tissue. The functional importance of this intron is demonstrated by complementation analyses in which a construct containing the first intron (12A) is able to fully complement the rhd3 mutant, whereas a construct lacking the first intron (12A-I) cannot (Fig. 7). Thus, the first intron is required for the high-level expression and appropriate function of RHD3.
This research has also yielded clues regarding the mechanism responsible for the effect of the RHD3 first intron. First, gene fusion experiments show that the intron cannot significantly increase reporter gene expression when placed upstream of the RHD3 native promoter or upstream of the heterologous 35S promoter, whether in the correct or the reverse orientation. This shows that the RHD3 first intron does not exhibit classic transcriptional enhancer-like properties. Second, the RHD3 first intron is unable to direct a high level of reporter gene expression or 35S::RHD3 cDNA expression when placed in the 5′-transcribed (non-coding) region, which implies that its normal location within the translated region is critical for its effect on RHD3 gene expression. Third, the 35S promoter is unable to drive appropriate expression of the RHD3 cDNA to enable complete complementation of the rhd3 mutant, despite the fact that the 35S::RHD3 rhd3 plants accumulate a greater amount of RHD3 RNA than do wild-type plants (Fig. 8). This result implies that the RHD3 first intron does not simply act by increasing the RHD3 transcript level; rather, it likely plays a regulatory role in RHD3 mRNA metabolism. Taken together, our results suggest that the RHD3 first intron acts posttranscriptionally to regulate the production of mature RHD3 mRNA.
An interesting possibility raised by our results is that the position or context of the RHD3 intron is critical for its ability to direct appropriate gene expression. Although several plant introns have been shown previously to influence gene expression (Callis et al., 1987; Dean et al., 1989; Vasil et al., 1989; Fu et al., 1995; Bolle et al., 1996; Rose and Last, 1997), only a few studies provide evidence for the importance of the intron position or context (Callis et al., 1987; Luehrsen and Walbot, 1991; Norris et al., 1993; Jeon et al., 2000), and these did not assess the functional importance of the intron position by conducting mutant complementation assays as in the present study. However, the position of an intron within a Xenopus laevis pre-mRNA transcribed in vivo has been shown to determine the translational efficiency of the mature mRNA in the cytoplasm (Matsumoto et al., 1998). Also, studies from several species indicate a connection between mRNA maturation (including 5′ capping, 3′ polyadenylation, and export to the cytosol) and splicing (Bentley, 1999). Together, these suggest that the particular pathway of mRNA maturation, which is influenced by intron position, may be a key determinant of gene expression. Further studies of RNA splicing and accumulation will be needed to precisely define the mechanism of action of the RHD3 first intron.
In addition to the first intron, another important RHD3 regulatory sequence was identified using the reporter gene fusions. This region is located in the upstream sequences between −1,500 and −600 bp and is required to confer gene expression in the vascular tissue of various Arabidopsis organs. Promoter constructs that lack this region and the intron are unable to direct detectable levels of reporter gene expression.
A second goal of the present study was to define the relationship between the RHD3 gene product and other possible regulators of cell expansion. Using the RHD3::GUS reporter gene and double-mutant tests, we have demonstrated that other root hair development genes (RHD1, RHD2, RHD4, and RHD6), the plant hormones auxin and ethylene, and hormone-related genes (AUX1, AXR2, ETO1, EIN2, and CTR1) do not significantly affect RHD3 gene expression. These data suggest that RHD3 functions in an independent pathway from these loci and hormones in the control of plant cell morphogenesis.
This RHD3 expression study supports the view that the RHD3 protein is involved in a fundamental cellular process that is required for regulated cell enlargement to occur during plant development. Whole-mount in situ hybridization and RHD3 promoter::GUS reporter gene fusion studies show that the RHD3 gene is highly expressed in most plant tissues, including the embryo, flower, shoot, and root tissues. These results are consistent with our previous data from a northern-blot analysis, which showed that RHD3 transcripts accumulate in all plant organs (Wang et al., 1997). Because rhd3 mutants do not exhibit a lethal phenotype (even rhd3 mutants bearing a nonsense mutation, like the rhd3-3 allele, are viable), the RHD3-associated cellular process is not essential or it is partially carried out by a redundant pathway. One possibility, suggested in earlier studies (Galway et al., 1997; Wang et al., 1997), is that RHD3 may control vacuole biogenesis, which is critical for cell enlargement. Further studies to determine the cellular location of the RHD3 product and RHD3-interacting proteins are likely to uncover new insights into the molecular basis of RHD3 function and plant cell morphogenesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rhd3 mutant alleles were described previously (Schiefelbein and Somerville, 1990; Wang et al., 1997). The source of the hormone-related mutants (aux1-7, axr1-3, axr2, eto1-1, ein2-1, and ctr1-2) was previously given (Masucci and Schiefelbein, 1996). Double mutants (rhd3-1 axr2, rhd3-1 eto1-1, and rhd3-1 ctr1-2) were constructed by crossing single mutant plants, examining the F2 progeny for putative double-mutant phenotypes, and confirming the double mutants in subsequent generations.
Unless otherwise described, Arabidopsis seeds were surface sterilized and grown on agarose-solidified medium in vertically oriented petri plates as described previously (Schiefelbein and Somerville, 1990). Media containing indole-3-acetic acid (Sigma, St. Louis), 1-aminocyclopropane-1-carboxylic acid (Sigma), or aminoethoxyvinyl-Gly (MAAG Agrochemicals, Vero Beach, FL) were prepared as described (Masucci and Schiefelbein, 1994; Wang et al., 1997).
Microscopy and Expression Assays
Root growth rate and epidermal cell length were determined as previously described (Wang et al., 1997) from at least 10 independent lines. The histochemical staining of plants containing the GUS reporter gene was performed essentially as described (Masucci et al., 1996). The RHD3::GUS (X1 and H2) were introduced into mutant backgrounds by genetic crosses, and histochemical staining was conducted with F2 seedlings exhibiting the mutant and wild-type phenotypes. Quantitative assay of the GUS activity was performed essentially as described by Jefferson et al. (1987). Whole-mount in situ hybridization with young Arabidopsis seedlings was performed as previously described (Masucci et al., 1996). RNA-blot (northern) analyses were conducted as described (Wang et al., 1997).
Constructs and Plant Transformations
The 12A construct was described by Wang et al. (1997). For the 12A-I construction, the BamHI-SacI (filling the SacI 5′-protruding end) fragment containing the GUS gene-coding region in the pBI101.2 vector was replaced with the BamHI-EcoRV fragment from the C3I clone of the RHD3 cDNA. Next, an XbaI-XhoI fragment containing the putative RHD3 promoter (1.6-kb 5′ non-coding sequences and 80-bp 5′-untranslated sequences) was subcloned (by filling the XhoI 3′-protruding end) from the genomic clone pR5.2 (Wang et al., 1997) into the XbaI and SmaI sites in the above modified pBI101.2 vector.
For the X1 construct, a XbaI-ClaI fragment containing the 1.6-kb 5′-non-coding sequences, the first exon, the first intron, and three nucleotides of the second exon was subcloned (by filling in the ClaI 3′-protruding end) into the XbaI and SmaI sites of the pBI101.2 vector to generate a translational fusion. The H2 construct was generated the same way as X1 except a HindIII-ClaI promoter fragment was subcloned, which lacks 100 bp from the non-coding 5′ end of the XbaI-ClaI fragment.
The H1 construct was generated by subcloning the HindIII-XhoI fragment containing the 1.5-kb 5′-non-coding sequences plus about 80 bp of 5′-untranslated sequences of the first exon from the plasmid pR5.2 into the HindIII and SalI sites of the pBI101.2 vector to generate transcriptional fusions. For the B2 and B4 constructs, a 690-bp BamHI-BglII fragment containing about 600 bp of 5′-non-coding sequences and 90 bp of 5′-untranslated sequences of the first exon from the plasmid pR5.2 was subcloned into the BamHI site of the pBI101.2 vector to generate transcriptional fusions, with the B2 construct having the promoter sequences in the correct orientation and B4 in the reverse orientation.
For construction of the H2-I clone, the T3 primer and another primer (5′ GTGGATCCCCCATTATCGCTCAACGAAACGC 3′) were used to amplify the RHD3 upstream sequences and the entire first exon using the genomic clone pR5.2 as the template. The PCR products were subcloned into the HindIII-BamHI sites of the pBI101.2 vector as a HindIII-BamHI fragment to generate a translational fusion. Two other primers (5′ TGGAAGCTTGTAATTTATATATCCATCTCTTC 3′ and 5′ CCTTCGAACTGCAACGTCC-AAGCAATAAAG 3′) were used to generate a PCR fragment containing the intron sequence using the genomic clone plasmid pR5.2 as the template and the PCR product was subcloned into the TA cloning vector PCR 2.1 (Invitrogen, Carlsbad, CA) to generate a clone called pR5.2E4. Next, the HindIII fragment containing the first intron sequences was subcloned into the HindIII sites of H1, B2, 35SM (35S 46-bp minimal promoter driving the GUS gene), and pBI121 clones to generate constructs H1Ia, H1Ib, B2I, 35SMI, 35SIa, and 35SIb. In addition, an XbaI-SacI (blunted end) fragment containing the intron sequence from the pR5.2E4 clone was subcloned into the XbaI-SmaI sites of the H1 and pBI121 clones to generate the clones H1Ic and 35SIc.
The 35S::RHD3 construct was made by replacing the BamHI-SacI (blunt ended) containing the GUS gene-coding region in the pBI121 vector by a BamHI-EcoRV fragment containing the full-length RHD3 cDNA from the cDNA clone C3I. For the 35SI::RHD3 construct, the XbaI-SacI (blunt ended) fragment containing the intron sequence from pR5.2E4 was subcloned into the XbaI-SmaI sites of the 35S::RHD3 clone.
For plant transformation, the above constructs were electroporated into the Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants using vacuum infiltration. The T1 seeds for each construct were collected in separate pools, and plated on selection media containing kanamycin (50 μg mL−1) plus timentin (100 μg mL−1). Positive transgenic plants from different pools were grown and considered as independent transformation events. For all experiments, T2 or T3 plants were used for phenotypic and GUS activity assays.
ACKNOWLEDGMENTS
We thank Dr. Mark Kinkema (Duke University, Durham, NC) for providing the A. tumefaciens strain GV3101 and helpful suggestions on plant vacuum infiltration transformation. We thank Dr. Hong Ma (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing the 35SM plasmid (35S minimal promoter-GUS construct).
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. IBN–9316409) and by the U.S. Department of Agriculture (grant nos. 97–35304–4580 and 01–35304–10134).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002675.
LITERATURE CITED
- Abeles FB, Morgan PW, Saltveit ME. Ethylene in Plant Biology. Ed 2. New York: Academic Press; 1992. [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Bolle C, Herrmann RG, Oelmuller R. Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J. 1996;10:919–924. doi: 10.1046/j.1365-313x.1996.10050919.x. [DOI] [PubMed] [Google Scholar]
- Callis J, Fromm M, Walbot V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P. Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell. 1989;1:201–208. doi: 10.1105/tpc.1.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organization of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Evans ML. Functions of hormones at the cellular level of organization. In: Scott TK, editor. Encyclopedia of Plant Physiology. Vol. 10. Berlin: Springer-Verlag; 1984. pp. 23–79. [Google Scholar]
- Fu H, Kim SY, Park WD. High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene requires 5′ and 3′ flanking sequences and the leader intron. Plant Cell. 1995;7:1387–1394. doi: 10.1105/tpc.7.9.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway ME, Heckman JW, Schiefelbein JW. Growth and ultrastructure of Arabidopsis root hairs: The rhd3 mutation alters vacuole enlargement and tip growth. Planta. 1997;201:209–218. doi: 10.1007/BF01007706. [DOI] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L. The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 1997;115:981–990. doi: 10.1104/pp.115.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J-S, Lee S, Jung K-H, Jun S-H, Kim C, An G. Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 2000;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Luehrsen KR, Walbot V. Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet. 1991;225:81–93. doi: 10.1007/BF00282645. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SR, Meyer SE, Callis J. The intron of Arabidopsis thaliana polyubiquitin genes is conserved in location and is a quantitative determinant of chimeric gene expression. Plant Mol Biol. 1993;21:895–906. doi: 10.1007/BF00027120. [DOI] [PubMed] [Google Scholar]
- Paquette AJ, Benfey PN. Axis formation and polarity in plants. Curr Opin Genet Dev. 2001;11:405–409. doi: 10.1016/s0959-437x(00)00210-0. [DOI] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 1990;94:1462–1466. doi: 10.1104/pp.94.3.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997;11:455–464. doi: 10.1046/j.1365-313x.1997.11030455.x. [DOI] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Masucci JD, Wang H. Building a root: the control of patterning and morphogenesis during root development. Plant Cell. 1997;9:1089–1098. doi: 10.1105/tpc.9.7.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge, UK: Cambridge University Press; 1989. [Google Scholar]
- Vasil V, Clancy M, Ferl RJ, Vasil IK, Hannah LC. Increased gene expression by the first intron of maize Shrunken-1 locus in grass species. Plant Physiol. 1989;91:1575–1579. doi: 10.1104/pp.91.4.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The Root Hair Defective3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wilson A, Pickett FB, Turner JC, Estelle M. A dominant mutant in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Mol Gen Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]