Abstract
We identified a new gene that is interrupted by T-DNA in an Arabidopsis embryo mutant called raspberry3. raspberry3 has “raspberry-like” cellular protuberances with an enlarged suspensor characteristic of other raspberry embryo mutants, and is arrested morphologically at the globular stage of embryo development. The predicted RASPBERRY3 protein has domains found in proteins present in prokaryotes and algae chloroplasts. Computer prediction analysis suggests that the RASPBERRY3protein may be localized in the chloroplast. Complementation analysis supports the possibility that the RASPBERRY3 protein may be involved in chloroplast development. Our experiments demonstrate the important role of the chloroplast, directly or indirectly, in embryo morphogenesis and development.
The molecular and cellular mechanisms that program the series of events leading to the development of a plant embryo are not well understood. Embryogenesis is a complex process that requires regulation of cell-specific and housekeeping genes within the embryo proper and neighboring seed tissues (e.g. endosperm) surrounding the embryo (Goldberg et al., 1994). In Arabidopsis, it has been estimated from genetic studies that there are about 4,000 essential genes and about 40 embryonic patterning genes that are required for normal plant embryogenesis (Jürgens et al., 1991). The regulation of these genes must be tightly coordinated and controlled in a spatially and timely manner starting from the zygote to the mature embryo in dormant seeds (Goldberg et al., 1989; West and Harada, 1993; Jürgens, 1995; Jürgens et al., 1995). How the plant embryo achieves this coordination is not yet known and is a major question of plant developmental biology.
A genetic approach is one of the strategies adopted to begin to understand the process of plant embryogenesis. There are more than 2,000 Arabidopsis embryo mutants that have been isolated by several laboratories over the last 10 years using ethyl methanesulfonate mutagenesis, T-DNA insertional inactivation, and transposon tagging (Errampalli et al., 1991; Mayer et al., 1991; Meinke et al., 1994; Schwartz et al., 1994; Yadegari et al., 1994; Altmann et al., 1995; Devic et al., 1996; McElver et al., 2001). The largest class of mutants within these collections are those that remain morphologically globular in shape (Errampalli et al., 1991; Mayer et al., 1991; Meinke et al., 1994; Schwartz et al., 1994; Yadegari et al., 1994). The raspberry (rsy) mutants, including rsy1 and rsy2, belong to this class (Yadegari et al., 1994).
Several embryo mutants have been characterized, leading to the identification of embryo genes and what their functions are during embryo development. Some genes encode proteins that are involved in transcription or that are associated with transcription factors (Aida et al., 1997; Hardtke and Berleth, 1998; Li and Thomas, 1998; Lotan et al., 1998; Stone et al., 2001). Others encode proteins that are important for cell division, cell polarity, differentiation (Berleth and Jürgens, 1993; Shevell et al., 1994; Lukowitz et al., 1996; Hardtke and Berleth, 1998; Shevell et al., 2000; Grebe et al., 2000; Schrick et al., 2000), or general metabolic functions (Patton et al., 1998).
Mutations in genes that are involved in chloroplast function can lead to defects in embryo development. For example, we demonstrated recently that a mutation in the nuclear-encoded chaperonin-60α protein required for proper folding of chloroplast-bound proteins leads to a defect in plastid development that results in cotyledon shortening and embryo arrest (Apuya et al., 2001). Likewise, mutations in nuclear-encoded ribosomal proteins S1 and S16, glycyl-tRNA synthetase, and EMB506 protein containing ankyrin repeats, all of which are also imported by the chloroplast, are associated with defects in embryo development (Tsugeki et al., 1996; Yadegari, 1996; Uwer et al., 1998; Albert et al., 1999; Despres et al., 2001). These studies suggest that the process of embryogenesis is linked to biochemical and developmental processes that occur in the chloroplast.
In this paper, we present results that characterize an embryo-defective mutant called rsy3. A T-DNA insertion in RSY3 causes the embryo to be morphologically arrested at the globular stage. Our analyses of the predicted protein encoded by the RSY3 gene indicate that it is a novel polypeptide. It has a signature motif characteristic of proteins capable of hydrolyzing ATP and has other motifs characteristic of proteins encoded by prokaryotic and photosynthetic algae genomes. Our experiments suggest that the RSY3 protein is localized in the chloroplast and its presence is required for chloroplast differentiation and embryonic development.
RESULTS
rsy3 Embryos Are Morphologically Arrested at the Globular Stage
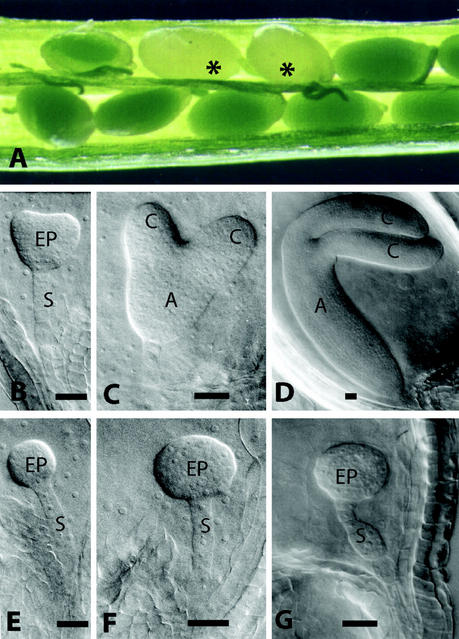
We analyzed the development of wild-type and mutant embryos using Nomarski microscopy to characterize the terminal phenotype of rsy3 embryos (see “Materials and Methods”). Within a heterozygous silique, wild-type embryos are distinguished from embryo-defective ones by seed color (Fig. 1A). Green seeds contained wild-type embryos, whereas white seeds contained mutant embryos, which segregated at a 3:1 ratio in rsy3/RSY3 siliques. This distinction was visible by the start of the early heart stage when greening occurs due to the initial development of the chloroplasts (Schulz and Jensen, 1968; Mansfield and Briarty, 1991). To understand the progression of the rsy3 mutant phenotype, normal and aborted seeds at different stages of development were isolated from rsy3/RSY3 siliques and were analyzed. The development of wild-type embryos progressed normally (Fig. 1, B–D), consistent with previously published analysis of Arabidopsis embryo development (Goldberg et al., 1994; Jürgens and Mayer, 1994). On the other hand, rsy3 embryos did not develop beyond the globular stage (Fig. 1, E–G). At the stage when the wild-type embryos were at the late-curling stage, typified by the bending of the two cotyledons (Fig. 1D), the rsy3 embryo proper did not show any indication of cotyledon formation. Instead, the embryo proper of rsy3 embryos exhibited “raspberry-like” cellular protuberances similar to that of rsy1 and rsy2 mutants (Yadegari et al., 1994). In addition, the rsy3 suspensor became enlarged (Fig. 1G) at the time when its counterpart in wild-type embryos was barely visible (Fig. 1D; see also Yadegari et al., 1994). The suspensor enlargement observed in rsy3 embryos was not as severe as that observed in rsy1 or rsy2 mutants (Yadegari et al., 1994).
Figure 1.
Developmental analysis of rsy3 mutant embryos. A, Typical heterozygous siliques containing wild-type and mutant (highlighted with asterisks) seeds. Nomarski images of wild-type embryos (B–D) were taken from the same siliques from which the corresponding mutant embryos (E–G) were taken (see “Materials and Methods”). A, Axis; C, cotyledon; EP, embryo proper; S, suspensor. Bars = 25 μm.
We attempted to rescue the rsy3 phenotype by allowing seeds with mutant embryos or mutant embryos dissected from seeds to grow in tissue culture medium containing Murashige and Skoog salt supplemented with vitamins and other components (e.g. growth regulators). The rsy3 embryos did not exhibit any response to the tissue culture treatments and eventually died (data not shown). This result indicates that the rsy3 genetic mutation could not be rescued by the components present in the tissue culture media used.
rsy3 Is Tagged with T-DNA
We performed genetic and molecular analyses to determine whether rsy3 was interrupted by a T-DNA insertion. The T-DNA vector used in these studies contains a neomycinphosphotransferase II gene that confers resistance to the antibiotic kanamycin (Errampalli et al., 1991; Feldmann, 1991). We tested a total of 928 F2 individuals and observed that kanamycin-resistant (Kan-R) individuals were segregating from kanamycin-sensitive individuals at a 2:1 ratio, respectively (data not shown). We also tested 100 randomly picked F2 Kan-R individuals and found that all of them segregated the embryo-defective rsy3 phenotype from wild type within their siliques in a 3:1 ratio (data not shown). From these results, we concluded that the rsy3 mutation was most likely due to a T-DNA insertion in a single locus.
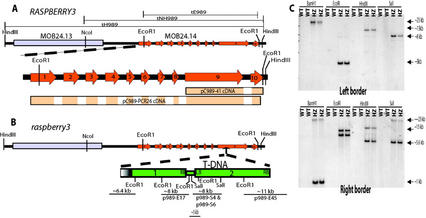
To identify the plant sequences flanking the inserted T-DNA, we used plasmid rescue (see “Materials and Methods”) and isolated three types of clones, p989-E17, p989-S4, and p989-E45, as diagrammed in Figure 2B. One of the rescued plasmids, fragment p989-E45 (Fig. 2B), contained a plant sequence that corresponded to a portion of a possible open reading frame (ORF; designated with gene ID MOB24.14 or At3g24560 in the National Center for Biotechnology Information [NCBI] database) found in a region of chromosome 3. This was consistent with mapping data (see “Materials and Methods”) that we obtained prior to completion of the Arabidopsis genome sequence that localized the rsy3 mutation to between positions 46.1 and 53.6 of chromosome 3 (data not shown). Based on sequence comparisons between rsy3 and RSY3 genes, we placed the T-DNA insertion in exon 9 of the predicted gene.
Figure 2.
Gene organization of RSY3 and T-DNA insertion in rsy3 mutant. A, Diagrammatic representation of a portion of the lambda genomic clone containing the RSY3 gene in chromosome 3. The predicted RSY3 gene (annotated for Columbia ecotype as MOB24.14) is expanded below the clone to highlight exons represented by solid arrows in orange and numbered accordingly. The genomic fragments tH989, tNH989, and tE989 used in the complementation analysis are outlined above the genomic clone. The cDNA clones are designated below the expanded region of RSY3 gene. Clone pC989–41 represents a partial cDNA isolated from a library, and clone pC989–41 represents the nearly full-length cDNA that were isolated using 5′- and 3′-RACE. Only the areas highlighted in colors within the rectangles represent the cDNA sequences. B, Diagrammatic representation of the T-DNA insertion in the rsy3 embryo mutant. Two T-DNAs that are arranged in concatemer are inserted in exon 9. Some of the EcoRI and SaI fragments, as revealed by plasmid rescue analysis, are highlighted with the approximate sizes written above the lines. Some of the restriction sites relevant to the DNA analysis shown in C are indicated. C, Restriction analysis of genomic DNAs isolated from wild-type (WT) and heterozygous (HZ) rsy3 individual segregants. DNAs were digested with restriction enzymes as indicated and were size separated by electrophoresis in a 1% (w/v) gel. The resulting blots were hybridized with a left or a right border probe as indicated in each panel. Restriction enzymes used are indicated. Note: Diagrams in A and B are not drawn to scale.
We were able to place the rescued plasmids as fragments of a concatemerized T-DNA inserted in the rsy3 gene as diagrammed in Figure 2B. The DNA sequences of the rescued plasmids were compared with the known sequence of the T-DNA used in the mutagenesis (Errampalli et al., 1991; Feldmann, 1991). We also performed genomic DNA-blot analyses using various fragments of the T-DNA as probes (Fig. 2C and data not shown). The results of these analyses indicated that there were two T-DNAs arranged in a concatemer in the rsy3 gene. One of the T-DNAs was defective (labeled no. 1 in Fig. 2B), with a deleted portion toward the left border region (Fig. 2B, highlighted in gradient gray color). Our plasmid rescue analysis also showed that there was a short extra partial fragment of the neomycinphosphotransferase II gene between numbers 1 and 2 T-DNAs (Fig. 2B, bottom panel). These results indicated that the T-DNA may have undergone rearrangements and deletions during insertion within the Arabidopsis genome.
We used the rescued fragment p989-E45 containing plant sequences as a probe to isolate clones from a cDNA library (see “Materials and Methods”). One of these cDNA clones was pC989–41, which covered only the predicted exons 9 and 10 (Fig. 2A). The same DNA sequence from pC989–41 was found in our nearly full-length cDNA clones (e.g. pC989-PCR26; Fig. 2A) that were isolated subsequently using 5′- and 3′-RACE (see “Materials and Methods”).
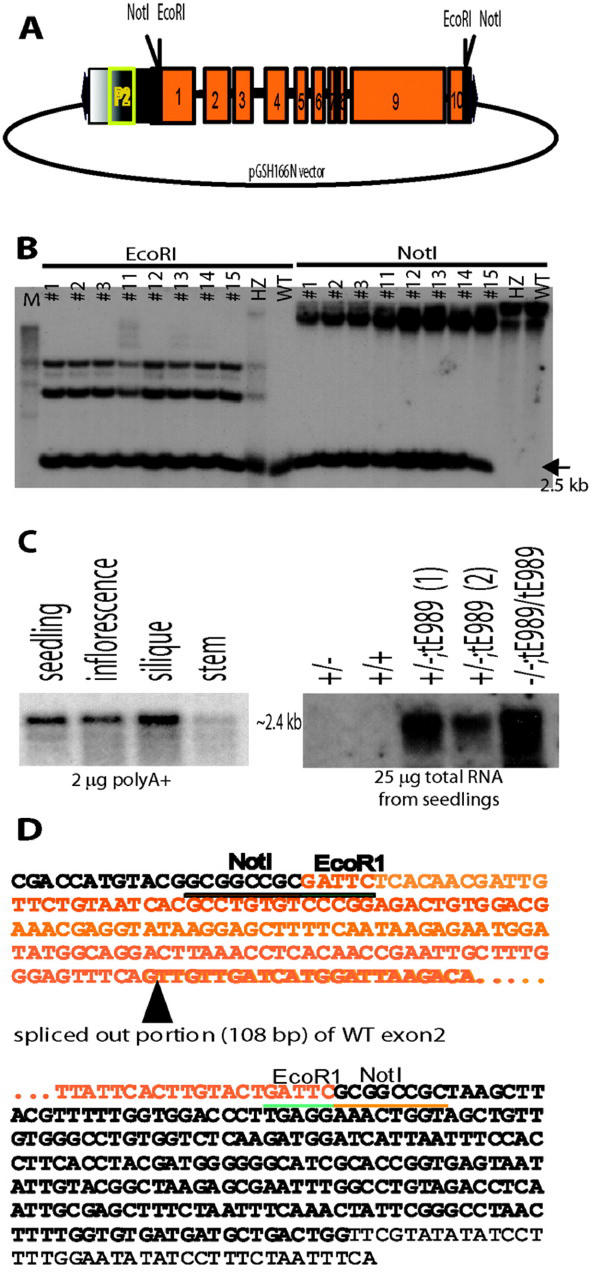
We performed genetic complementation to determine that the predicted ORF (MOB24.14) was the corresponding gene mutated in rsy3. We isolated RSY3 genomic clones from a wild-type genomic phage library using fragment p989-E45 as a probe (see “Materials and Methods”). Several clones were isolated and one of them contained a 9-kb HindIII fragment, as shown in Figure 2A. This HindIII fragment (designated as tH989) and its subfragments (Fig. 2A, designated as tNH989 and tE989) were used in the genetic complementation. Fragments tH989 and tNH989 were subcloned into the pHYG-A vector, and fragment tE989 was subcloned into the pGSH166N vector (Fig. 2A; see “Materials and Methods”). Both of the vectors contained hygromycin-resistant (Hyg-R) markers that were used to select for Arabidopsis transformants.
Three types of transformants were generated with each of the three genomic fragments (tH989, tNH989, or tE989). Transformants from each type were genetically crossed to heterozygous rsy3/RSY3 plants (Kan-R), and the progenies were analyzed following our previous strategy for genetic and molecular complementation analyses of embryo-defective phenotypes (Apuya et al., 2001). Our genetic crosses to each type of transformant gave rise to complemented heterozygous F2 individuals (Kan-R and Hyg-R) that produced about 6.25% mutant seeds instead of the 25% mutant seeds produced by selfing rsy3/RSY3 uncomplemented plants (data not shown; see Table I for representative data). We also found complemented homozygous rsy3/rsy3 plants, which were otherwise dead if not complemented, that produced wild-type seeds instead of rsy3 mutant seeds (example shown in Fig. 4 and data not shown). By testcross analysis, complemented homozygous rsy3/rsy3 individuals were determined to contain homozygous copies of the transgene (i.e. tH989/tH989, tNH989/tNH989, or tE989/tE989, depending on the type of transgene; data not shown). From these results, we concluded that the ORF corresponding to the interrupted gene in rsy3 is MOB24.14 (Fig. 2A, highlighted in orange).
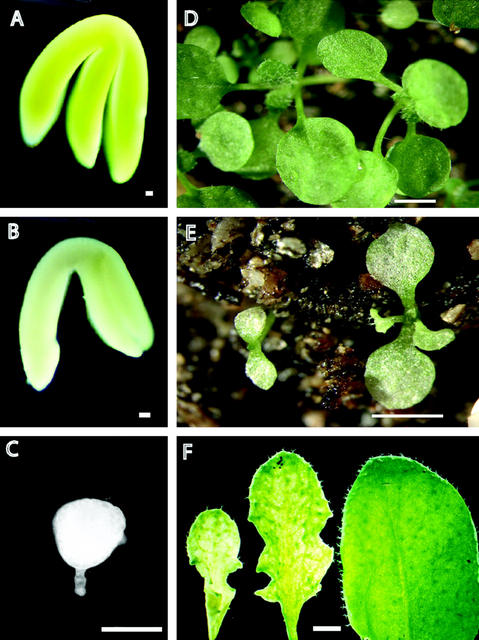
Figure 4.
The embryo-defective morphology of the rsy3 mutant can be rescued by a partial RSY3 genomic fragment. A through C, Three types of embryos produced by a heterozygous rsy3/RSY3 plant containing one copy of tE989 transgene. A, Morphologically wild-type green embryo (RSY3/RSY3;tE989). B, Partially rescued rsy3 pale-green embryo (rsy3/rsy3;tE989/tE989). C, Mutant rsy3 embryo (rsy3/rsy3). D, Seedlings generated from morphologically wild-type green embryos with genotype rsy3/RSY3; tE989. E, Seedlings generated from partially rescued rsy3 pale-green embryos with genotype rsy3/rsy3; tE989/tE989. F, Close-up view of rosette leaves taken from partially rescued rsy3 (rsy3/rsy3; tE989/tE989) plants (first two leaves from left; approximately 50 d postgermination) and from wild-type plants (right leaf; approximately 30 d postgermination). Bars = 50 μm in A through C; 3 mm in D and E; and = 2 mm in F.
RSY3 Encodes a Novel Protein
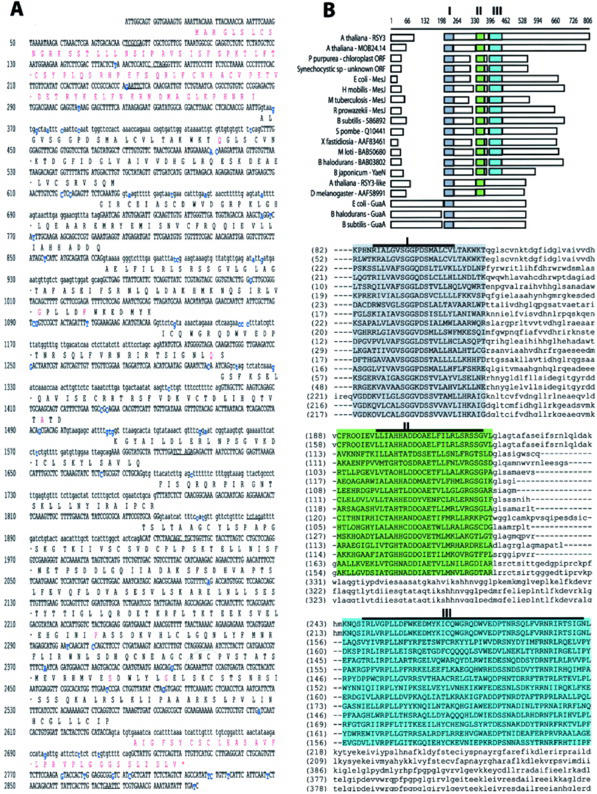
We sequenced the RSY3 genomic HindIII fragment and the pC989–41 and pC989-PCR26 cDNA clones (Fig. 2A), and predicted an ORF from the contigated DNA sequences generated from both cDNA clones. The RSY3 exons and introns were identified by comparing the genomic and cDNA sequences as shown in Figure 3A. On the basis of these comparisons, we to identified 10 RSY3 exons instead of the computer-predicted nine exons for the MOB24.14 gene as annotated in the NCBI database. The identified introns had the canonical GT at the 5′ end and the canonical AG at the 3′ end, except for the 5th intron, which had GC at the 5′end (Fig. 3A). Our sequencing revealed that there were minor differences (Fig. 3A, highlighted in blue) between RSY3 and the MOB24.14 locus, as reported in the NCBI database. These differences can be attributed to ecotypic variation—our RSY3 clones were isolated from the Wassilewskija (WS) ecotype, and the sequence reported for MOB24.14 was based on the Columbia ecotype. However, these differences introduced some variations (Fig. 3A, highlighted in red) in the predicted amino acid sequences, especially in the amino- and carboxy-terminal regions. Our predicted RSY3 protein had 663 amino acid residues (see accession no. AY077630), whereas the predicted MOB24.14 protein has only 614 amino acids (see accession no. BAB02008). We ran the predicted RSY3 protein through the P-sort software (Nakai, 2000) to determine its possible cellular localization. The results of this analysis indicated that RSY3 protein had 74% probability of being sorted into the chloroplast thylakoid membrane.
Figure 3.
RSY3 genomic DNA sequence and its predicted protein. A, Portion of the genomic DNA sequence of the RSY3 gene. Introns are in lowercase letters, and the exons and the untranslated sequences are in uppercase letters. Nucleotides highlighted in blue are missing or are different from the reported sequence based on a Columbia background (see sequence of predicted gene MOB24.12 in accession AB020746). The predicted amino acids are given above the coding sequence. Amino acids highlighted in red are those that differ from the predicted amino acid sequence in the RSY3 locus of the Columbia ecotype. B, Alignment of the RSY3 protein to other proteins with similar domains. The amino acid sequences derived from the predicted coding sequences were aligned using the AlignX program of Vector NTI software. The highlighted domains (I, II, and III) were subsequently found using the AlignX Block program of the same software. Domain I, highlighted in gray, is the putative ATP-binding domain. Consensus core regions (as cited in the text) within the three domains are highlighted with a bold line above the sequences. The proteins included in the above alignment have the following accession numbers: Porphyra purpurea (AAC08269), Synechocystis sp (BAA10210), Escherichia coli (BAA77863), Heliobacillus mobilis (AAC84036), Mycobacterium tuberculosis (AAK48088), Rickettsia prowazekii (CAA14513), Bacillus subtilis (BAA05302), Xylella fastidiosia (AAF83469), Mesorhizobium loti (BAB50680), Bacillus halodurans (BAB03802), Bradyrhizobium japonicum (BAB50680), Saccharomyces pombe (CAA94698), E. coli GuaA (AAG57618), B. halodurans GuaA (Q9KF78), B. subtilis GuaA (P29727), Arabidopsis RSY3-like (AAC16077), and fruit fly (Drosophila melanogaster) predicted protein (AAF58991). The accession number for the RSY3 genomic sequence is AY077630.
Our psi-blast analysis (Altschul et al., 1997), at a minimum of three iterations, revealed that RSY3 protein has distinct domains present in other putative proteins predicted from ORFs (Fig. 3B). Most of these predicted proteins are prokaryotic in origin. Among these are ORFs from the chloroplast genome of P. purpurea (accession no. AAC08269) and the genome of cyanobacterium Synechocystis sp. (accession no. BAA10210). The similarity of the RSY3 protein to these proteins is between 8% and 15% in the overall alignment, but the presence of three distinct domains suggests a possible similarity in their functions.
Three identified domains, designated as I, II, and III, are shown in Figure 3B, highlighting the optimum alignment among these proteins using the AlignX-Block program of Vector NTI software. Domain I has a core consensus sequence of RILVANSGG-DSMALLHLL and potentially corresponds to an ATP-binding site (Tiedeman et al., 1985; Zalkin et al., 1985; Tesmer et al., 1996). The core sequence SGG-DS within domain I is identical to the ATP pyrophosphatase domain present in the GMP synthetase class of Gln amidotransferase (GuaA) and other synthetase enzymes (e.g. NAD synthetase and Asn synthetase) that have ATP-hydrolyzing activity (Tesmer et al., 1996). Domain II has the LLLAHHADDQAETILLRL-RGSG as a consensus core sequence, whereas domain III has the I/L-LVRPLL-I-K/R-EL—YCK—L-W-ED-SN—Y-RNRI/LR–I/LLP sequence (Fig. 3B). Domains II and III were not identified in the GuaA proteins even when we use less stringent parameters in our alignment procedures.
Another putative protein encoded by a gene within the Arabidopsis genome shows similarity to the RSY3. This is designated as RSY3-like protein in Figure 3B and is annotated as a hypothetical protein in the database (accession no. AAC16077). It is more closely related (79% similar) to a predicted fruit fly protein (accession no. AAF58991) than to the RSY3 protein (12% similar) considering the overall alignment. The RSY3-like and the fruit fly proteins have domains I and II, but not domain III (see Fig. 3B).
Taken together, the presence of these domains in the RSY3 protein suggests that its function may be similar to the prokaryotic and chloroplast proteins predicted from the algae chloroplast and bacterial genomes.
Partial Complementation of rsy3
Results from our complementation experiments using less than a full-length RSY3 gene (i.e. tE989) provided clues about its function. Transgene tE989 covered RSY3 gene regions between the first exon and the 3′-untranslated region (see diagram in Fig. 2A and sequence in Fig. 3A). We observed that some of the progeny seedlings resulting from the genetic cross between transformants containing the tE989 transgene exhibited abnormal coloration, although they were morphologically normal (Fig. 4). We observed that seeds from self-pollinated heterozygous plants with the tE989 transgene (rsy3/RSY3;tE989) produced three types of embryos: morphologically normal green embryos (Fig. 4A), morphologically normal pale-green embryos (Fig. 4B), and rsy3-type mutant embryos (Fig. 4C) in a 12:3:1 ratio, respectively (data not shown). This ratio was observed using 12 independent transformants with the same tE989 transgene serving as parents for the complementation genetic cross (data not shown). The normal seeds represented wild-type (RSY3/RSY3, RSY3/RSY3;tE989, and RSY3/RSY3;tE989/tE989) and heter-ozygous segregants (rsy3/RSY3, rsy3/RSY3;tE989, and rsy3/RSY3;tE989/tE989), the pale-green seeds represented the homozygous segregants containing the transgene (rsy3/rsy3;tE989 and rsy3/rsy3;tE989/tE989), and the rsy3-type seeds represented the homozygous rsy3/rsy3 segregants without the transgene (data not shown).
We also observed that the development of the rescued embryos with the tE989 transgene was slower than that of normal embryos. At the stage in which normal embryos were already at the late curling stage, the pale-green embryos (rsy3/rsy3;tE989 and rsy3/rsy3;tE989/tE989) isolated from the same silique were still at the torpedo stage (data not shown). This is also evident from the embryos shown in Figure 4, A and B (taken from the same siliques), where the pale-green embryo (rsy3/rsy3;tE989; Fig. 4B) was not at the same mature stage as the normal embryo (RSY3/RSY3;tE989; Fig. 4A).
Seeds containing pale-green embryos were able to germinate. However, unlike the green seedlings that developed from wild-type seeds (as shown in Fig. 4D) containing the same transgene (i.e. RSY3/RSY3; tE989 or rsy3/RSY3;tE989), the seedlings from seeds containing pale-green embryos (i.e. rsy3/rsy3;tE989 or rsy3/rsy3;tE989/tE989) developed slower and had a pale-green color as well (Fig. 4E). To increase the viability of the pale-green seedlings, they were placed in a shaded area of our greenhouse to prevent “bleaching.” The overall color of the leaves (rosette and cauline) was pale-green. As the plants became older (approximately 6 weeks), some portion of the leaves turned greener (Fig. 4F). However, the leaves still had a curly and mottled appearance, especially on their abaxial side (Fig. 4F, compare the wild-type and pale-green leaves).
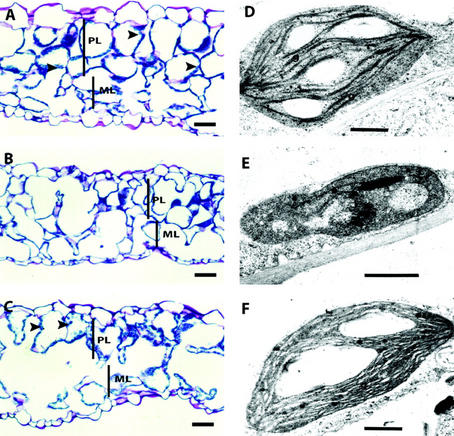
To determine whether there were tissue and cellular abnormalities within the pale-green leaves, we did a histological analysis of the leaf samples taken from pale-green leaves of partially complemented rsy3 plants (Fig. 5). The tissue section of the leaves from normal-green plants showed highly organized palisade and spongy mesophyll layers (Fig. 5A). The chloroplasts were ubiquitously present in the cells of both layers (Fig. 5A, highlighted with arrows). The palisade layer within the yellowish area of leaves taken from partially complemented rsy3 plants was not as organized as that in wild-type leaves (Fig. 5B). The palisade cells were smaller, not properly stacked, and did not contain as many chloroplasts as that observed from a normal leaf. Tissue sections taken from the greenish area (see Fig. 4F) of leaves from the partially complemented rsy3 plants also showed improperly stacked palisade cells (Fig. 5C). However, unlike the section from the yellowish area, the palisade cells within the greenish area showed presence of chloroplasts similar to those in wild-type leaves (Fig. 5C, highlighted with arrows).
Figure 5.
Histological analysis of partially rescued rsy3 leaves. Separate sections of leaves shown in Figure 4E were taken for histological analysis (A–C) and for transmission electron microscopic (TEM) analysis of chloroplasts (D–F). Tissue section (A) and TEM of chloroplast (D) from morphologically wild-type green plants (RSY3/RSY3;tE989). Leaf tissue section (B) and TEM of a chloroplast (E) from a pale-yellow region of partially rescued rsy3 mutant plants (rsy3/rsy3; tE989/tE989). Leaf tissue section (C) and TEM of a chloroplast (F) from a green region of partially rescued rsy3 mutant plants (rsy3/rsy3; tE989/tE989). Arrowheads point to chloroplasts. PL, Palisade mesophyll layer; ML, spongy mesophyll layer; g, grana. Bars = 100 μm in A through C and = 0.15 μm in D through F.
We performed TEM analysis of leaf samples taken from the same leaves as those shown in Figure 5, A through C, to determine whether there were differences in the morphology of chloroplasts formed in the partially complemented mutant. We found that the chloroplasts in wild-type leaves did not show any ultrastructure abnormalities, as shown by the proper stacking of the grana layers and the apparent formation of starch (Fig. 5D). On the other hand, the chloroplasts in the yellowish section of pale-green leaves (shown in Fig. 4B) were smaller (see sample in Fig. 5E). Although there was some evidence of grana stacking, this was not to the same degree as what we observed for a normal chloroplast. The chloroplasts from the greenish region, as shown in Figure 5C, showed normal grana stacking to the same extent as in wild-type leaves (Fig. 5F).
Taken together, our partial complementation analyses suggest that the EcoRI fragment from the RSY3 locus (see Fig. 2A) is sufficient to restore the normal morphology of rsy3 embryos, but is not sufficient to rescue the full green color of the resulting seedling or plant.
Molecular Analysis of Partially Complemented rsy3
To determine the molecular basis of the partial complementation, we performed several analyses. To rule out the possibility that the pale-green embryo phenotype was due to the T-DNA insertion in plants carrying the tE989 transgene, we analyzed the parental transformants that were homozygous for the transgene (i.e. RSY3/RSY3;tE989/tE989) and found that they produced normal embryos (data not shown). To determine that the transgene was present in these partially rescued rsy3 segregants, we performed genomic DNA analysis. Our results showed that the predicted 2.5-kb NotI fragment from the construct (Fig. 6A) was present in the partially complemented F2 individuals with the transgene tE989 in a hemizygous state (e.g. individual no. 5) or in a homozygous state (e.g. individuals nos. 1 and 7). The NotI fragment was absent in a wild-type line and in an uncomplemented heterozygous rsy3 line (Fig. 6B), both of which were expected not to contain the NotI sites in their wild-type genomic fragment.
Figure 6.
Molecular analysis of partially rescued rsy3 plants. A, Diagram of the construct used in the partial complementation. The EcoRI fragment (see Fig. 2A) was blunt-end ligated into the NotI site of the T-DNA vector pGHS166N (see “Materials and Methods”). P2 is the mannopine synthase promoter contained within pGSH166N vector. B, Genomic DNA restriction analysis of some F2 segregants generated from the complementation cross between rsy3/RSY3 and rsy3/RSY3;tE989 lines. Two separate sets of genomic DNAs were digested with EcoRI or NotI and were size fractionated by electrophoresis in a 1% (w/v) agarose gel; the resulting blots were hybridized with the 2.5-kb EcoRI fragment of the RSY3 genomic clone (see Fig. 2A). C, RNA analysis of RSY3 transcripts in different tissues of wild-type (left panel) and in seedlings generated from partially complemented rsy3 mutant (rsy3/rsy3;tE989) plants (right panel). The length of exposure for the left panel was 3 d, and for the right panel was 1 d. D, Portion of the cDNA sequence generated from the reverse transcriptase-PCR analysis using mRNA samples from partially rescued rsy3 mutant (rsy3/rsy3;tE989) plants (see “Materials and Methods”). The series of dots (…) represents internal sequences identical to the sequence shown in Figure 3A (see also accession no. AY077630).
To determine that the transgene was transcribed in the partially complemented plants, we performed RNA-blot analysis (see “Materials and Methods”). The insert fragment was suspected to be driven by the mannopine synthase P2 promoter contained upstream of the NotI site of the pGSH166N vector (see diagram in Fig. 6A; Fox et al., 1992; Kim and Farrand, 1996). Total RNAs were isolated from green (as shown in Fig. 4E) and pale-green seedlings (as shown in Fig. 4F), which contained the tE989 transgene. Our RNA-blot hybridization analysis showed that these seedlings transcribed the tE989 transgene (Fig. 6C, right panel). The level of tE989 transcripts in partially rescued seedlings was higher than the level of the endogenous RSY3 transcripts present in wild-type seedlings (Fig. 6C, right panel). The endogenous RSY3 mRNA was barely detectable when total RNA was used (Fig. 6C, wild-type lane), but was enhanced when poly(A)+ mRNAs were used in the RNA-blot analysis (Fig. 6C, left panel).
To determine whether the transgene was properly transcribed in the partially complemented rsy3 (rsy3/rsy3;tE989 or rsy3/rsy3;tE989/tE989) line, we performed 5′- and 3′-RACE analyses and subsequently sequenced the resulting RACE clones (see “Materials and Methods”). We found that tE989 transgene transcripts contained nucleotides due to the transcription of a portion of the T-DNA vector upstream and downstream of the EcoRI insert (Fig. 6D, highlighted in black letters). In addition, a portion of exon 2 with the canonical nucleotides GT in the 5′ end and AG in the 3′ end was cryptically spliced out of the transcripts (Fig. 6D, highlighted with a triangle). All other sequences were identical to the wild-type cDNA (see Fig. 3A). However, the truncated RSY3 protein, predicted from the tE989 transcript resulting from truncation of exon 1 in the tE989 fragment and from the cryptic splicing of exon 2 region, would still contain the three identified domains as shown in Figure 3B. Taken together, these results indicate that an RSY3 protein missing the N terminus encoded by exons 1 and 2 of the RSY3 gene can complement the embryo phenotype, but can lead to mosaic green plants with defective and normal chloroplasts.
DISCUSSION
In this study, we describe the mutant rsy3 that is defective in embryo development. rsy3 remains morphologically arrested at the globular stage and fails to differentiate cotyledons and axis. The rsy3 embryo-proper resembles that of a wild-type embryo at the globular stage and has raspberry-like protuberances. By contrast, the rsy3 suspensor is enlarged compared with that in wild-type embryos (Fig. 1). We previously demonstrated that rsy1 and rsy2 embryos fail to undergo morphogenesis, but that their embryo-proper cells undergo a normal cell differentiation pathway (Yadegari et al., 1994). The rsy1 and rsy2 suspensors, on the other hand, enter an embryogenic pathway. Because the phenotype of rsy3 embryos is similar to that of rsy1 and rsy2 (Yadegari et al., 1994), the rsy3 suspensor probably enters an embryogenic pathway as well and the embryo-proper cells continue to differentiate like wild-type embryos (Fig. 1). Here, we demonstrate by molecular and genetic complementation analyses that the embryo defects in rsy3 are caused by a T-DNA insertion in a novel gene that has not been described previously (Figs. 2 and 3). Our experiments suggest that the RSY3 protein is localized in the chloroplast and that the defect in rsy3 embryos is caused indirectly by a failure to produce normal chloroplasts during embryo development.
RSY3 Is a Novel Protein
The precise function of the RSY3 protein is unknown; however, the RSY3 protein contains features that suggest that it is localized within the chloroplast and is important for chloroplast differentiation. First, analysis of the 50 amino-terminal amino acids of the RSY3 protein indicates that it has a putative transit peptide specific for importing proteins into the chloroplasts (Hand et al., 1989; Ko and Cashmore, 1989; Archer and Keegstra, 1993; Rolland et al., 1993). The presence of Ala as the second amino acid and the positively charged Lys and Arg residues are features of proteins imported by chloroplast (Hand et al., 1989; Ko and Cashmore, 1989; see Fig. 3A). Second, the presence of the SNGRKS motif in its amino terminus is similar to the motif identified in the carboxy terminus of the chloroplast-localized Rubisco small-subunit protein (Archer and Keegstra, 1993). Third, our P-sort analysis (Nakai, 2000) shows that RSY3 has a 75% probability of being localized in the chloroplasts. Although all of the above are computer predictions, the results of our partial complementation analyses (Figs. 4–6) support the possibility that the RSY3 protein is localized in the chloroplast and that it is required for chloroplast development. We observed that a partial RSY3 gene that excludes portions of the first and second exons gives rise to pale-green plants (Fig. 4E) with leaf chloroplasts that are not fully developed (Fig. 5E).
Other domains found in the RSY3 protein suggest a possible function. There are three distinct domains, designated as domains I, II, and III (Fig. 3B), that are characteristic of other known proteins or proteins predicted from the ORFs of different sequenced genomes. Domain I has a conserved core sequence, SGG-DS, that is a signature motif for a P-loop domain present in a number of enzymes that have “N-type” ATP pyrophosphatase activity (Tesmer et al., 1996). These enzymes include NAD synthetase, Arg synthetase, Gln synthetase, and arginosuccinate synthetase (Tesmer et al., 1996). The general reaction catalyzed by these synthetases includes the activation of carboxyl or carbonyl groups by adenylation, resulting in an adenylated intermediate that is reactive to a nitrogen nucleophile (Tesmer et al., 1996). Using domain I as a clue, it is tempting to speculate that the RSY3 protein is capable of ATP binding and that ATP hydrolysis is one of its functional activities. How this occurs and what pathway within the chloroplast uses the RSY3-mediated ATP hydrolysis are questions that remain to be answered.
RSY3 Protein and Chloroplast and Embryo Development
If the RSY3 protein is localized in the chloroplast, what is its role in chloroplast and embryo development? Chloroplasts develop from progenitor proplastids contained in the zygote as maternally inherited organelles (Schulz and Jensen, 1968; Kirk and Tilney-Bassett, 1978; Mansfield and Briarty, 1991). It is possible that early in embryo development, the chloroplast may synthesize biosynthetic products that are required directly or indirectly by the embryo to initiate and undergo morphogenesis. For example, fatty acids are synthesized in the chloroplast as well as precursors for the plastid-dependent synthesis of isoprenoid geranylgeranyl diphosphate, which is a key substrate for gibberellin biosynthesis (Hedden and Kamiya, 1997). A defect in chloroplast differentiation during early embryo formation may prevent the formation of important biosynthetic precursors that are required in subsequent metabolic steps for the production of embryo signaling molecules. Absence of these signaling molecules might then result in a mutant embryo with a rsy3 phenotype.
The rsy3 embryo phenotype (Fig. 1) and the defective chloroplasts in the partially rescued rsy3 plants (Fig. 5) are consistent with the view that RSY3 may be one of the components that are required during early chloroplast development. Mutations in genes encoding chloroplast ribosomal protein S16 (Tsugeki et al., 1996), chloroplast ribosomal protein S1 (Yadegari, 1996), chloroplast-localized EMB506 (Albert et al., 1999; Despres et al., 2001), and plastid glycyl-tRNA-synthetase (Uwer et al., 1998) have phenotypes similar to that of rsy3. This suggests that the proteins encoded by these genes, acting in different plastid processes, are required within the same developmental timeframe of embryo chloroplast biogenesis, and are prerequisites for the synthesis of biosynthetic products leading to embryo signaling molecules. By contrast, SCHLEPPERLESS (Apuya et al., 2001), a gene that encodes a chaperonin-60α-subunit, may be required in processes that occur later in chloroplast biogenesis. Mutations in these genes lead to defective embryos that are morphologically at a more advanced stage than the rsy3 embryos. Although the plastids in schlepperless embryos are abnormal (Apuya et al., 2001), they have probably differentiated to a greater extent than those in rsy3 embryos and have the capacity to synthesize precursor molecules necessary for normal morphogenesis to occur. Not all mutations that affect chloroplast development lead to embryo arrest or lethality. For example, albino mutants like cla1 (Mandel et al., 1996) and albino3 (Sundberg et al., 1997) produce defective chloroplasts within embryos that have wild-type morphology. The products of these genes may be required for other aspects of plastid function (e.g. chlorophyll formation) after the chloroplasts have differentiated and are competent to synthesize products essential for embryo morphogenesis.
The results of our partial complementation analysis indicate that a partial RSY3 protein is able to rescue the morphological defect of rsy3 mutant embryos (Figs. 4 and 5). The truncation in the partial RSY3 transgene (Fig. 2A, referred as fragment tE989; see also Fig. 6A) removes most of the chloroplast transit peptide predicted for RSY3 (Fig. 3A). In addition, a cryptic splicing within the second exon (Fig. 6D) further truncates the predicted protein encoded by the transgene. These data suggest that the remaining regions (including the putative ATP-binding region in domain I; see Fig. 3B) within the predicted truncated RSY3 protein are partly active and capable of rescuing embryogenesis. How might this occur? The partially complemented embryos are pale green (Fig. 4B) and probably contain a mix of abnormal and fully developed chloroplasts similar to those that we observed within leaves (Fig. 5E). If so, it is possible that truncation of the chloroplast transit peptide causes inefficient targeting of the partial RSY3 protein to embryo chloroplasts. Although inefficient, perhaps a sufficient number of embryo proplastids import the truncated RSY3 protein to undergo differentiation. This will allow the differentiated chloroplasts to produce enough biosynthetic precursors of signaling molecules so that rescued rsy3 embryos can undergo normal morphogenesis. However, the accumulation of sufficient precursors to reach the critical threshold may take longer, which might explain the slower development of partially rescued embryos and seedlings (Fig. 4, B and E).
What about the phenotype of the partially rescued rsy3 plants? Following germination of rescued rsy3 seeds, the accumulation of truncated RSY3 in a subset of chloroplasts might allow these chloroplasts to develop to the point of being able to assemble a functional photosynthetic apparatus. If so, this would explain the random appearance of developed and undeveloped chloroplasts that we observe in the pale-green leaves of partially rescued rsy3 plants (Figs. 4F and 5F).
In conclusion, our results suggest that the RSY3 gene is required for embryo development and for normal development of the chloroplast. RSY3 and many other genes may be involved in a series of chloroplast-mediated developmental processes that are required for embryogenesis. How this occurs and the precise function of RSY3 remain to be determined.
MATERIALS AND METHODS
Mutant Isolation and Genetic Analysis
The rsy3 embryo mutant corresponds to line A989, one of the 5,822 T-DNA-mutagenized lines of Arabidopsis ecotype WS that was screened at the DuPont Experimental Station (Wilmington, DE) in November 1990 and at the University of Arizona (Tucson, AZ) in November 1991 (Feldmann and Marks, 1987; Errampalli et al., 1991; Feldmann, 1991; Castle et al., 1993; Yadegari et al., 1994). The recessive embryo-defective mutation was maintained in heterozygous plants (rsy3/RSY3), which produced wild-type and mutant seeds in a 3:1 ratio. The cosegregation analysis of T-DNA and the embryo-defective phenotype, and the mapping of the chromosomal location of the T-DNA insertion were done following the procedures outlined by Apuya et al. (2001).
Seed Germination in Tissue Culture
For experiments that required seed sterilization for aseptic germination, the procedures outlined by Apuya et al. (2001) were followed. For Kan-R and/or Hyg-R assay, seeds were germinated in Murashige and Skoog germination medium in the presence of 50 μg mL−1 kanamycin sulfate and/or 20 μg mL−1 hygromycin. Wild-type and partially complemented seedlings used as source of RNA samples were allowed to grow in Murashige and Skoog plates for about 3 weeks prior to harvest at standard conditions in an incubator (I-60LLVL; Percival Scientific, Perry, IA).
Microscopy
Bright-Field Microscopy
The procedures of Yadegari et al. (1994) for fixation and embedding of plant tissue samples in paraffin were followed for the preparation of 3-mm leaf samples collected from heterozygous and partially complemented mutant plants. The embedded samples were sectioned (5 μm thick) using a microtome and were appropriately placed on microscope slides. Sections were hydrated after removal of the paraffin and were subsequently stained with 0.5% (w/v) toluidine blue in 0.1% (w/v) borate solution. Bright-field photographs were taken with film (Gold 100; Eastman-Kodak, Rochester, NY; ISO 100/21°) using a compound microscope (Olympus BH-2; Olympus, Lake Success, NY).
Nomarski Microscopy
Mutant and wild-type seeds were fixed in ethanol:acetic acid (9:1) solution overnight and were successively washed in 90% and 70% (v/v) ethanol for at least 30 min each. Seeds were cleared with 72.7% (w/v) chloral hydrate in 50% (w/v) glycerol for at least 2 h prior to microscopy (Berleth and Jürgens, 1993). Embryos were visualized using Nomarski optics on a Zeiss Axiophot (Carl Zeiss, Oberkochlen, Germany). Photographs were taken using TMAX 100 (E.I. 100/21°) film (Eastman-Kodak).
TEM
The procedures of Yadegari et al. (1994) were followed except that L.R. White plastic resin was used as the embedding medium.
Whole-Mount Photography
Bright-field and dark-field photographs of dissected embryos and germinating seedlings in culture and in soil were taken using Olympus SZH (Olympus) and StemiSV11-Apo (Carl Zeiss) dissecting microscopes.
Genomic DNA Isolation, Restriction Analysis, DNA Blotting, and Labeling
Materials and procedures related to genomic DNA isolation, restriction analysis, and Southern-blot analysis were those of Apuya et al. (2001).
Isolation of Mutant and Wild-Type Genomic Clones
Plasmid rescue was done using the protocol of Behringer and Medford (1992). Procedures followed to isolate wild-type and mutant genomic clones are cited by Apuya et al. (2001) using rescued plasmid p989-E45 (Fig. 2B) and pC989–41 cDNA (Fig. 2A) as probes.
Isolation of cDNA Clones
A λZAP cDNA library, constructed from poly(A)+ mRNA from wild-type Arabidopsis siliques, was used to isolate RSY3 cDNA clones. The procedures of Apuya et al. (2001) were followed. The plasmid p989-E45, isolated using the plasmid rescue, was used as a probe for the cDNA library screening. 5′- and 3′-RACE (Frohman, 1993) was used to isolate cDNA clones corresponding to RSY3 and to tE989 transgene from partially rescued mutant lines (Fig. 6) using the Marathon cDNA Amplification kit following the manufacturer's recommendations (CLONTECH Laboratories, Palo Alto, CA).
DNA Sequencing
The sequencing of cDNA clones and subclones of rescued plasmids and phage genomic clones was carried out following the dideoxy-sequencing procedures recommended by United States Biochemicals (Cleveland). Sequence analysis was done using the Genetics Computer Group (Madison, WI) software, the NCBI BLAST e-mail server, and the Vector NTI software package (InforMax, Bethesda, MD).
RNA Techniques
Polysomal RNAs from different tissues used for RNA-blot analysis were isolated according to the procedures described by Cox and Goldberg (1988). Total RNAs from transgenic seedlings used for RNA-blot analysis were isolated using the Qiagen Extraction kit (Qiagen, Ventura, CA) following the manufacturer's recommendations. Poly(A)+ mRNAs, used for RNA-blot and RACE analyses, were isolated using the Poly-AT Tract mRNA Isolation System (Promega, Madison, WI) following the recommended protocol by the manufacturer. The isolated mRNAs were separated by size using formaldehyde-agarose gel electrophoresis, transferred to a Nytran membrane (Schleicher & Schuell, Keene, NH), and hybridized with 32P-labeled DNA according to the procedures recommended by Ausubel et al. (1992).
Plant Transformation and Complementation Analysis
The 9-kb HindIII genomic fragment (designated as tH989 in Fig. 2A) was ligated with HindIII-Not linker, and the 5-kb-NcoI/HindIII fragment (designated as tNH989) was blunt-ended by Klenow treatment prior to subcloning into the NotI site of pHYG-A vector (Honma et al., 1993; Klucher et al., 1996). Fragment tE989 was ligated with EcoRI-NotI linker prior to subcloning into the NotI site of pGSH166N vector (courtesy of Plant Genetic Systems, Gent, Belgium). The resulting recombinants were subsequently transferred to Agrobacterium tumefaciens to be used for root transformation. The original procedures established by Valvekens et al. (1988) for A. tumefaciens-mediated transformation of Arabidopsis root explants were followed. Wild-type Arabidopsis (ecotypes WS, C24, and Nossen) were used as recipients. The procedures and the strategies for genetic and molecular analyses outlined by (Apuya et al., 2001) were followed for rsy3 gene complementation.
Table 1.
Summary of P2 segregation for kanamycin and hygromycin resistance resulting from one of the complementation crosses between raspberry3 mutant and transformant containing the tE989 fragment (see map in Fig. 2A)
| Class of Seedlingsa | Genotypes of the Classb | Nos. Observed (n = 609) | Observed Frequency | Expected Frequency |
|---|---|---|---|---|
| Kan-R/Hygro-R (green seedlings) | rsy3/RSY3;tE989 (4) | |||
| rsy3/RSY3;tE989/tE989 (2) | 259 | 42.5% | 37.5% | |
| Kan-R/Hygro-R (pale-green seedlings) | rsy3/rsy3; tE989 (2) | |||
| rsy3/rsy3;tE989/tE989 (1) | 114 | 18.7% | 18.75% | |
| Kan-S/Hygro-R | RSY3/RSY3;tE989 (2) | |||
| RSY3/RSY3;tE989/tE989 (1) | 92 | 15.1% | 18.75% | |
| Kan-S/Hygro-S | RSY3/RSY3 (1) | |||
| Kan-R/Hygro-S | rsy3/RSY3 (2) | 116 | 19.1% | 18.75% |
| Aborted seeds (not germinating) | rsy3/rsy3 (1) | 28 | 4.6% | 6.25% |
Chi-square analysis: calculated χ2 = 5.85 < tabular χ2 = 9.49 (at 0.05 level at degrees of freedom = 4).
See “Materials and Methods” for testing for kanamycin and hygromycin resistance.
Nos. in parentheses indicate the no. of individuals out of 16 expected progenies to have the particular genotypes.
ACKNOWLEDGMENTS
We would like to acknowledge Birgitta Sjostrand (University of California, Los Angeles) for help with electron microscopy. We would like to thank Ken Feldmann (Ceres Inc.) for allowing us to screen the T-DNA mutants while he was at the University of Arizona and for helpful suggestions regarding the manuscript. We also would like to acknowledge Shing Kwok (Ceres Inc.) for critical reading of the manuscript. We extend our gratitude to all the individuals within our Seed Institute collaboration for incisive discussion and help in carrying out this research.
Footnotes
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.004010.
LITERATURE CITED
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Despres B, Guilleminot J, Bechtold N, Pelletier G, Delseny M, Devic M. The EMB 506 gene encodes a novel ankyrin repeat containing protein that is essential for the normal development of Arabidopsis embryos. Plant J. 1999;17:169–179. doi: 10.1046/j.1365-313x.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- Altmann T, Felix G, Jessop A, Kauschmann A, Uwer U, Peña-Cortés H, Willmitzer L. Ac/Ds transposon mutagenesis in Arabidopsis thaliana: mutant spectrum and frequency of Ds insertion mutants. Mol Gen Genet. 1995;247:646–652. doi: 10.1007/BF00290357. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB. The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiol. 2001;126:717–730. doi: 10.1104/pp.126.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer EK, Keegstra K. Analysis of chloroplast transit peptide function using mutations in the carboxyl-terminal region. Plant Mol Biol. 1993;23:1105–1115. doi: 10.1007/BF00042345. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocol in Molecular Biology. New York: Greene Publishing and Wiley-Interscience; 1992. [Google Scholar]
- Behringer FJ, Medford JI. A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol Biol Rep. 1992;10:190–198. [Google Scholar]
- Berleth T, Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- Castle LA, Errampalli D, Atherton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1988. pp. 1–34. [Google Scholar]
- Despres B, Delseny M, Devic M. Partial complementation of embryo defective mutations: a general strategy to elucidate gene function. Plant J. 2001;27:149–159. doi: 10.1046/j.1365-313x.2001.01078.x. [DOI] [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M. Induction and expression of seed-specific promoters in Arabidopsis embryo-defective mutants. Plant J. 1996;9:205–215. doi: 10.1046/j.1365-313x.1996.09020205.x. [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke DW. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell. 1991;3:149–157. doi: 10.1105/tpc.3.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann K. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Feldmann K, Marks MD. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis: a non-tissue culture approach. Mol Gen Genet. 1987;208:1–9. [Google Scholar]
- Fox PC, Vasil V, Vasil IK, Gurley WB. Multiple ocs-like elements required for efficient transcription of the mannopine synthase gene of T-DNA in maize protoplasts. Plant Mol Biol. 1992;20:219–233. doi: 10.1007/BF00014490. [DOI] [PubMed] [Google Scholar]
- Frohman MA. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L. Regulation of gene expression during plant embryogenesis. Cell. 1989;56:149–160. doi: 10.1016/0092-8674(89)90888-x. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jürgens G. A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand JM, Szabo LJ, Vasconcelos AC, Cashmore AR. The transit peptide of a chloroplast thylakoid membrane protein is functionally equivalent to a stromal-targeting sequence. EMBO J. 1989;8:3195–3206. doi: 10.1002/j.1460-2075.1989.tb08478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Honma MA, Baker BJ, Waddell CS. High-frequency germinal transposition of DsALS in Arabidopsis. Proc Natl Acad Sci USA. 1993;90:6242–6246. doi: 10.1073/pnas.90.13.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Axis formation in plant embryogenesis: cues and clues. Cell. 1995;81:467–470. doi: 10.1016/0092-8674(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard JBL, editor. Embryos, Color Atlas of Development. London: Wolfe Publishing; 1994. pp. 7–21. [Google Scholar]
- Jürgens G, Mayer U, Busch M, Lukowitz W, Laux T. Pattern formation in the Arabidopsis embryo: a genetic perspective. Philos Trans R Soc Lond B Biol Sci. 1995;350:19–25. doi: 10.1098/rstb.1995.0132. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Mayer U, Torres Ruiz RA, Berleth T, Misera S. Genetic analysis of pattern formation in the Arabidopsis embryo. Development Suppl. 1991;1:27–38. [Google Scholar]
- Kim K, Farrand S. Ti plasmid-encoded genes responsible for catabolism of the crown gall opine mannopine by Agrobacterium tumefaciensare homologs of the T-region genes responsible for synthesis of this opine by the plant tumor. J Bacteriol. 1996;178:3275–3284. doi: 10.1128/jb.178.11.3275-3284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk JTO, Tilney-Bassett RAE. The Plastids: Their Chemistry, Structure, Growth, and Inheritance. Ed 2. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1978. [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K, Cashmore AR. Targeting of proteins to the thylakoid lumen by the bipartite transit peptide of the 33-kD oxygen-evolving protein. EMBO J. 1989;8:3187–3194. doi: 10.1002/j.1460-2075.1989.tb08477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Thomas TL. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell. 1998;10:383–398. doi: 10.1105/tpc.10.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana: the developing embryo. Can J Bot. 1991;69:461–476. [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Misera S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA et al. Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics. 2001;159:1751–1763. doi: 10.1093/genetics/159.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K. Protein sorting signals and prediction of subcellular localization. Adv Protein Chem. 2000;54:277–344. doi: 10.1016/s0065-3233(00)54009-1. [DOI] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 1998;116:935–946. doi: 10.1104/pp.116.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland N, Job D, Douce R. Common sequence motifs coding for higher-plant and prokaryotic O-acetylserine (thiol)-lyases: bacterial origin of a chloroplast transit peptide? Biochem J. 1993;293(Pt 3):829–833. doi: 10.1042/bj2930829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K, Mayer U, Horrichs A, Kuhnt C, Bellini C, Dangl J, Schmidt J, Jurgens G. FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 2000;14:1471–1484. [PMC free article] [PubMed] [Google Scholar]
- Schulz SR, Jensen WA. Capsella embryogenesis: the egg, zygote, and young embryo. Am J Bot. 1968;55:807–819. [Google Scholar]
- Schwartz B, Yeung E, Meinke D. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Dev Suppl. 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- Shevell DE, Kunkel T, Chua NH. Cell wall alterations in the Arabidopsis emb30 mutant. Plant Cell. 2000;12:2047–2060. doi: 10.1105/tpc.12.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH. EMB30is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Klem TJ, Deras ML, Davisson VJ, Smith JL. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat Struct Biol. 1996;3:74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
- Tiedeman AA, Smith JM, Zalkin H. Nucleotide sequence of the guaA gene encoding GMP synthetase of Escherichia coliK12. J Biol Chem. 1985;260:8676–8679. [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell. 1998;10:1277–1294. doi: 10.1105/tpc.10.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis root explants using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Harada JJ. Embryogenesis in higher plants: an overview. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R. Regional specification and cellular differentiation during early plant embryogenesis. PhD thesis. Los Angeles: University of California; 1996. [Google Scholar]
- Yadegari R, de Paiva G, Laux T, Koltunow AM, Apuya NR, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell. 1994;6:1713–1729. doi: 10.1105/tpc.6.12.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalkin H, Argos P, Narayana SV, Tiedeman AA, Smith JM. Identification of a trpG-related glutamine amide transfer domain in Escherichia coliGMP synthetase. J Biol Chem. 1985;260:3350–3354. [PubMed] [Google Scholar]