Abstract
AtWRKY18 is a pathogen- and salicylic acid-induced Arabidopsis transcription factor containing the plant-specific WRKY zinc finger DNA-binding motif. In the present study, we have transformed Arabidopsis plants with AtWRKY18 under control of the cauliflower mosaic virus 35S promoter. Surprisingly, transgenic plants expressing high levels of AtWRKY18 were stunted in growth. When expressed at moderate levels, AtWRKY18 potentiated developmentally regulated defense responses in transgenic plants without causing substantial negative effects on plant growth. As they grew from seedling to mature stages, transgenic AtWRKY18 plant showed marked increase in the expression of pathogenesis-related genes and resistance to the bacterial pathogen Pseudomonas syringae, whereas wild-type plants exhibited little enhancement in these defense responses. Potentiation of developmentally regulated defense responses by AtWRKY18 was not associated with enhanced biosynthesis of salicylic acid but required the disease resistance regulatory protein NPR1/NIM1. Thus, AtWRKY18 can positively modulate defense-related gene expression and disease resistance. To study the regulated expression of AtWRKY18, we have identified a cluster of WRKY binding sites in the promoter of the gene and demonstrated that they acted as negative regulatory elements for the inducible expression of AtWRKY18. These negative cis-acting elements may prevent overexpression of AtWRKY18 during the activation of plant defense responses that could be detrimental to plant growth as inferred from the transgenic plants ectopically expressing the transgene.
Like many other complex biological processes, plant defense responses upon pathogen infection involve transcriptional regulation of a large number of plant host genes (Rushton and Somssich, 1998). Many of these differentially regulated genes encode enzymes in a variety of primary and secondary metabolic pathways and the change of their synthesis may result in reprogramming of cellular metabolism. Some of the differentially induced genes encode pathogenesis-related (PR) proteins including antimicrobial chitinases and glucanases that are capable of degrading cell wall components of microbial pathogens. Other differentially regulated plant host genes encode regulatory factors that are involved in the activation, suppression, and modulation of various signaling pathways in plant cells upon pathogen infection. Thus, transcriptional regulation of plant host genes is an integral part of plant defense responses with a critical role in induced plant disease resistance.
Transcriptional regulation of gene expression is mediated largely by the change in the level and/or activity of sequence-specific DNA-binding transcription factors. To understand the molecular mechanisms by which plant host genes are transcriptionally regulated during plant defense responses, we have been studying a novel class of DNA-binding factors containing WRKY domains (Wang et al., 1998; Yang et al., 1999; Chen and Chen, 2000; Du and Chen, 2000; Yu et al., 2001). WRKY domains are defined by the conserved amino acid sequence WRKYGQK at their N-terminal ends and a novel zinc finger motif at the C termini (Eulgem et al., 2000). WRKY proteins have been identified from a number of plants and appear to be encoded by a large gene family, with more than 70 members in Arabidopsis (Eulgem et al., 2000; Z. Chen, unpublished data).
A number of studies have shown that WRKY proteins have regulatory functions in plant response to pathogen infection. First, several WRKY genes from a number of plants are rapidly induced by pathogens, pathogen elicitors, or treatment of salicylic acid (SA) (Eulgem et al., 1999; Chen and Chen, 2000; Dellagi et al., 2000; Hara et al., 2000; Kim et al., 2000). Second, a number of defense-related genes, including the well-studied PR genes, contain W box elements in their promoter regions (Rushton et al., 1996; Yang et al., 1999). A number of studies have shown that these W box sequences are specifically recognized by WRKY proteins and are necessary for the inducible expression of these genes (Rushton et al., 1996; Yang et al., 1999). More recently, we have shown that pathogen- and SA-induced WRKY proteins also regulate genes encoding proteins with regulatory functions (e.g. receptor protein kinases and NPR1; Du and Chen, 2000; Yu et al., 2001). The involvement of WRKY protein in regulating plant defense responses is further substantiated by a recent study of gene expression changes in Arabidopsis under 14 different systemic acquired resistance (SAR)-inducing or SAR-repressing conditions using a DNA microarray with 10,000 expression sequence tags (Maleck et al., 2000). A group of 26 genes including PR-1 was identified to be coordinately induced by various pathogens and defense-inducing conditions. Within the 1.1-kb regions upstream of the predicated translation start sites, only the binding site for WRKY proteins (W boxes; TTGAC) were found in all 26 promoters. Although these 26 genes contained an average of 4.3 copies of the W box per promoter that are often organized in clusters, a randomly selected set of genes contained, on average, fewer than two W boxes per promoter (Maleck et al., 2000).
Although there is a large body of indirect evidence that implicates WRKY proteins in the regulation of defense-related genes, no reported study has provided direct evidence for important roles of individual WRKY proteins in plant defense. The direct evidence may come from “knockout” or gene-silencing mutants for individual WRKY genes. This loss-of-function approach, however, has a major disadvantage for genes that belong to large gene families with possible overlapping functions. For these genes, it may be necessary to disrupt multiple genes to produce an altered phenotype. An alternative approach is to use gain-of-function mutants to infer biological functions of a gene. In plants, gain-of-function mutants are often generated through overexpression of a gene in transgenic plants. The biological functions of a gene inferred from the phenotypes of overexpression experiments sometimes may not be consistent with those determined from loss-of-function mutants and this has often raised the issue about the value of overexpression experiments in determining the biological functions of a gene. The discrepancy means that the functions of a gene are not unchanged; instead, they are variable, depending on their expression levels and/or biological context. The loss-of-function approaches reveal the functions of a gene when it is expressed at the patterns found in wild-type plants, whereas the overexpression reveals potential functions of the gene when it is expressed at the patterns found in transgenic lines. The dynamic functions of a gene associated with its expression levels might play an important role during natural evolution of important biological traits. For example, alterations in tomato (Lycopersicon esculentum) fruit size, imparted by the fw2.2 quantitative trait locus, are most likely due to changes in expression regulation rather than in the sequence and structure of the encoded protein key to the evolution of fruit size (Frary et al., 2000). In addition, one of the major goals of fundamental plant biological research is to develop necessary knowledge and tools to improve agronomic traits of crop plants. To do so, we must also explore the potential functions of a gene associated with the expression levels or biological context not normally found in wild-type plants. There are obviously caveats associated with the approach of overexpression for functional analysis of a gene, particularly when the gene that is highly overexpressed could lead to altered phenotypes that are grossly pleiotropic but not necessarily biologically significant.
In the present study, we take a gain-of-function approach to analyze the function of one of the SA-induced WRKY genes, AtWRKY18, through constitutive expression in transgenic plants. Analysis of the transgenic plants showed that AtWRKY18 was able to potentiate developmentally regulated PR gene expression and resistance to the bacterial pathogen Pseudomonas syringae. When expressed at high levels, however, AtWRKY18 also caused severe abnormality in plant growth. These results suggest that properly regulated expression of AtWRKY18 is important for enhancing plant defense response without severe detrimental effects on plant growth. A cluster of WRKY binding sites (W boxes) in the promoter of AtWRKY18 act as negative regulatory elements for its inducible expression and, therefore, may play a role in preventing overexpression of the gene during the activation of plant defense response.
RESULTS
Transgenic Plants Constitutively Expressing AtWRKY18
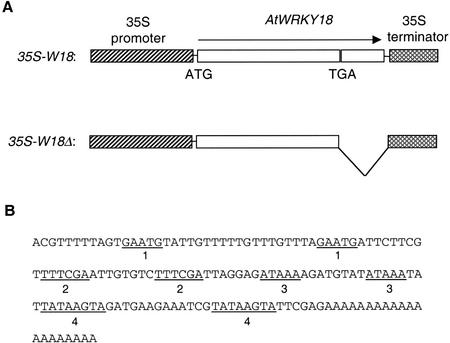
AtWRKY18 was the first WRKY gene identified in our search for SA-induced WRKY genes in Arabidopsis (Yu et al., 2001). It is rapidly induced in Arabidopsis plants upon treatment with SA (Yu et al., 2001). To determine the biological impacts of its induced expression, we attempted to express AtWRKY18 constitutively in transgenic Arabidopsis plants. A cDNA with both the full-length coding region and the 3′-untranslated region of AtWRKY18 was placed behind the cauliflower mosaic virus 35S promoter (35S-W18; Fig. 1A) and transformed into Arabidopsis. An examination of the 3′-untranslated region of AtWRKY18 revealed the presence of an unusually large number of short direct repeats (Fig. 1B). A second construct was made, therefore, that contained only the coding region of the cDNA clone behind the 35S promoter (35S-W18Δ, Fig. 1A) to determine whether these repeats affected gene expression in Arabidopsis.
Figure 1.
Overexpression constructs of AtWRKY18. A, Schematic diagrams of the construct 35S-W18 (the full-length AtWRKY18 cDNA sequence placed between the cauliflower mosaic virus 35 promoter and 35S terminator) and the construct 35S-W18Δ (the 3′-untranslated sequence of AtWRKY18 deleted). B, Sequence of the AtWRKY18 3′-untranslated region. The four direct repeats are highlighted (underlined).
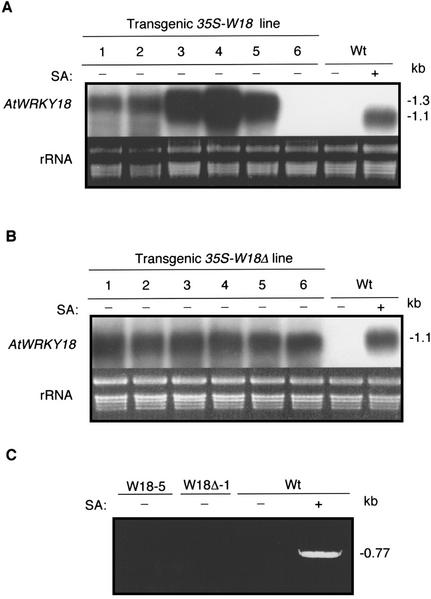
Northern blotting showed a substantial variation in the level of AtWRKY18 transcripts in the transgenic plants harboring the 35S-W18 construct (Fig. 2A). Among the six lines examined, two expressed AtWRKY18 at the levels similar to that detected in SA-treated wild-type plants, three at levels substantially higher than the physiological levels of the endogenous gene, and one expressed no detectable AtWRKY18 transcripts (Fig. 2B). In contrast, the transformants harboring the 35S-W18Δ construct expressed the transgene rather uniformly, at levels similar to that of the endogenous AtWRKY18 in SA-treated control plants (Fig. 2B). Thus, although inclusion of the 3′-untranslated sequence of AtWRKY18 could increase the overall level of transgene expression, it also appeared to increase the variability among independent transformants.
Figure 2.
Expression of AtWRKY18 in transgenic plants. A, Total RNA was isolated from 4-week-old wild-type plants (Wt) with (+) or without (−) SA treatment (2 mm for 3 h) and untreated T3 progeny of transgenic lines harboring the 35S-W18 construct, separated on a 1.2% (w/v) agarose-formaldehyde gel, and probed with an AtWRKY18 cDNA fragment. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading. B, Total RNA was isolated from 4-week-old wild-type plants (Wt) with (+) or without (−) SA treatment (2 mm for 3 h) and untreated T3 progeny of transgenic lines harboring the 35S-W18Δ construct, separated on a 1.2% (w/v) agarose-formaldehyde gel, and probed with an AtWRKY18 cDNA fragment. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading. C, Total RNA was isolated from 4-week-old wild-type plants (Wt) with (+) or without (−) SA treatment (2 mm for 3 h) and untreated T3 progeny of two transgenic lines. Reverse transcription and PCR were performed as described in “Materials and Methods.”
To distinguish expression between the endogenous gene and transgene of AtWRKY18, we performed reverse transcriptase (RT)-PCR using a pair of AtWRKY18-specific primers. One of the primers corresponds to a 5′-transcribed but untranslated region of AtWRKY18 that is not present in the AtWRKY18 transgene and, as a result, RT-PCR will detect expression of only the endogenous AtWRKY18 gene. As shown in Figure 2C, RT-PC detected expression of the endogenous AtWRKY18 gene in SA-treated wild-type plants, but not in untreated wild-type or transgenic plants. Thus, the transcripts of AtWRKY18 in the transgenic plants detected by northern blotting (Fig. 2, A and B) resulted from expression of the introduced transgene of AtWRKY18.
Growth and Morphology of Transgenic Plants
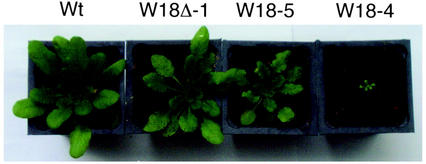
As with the levels of transcripts, there was a substantial variation in the growth and morphology among different transformants. 35S-W18-L4, the highest expresser of AtWRKY18, was severely stunted when the plant was transferred to soil (Fig. 3) and the seed set was also markedly reduced. Transgenic lines 35S-W18-L3 and L5 also expressed AtWRKY18 at high levels, but substantially lower than that in 35S-W18-L4 (Fig. 2A). These two lines also showed substantial growth inhibition (Fig. 3). Unlike the progeny from 35S-W18-L4, however, these plants had normal survival rates and set a normal number of seeds. 35S-W18-L1, 35S-W18-L2, and all the lines harboring 35S-W18Δ expressed AtWRKY18 at levels like those found in SA-treated wild-type plants (Fig. 2) and grew and developed largely normally, although their leaves appeared to be slightly smaller and more serrated (Fig. 3). Altered growth and morphology of these transgenic plants were observed at all growth stages. The amount of AtWRKY18 transcripts in the transgenic line 35S-W18-L6 was below the level of detection, and plants exhibited no alterations in growth or morphology (data not shown).
Figure 3.
Growth of transgenic lines in soil. Wild-type plants, T3 progeny of transgenic AtWRKY18Δ line 1 (W18Δ-1), and transgenic AtWRKY18 lines 4 and 5 (W18-4 and W18-5) were germinated and grown in a growth chamber. The plants were photographed 32 d after germination.
PR Gene Expression and Disease Resistance
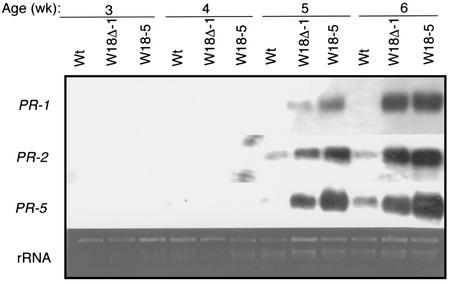
Because of the highly stunted growth and limited number of seeds, we were unable to analyze possible changes in disease resistance in the progeny of 35S-W18-L4 that expressed very high levels of AtWRKY18. To analyze defense responses in other lines, we chose homozygous progeny of 35S-W18Δ-L1 and 35S-W18-L5. These lines contained a single T-DNA locus in their genome (as determined from the ratio of antibiotic resistance phenotypes). First, we examined expression of PR genes in the transgenic plants at different stages of their life cycle. Wild-type plants produced no detectable mRNA for PR-1, the reliable molecular marker of SAR, at either young (3–4 weeks old) or mature (5–6 weeks old) stages of development (Fig. 4). For PR-2 and PR-5, no transcript was detected at young stages but significant levels of expression were consistently found at mature stages in wild-type plants (Fig. 4). Thus, there was a marginal enhancement in PR gene expression in wild-type plants as they grew from young seedlings to mature plants. In contrast, although there was no significant expression of the PR genes in 3- to 4-week-old transgenic lines 35S-W18Δ-L1 and 35S-W18-L5, transcripts for these defense-related genes accumulated to high levels in the 5- to 6-week-old transgenic plants (Fig. 4). These results indicated that constitutive expression of AtWRKY18 did not induce, but rather markedly potentiated, the developmentally regulated PR gene expression.
Figure 4.
Potentiation of PR gene expression in transgenic plants. Total RNA was isolated at indicated times after germination from wild type (Wt) and T3 progeny of two transgenic lines harboring the 35S-W18 or 35-W18Δ construct, separated on a 1.2% (w/v) agarose-formaldehyde gel, and probed with a PR-1 fragment. The blot was subsequently stripped and reprobed with PR-2 and PR-5. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading.
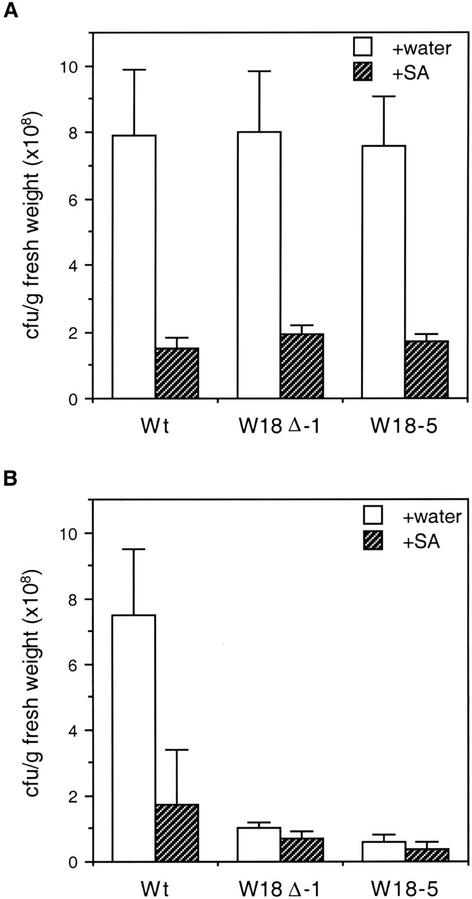
To determine whether there was a correlation between PR gene expression and disease resistance, we examined the response of both wild-type plants and transgenic AtWRKY18 plants at young (3.5 weeks old) and mature stages (5.5 weeks old) to P. syringae pv tomato DC3000, which is virulent on Arabidopsis ecotype Columbia (Whalen et al., 1991). Plants were inoculated with the bacteria and the growth of the pathogen was monitored 3 d later. As shown in Figure 5, wild-type plants showed little difference in bacterial growth between young and mature stages. On the other hand, although there was no significant increase in resistance to the bacterial pathogen during the young stages (Fig. 5A), a marked decrease (8–10-fold) in the bacterial growth was observed during the mature stages in the progeny of two transgenic AtWRKY18 lines (Fig. 5B). These levels of resistance were similar to that observed in SA-treated wild-type plants (2 mm for 3 d; Fig. 5B). Previously, treatment of Arabidopsis plants with two synthetic disease resistance inducers, 2,6-dichloroisonicotinic acid and benzothiadiazole, has been shown to induce similar levels of reduction (5–10-fold) in the growth of the same strain of the bacterial pathogen (Uknes et al., 1992; Lawton et al., 1996). It has also been reported previously that after local inoculation with an avirulent strain of P. syringae carrying the avrRpt2 avirulent gene, other leaves become resistance to the challenge inoculation of the P. syringae DC3000 virulent strain with a 5- to 10-fold reduction in bacterial growth (Cameron et al., 1994). Thus, the levels of resistance to the P. syringae DC3000 virulent strain in the mature transgenic AtWRKY18 plants were similar to those observed in chemically or biologically induced SAR.
Figure 5.
Enhanced disease resistance in transgenic plants. Wild type (Wt) and T3 progeny of transgenic line W18Δ-1 and W18-5 at 3.5-week (A) and 5.5-week (B) ages were treated with water or SA (2 mm). Three days after the treatment, the plants were inoculated with P. syringae DC3000 (OD600 = 0.001). Samples were taken 3 d after inoculation to determine the growth of the bacterial pathogen. The means and ses were calculated from triplicate determinations. The experiment was repeated three times with similar results.
NPR1 Dependence
Increased biosynthesis of endogenous SA or application of exogenous SA is known to induce expression of PR genes as well as resistance to a wide range of microbial pathogens (Uknes et al., 1992; Lawton et al., 1995). Overexpression of NPR1 can also stimulate PR gene expression and enhance resistance to a broad spectrum of pathogens (Cao et al., 1998). In addition, we recently have shown that WRKY proteins are important in the inducible expression of NPR1 (Yu et al., 2001). Thus, constitutive expression of PR genes and increased disease resistance in the mature transgenic AtWRKY18 plants may result from the positive effects of AtWRKY18 on the biosynthesis of SA and/or the expression of NPR1. To test these possibilities, we compared the levels of SA and the expression of NPR1 between the control plants and the transgenic AtWRKY18 plants. These analyses revealed no significant alteration in the levels of SA or the expression of NPR1 between wild-type and transgenic plants at mature stages when the two types of plants exhibited marked difference in PR gene expression and resistance to a bacterial pathogen (data not shown).
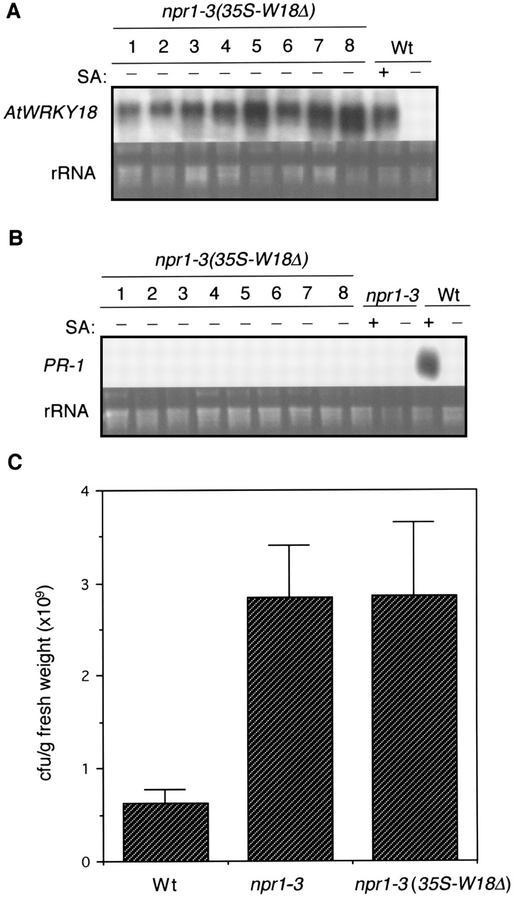
To further examine the role of NPR1 in the potentiation of plant defense response by AtWRKY18, we transformed an Arabidopsis npr1 mutant (npr1-3) with the 35S-W18Δ construct. The npr1-3 mutant contains a nonsense codon at residue 400, resulting in a truncated protein lacking 194 amino acids of the C-terminal end of the NPR1 protein (Cao et al., 1997). The transgenic npr1-3 mutant transformed with AtWRKY18 expressed the transgene at levels similar to those found in wild-type transgenic plants (Fig. 6A). Unlike wild-type plants, however, the npr1-3 mutants transformed with AtWRKY18 exhibited no potentiation of PR gene expression (Fig. 6B) or disease resistance to the bacterial pathogen P. syringae (Fig. 6C). Thus, the functional activity of AtWRKY18 in plant defense response is dependent on NPR1. Despite absence of PR gene expression and enhanced disease resistance, transgenic npr1-3 mutant plants expressing AtWRKY18 still exhibited altered changes of leaf morphology (slightly smaller and more serrated leaves) as found in transgenic AtWRKY18 wild-type plants (data not shown).
Figure 6.
NPR1 dependency of AtWRKY18-potentiated defense response. A, Total RNA was isolated from wild-type plants (Wt) with (+) or without (−) SA treatment (2 mm for 3 h) and untreated F2 progeny of npr1-3 independent transformant lines harboring the 35S-W18Δ construct, separated on a 1.2% (w/v) agarose-formaldehyde gel, and probed with an AtWRKY18 cDNA fragment. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading. B, Total RNA was isolated from 5.5-week-old wild-type (Wt), npr1-3 mutant with (+) or without (−) SA treatment (2 mm for 3 h), and untreated T2 progeny of npr1-3 independent transformant lines harboring the 35-W18Δ construct, separated on a 1.2% (w/v) agarose-formaldehyde gel, and probed with a PR-1 fragment. The ethidium bromide stain of rRNA is shown for each lane to allow assessment of equal loading. C, Wild type (Wt), npr1-3 mutant, and T2 progeny from eight npr1-3 independent transformant lines harboring the 35S-W18Δ construct were inoculated with P. syringae DC3000 (OD600 = 0.001) 5.5 weeks after germination. Samples were taken 3 d after inoculation to determine the growth of the bacterial pathogen. The means and ses were calculated from triplicate determinations. The experiment was repeated three times with similar results.
W Box Sequences in the AtWRKY18 Promoter
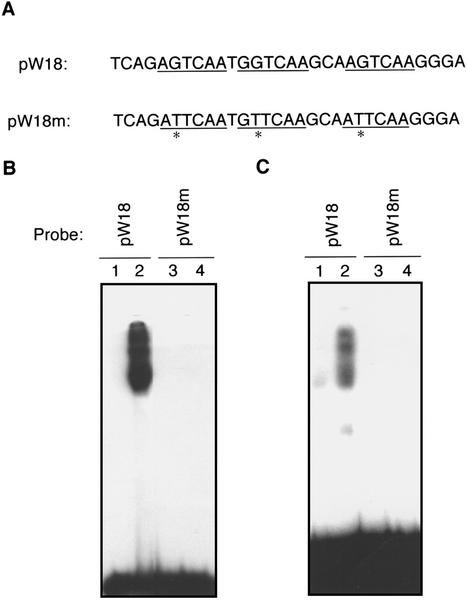
Results from transgenic plants indicated that AtWRKY18 could positively modulate both PR gene expression and disease resistance to a bacterial pathogen (Figs. 4 and 5). However, if expressed at very high levels, the same transcription factor caused severe abnormality in plant growth (Fig. 3). Thus, regulated expression of AtWRKY18 may be important during the activation of plant defense response. To identify the regulatory components and study the mechanisms involved in the regulated expression of AtWRKY18, we analyzed the promoter of the pathogen-induced gene. Interestingly, AtWRKY18 contains multiple W box sequences in its promoter region (Fig. 7A). The presence of W box elements in a pathogen- and SA-induced WRKY gene suggests that it is either autoregulated by itself or cross regulated by other WRKY proteins. To test how either type of control might affect the AtWRKY18 gene expression, we first examined the recognition of these W box elements by purified recombinant AtWRKY18 protein and by SA-induced WRKY DNA-binding activities isolated from SA-treated Arabidopsis plants. When incubated with recombinant AtWRKY18 protein, the probe containing the three W boxes of the AtWRKY18 gene promoter produced retarded bands in electrophoretic mobility shifting assays (EMSA; Fig. 7B). The binding was sequence specific because mutations of these W boxes from TTGAC to TTGAA (Fig. 7A) abolished the retarded bands (Fig. 7B). When incubated with nuclear extracts, the same probe containing the W box sequences also detected a number of SA-induced DNA-binding activities (Fig. 7C). Again, when these W boxes were mutated from TTGAC to TTGAA, the intensities of the retarded bands were drastically decreased (Fig. 7C). These results indicated that the W boxes in the AtWRKY18 gene promoter were specifically recognized by AtWRKY18 and by SA-induced WRKY DNA-binding activities in plant nuclear extracts.
Figure 7.
Recognition of W box elements in the AtWRKY18 gene promoter by WRKY DNA-binding proteins. A, Sequences of the PW18 probe (the −1301 to −1330 region of the AtWRKY18 gene promoter) and the mPW18 probe with the TTGAC repeats mutated into TTGAA repeats. B, Sequence-specific binding of PW18 by the AtWRKY18 protein (lane 2). Change of the TTGAC repeats to TTGAA repeats in the mPW18 probe abolished the intensities of retarded bands (lane 4). No retarded bands were detected in the absence of proteins (lanes 1 and 3). C, The PW18 probe recognized DNA-binding activities present at low levels in untreated plants (lane 1) and induced in SA-treated plants (lane 2). The mPW18 probe failed to recognize these DNA-binding activities from untreated (lane 3) or SA-treated plants (lane 4).
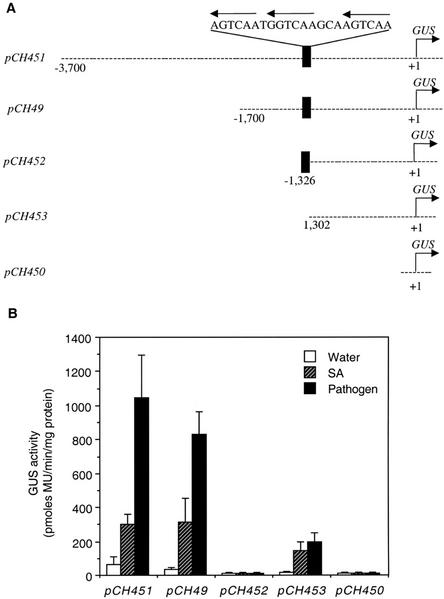
To determine the role of the W box elements in the inducible expression of AtWRKY18, we first performed promoter deletion experiments in transgenic Arabidopsis plants harboring the promoter fused with the reporter gene GUS. The AtWRKY18 gene promoter was isolated as a 3.7-kb fragment 5′ to the translation start site and fused with the reporter gene (Fig. 8A). Analysis of transgenic plants harboring the construct showed induction of GUS expression after SA treatment or infection by the bacterial pathogen P. syringae (Fig. 8B). Deletion of the promoter to 1.7 kb had no significant effect on the inducible expression of the reporter gene (Fig. 8B). Further deletion to the region immediately upstream of the W box elements drastically reduced the GUS activities (Fig. 8B), suggesting that these W box elements were not sufficient for the inducible expression of the downstream gene. Interestingly, deletion of a 24-bp region encompassing the three W boxes increased the inducible GUS expression (Fig. 8B). These results suggested that the W box elements in the AtWRKY18 gene promoter acted as negative regulatory elements.
Figure 8.
Deletion analysis of the AtWRKY18 gene promoter. A, Fusion constructs of different lengths of the AtWRKY18 gene promoter and the GUS reporter gene. Lengths of promoter deletion fragments are indicated from 5′ ends to the translation initiation site. The GUS reporter gene with no upstream promoter sequence (pCH450) was included as a negative control. B, GUS activities in 4-week-old T2 progeny from 10 independent T1 transgenic Arabidopsis lines harboring the above promoter deletion constructs 24 h after the treatments. GUS activity on a pool of four T2 plants from each T1 line was determined for each treatment and the mean and se were calculated from the 10 independent T1 lines.
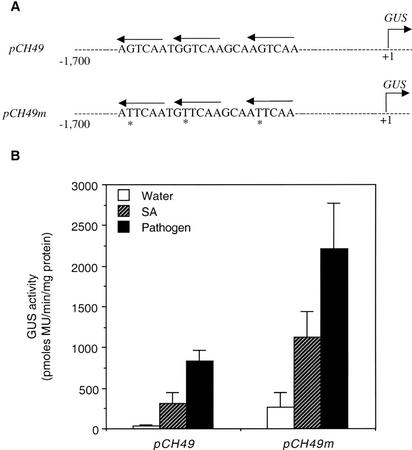
To confirm the negative regulatory role of the W boxes in the control of AtWRKY18 gene expression, we also performed promoter mutation experiments to examine the specific effects of these W boxes on the promoter activity (Fig. 9). In this case, we compared the GUS activity of the 1.7-kb promoter fragment with that of the same promoter fragment with mutated W boxes (Fig. 9A). As shown in Figures 8 and 9, the 1.7-kb promoter fragment conferred a substantial level of SA- and pathogen-inducible expression of the reporter gene GUS. Importantly, mutations of the three W boxes from TTGAC to TTGAA substantially increased the GUS activities in healthy, SA-treated and pathogen-infected transgenic Arabidopsis plants (Fig. 9B). These results supported that the W-box elements in the AtWRKY18 gene promoter acted as negative regulatory elements.
Figure 9.
Functional analysis of W box elements in the AtWRKY18 gene promoter. A, Constructs of pCH49 (the +1 to −1,700 promoter sequence of the AtWRKY18 gene fused with the GUS reporter gene) and pCH49 m (the +1 to −1,700 promoter sequence with mutated TTGAA repeats fused with the GUS reporter gene). B, GUS activities in 4-week-old T2 progeny of 10 independent T1 transgenic Arabidopsis lines harboring the above promoter constructs 24 h after the treatments. GUS activity on a pool of four T2 plants from each T1 line was determined for each treatment and the mean and se were calculated from the 10 independent T1 lines.
DISCUSSION
In the present study, we have attempted to analyze the biological impacts of constitutive expression of a pathogen-induced gene encoding Arabidopsis WRKY18 DNA-binding transcription factor. Among 12 independent transgenic lines generated from two constructs, we observed a range of altered phenotypes that were correlated with the levels of the transgene expression. When expressed at the physiological levels observed in SA-treated wild-type plants, AtWRKY18 was able to activate PR gene expression and enhance resistance to the bacterial pathogen P. syringae in mature transgenic plants, but not in young seedlings (Figs. 4 and 5). The levels of resistance to the bacterial pathogen in these transgenic plants were similar to those observed in chemically and biologically induced SAR to the same strain of the bacterial pathogen (Uknes et al., 1992; Cameron et al., 1994; Lawton et al., 1996). The ability of the transcription factor to potentiate defense response at mature stages of plants made it a potential target of molecular manipulation for controlling diseases at the critical stages of yield production for many crop plants.
The observed enhancement of PR gene expression and disease resistance in transgenic AtWRKY18 plants at mature stages but not during the younger stages raises an intriguing question about the mode of action of this transcription factor in transgenic plants. It appears that constitutive expression of AtWRKY18 alone is not sufficient to activate PR gene expression or enhance disease resistance. Rather, the transcription factor may require coordination with developmentally regulated components to activate plant defense responses. Because the marked increase in PR gene expression and disease resistance in the transgenic plants occurred just before or at the onset of flower formation (Figs. 4 and 5), certain reproduction-associated signaling pathways may be involved in the AtWRKY18-enhanced defense response. Components of these pathways may be activated or induced by ethylene, jasmonic acid, and/or other defense-associated signaling molecules whose levels may be elevated during the transition from vegetative to reproductive stages. These components, in turn, may modify the AtWRKY18 protein, or alternatively, interact with AtWRKY18 to activate plant defense gene expression. It should be noted that in tobacco (Nicotiana tabacum), developmentally regulated PR gene expression occurs in leaves and certain flower parts of flowering plants (Lotan et al., 1989; Uknes et al., 1993). Moreover, we have recently observed induction of a number of pathogen-activated WRKY genes in healthy plants as they grow into mature/reproductive stages (Z. Chen, unpublished data), suggesting the presence of developmentally regulated basal defense responses in Arabidopsis as well. However, these basal levels of defense responses did not appear to be sufficient to activate PR genes and increase disease resistance at high levels in Arabidopsis. Boosting the production of AtWRKY18 allowed the plants to benefit from these developmentally regulated basal defense responses and become more disease resistant. Interestingly, we observed no further substantial enhancement of PR gene expression in mature transgenic plants after pathogen infection (data not shown). This observation would suggest that AtWRKY18 is capable of enhancing basal levels of plant defense response but not necessarily activating defense responses against pathogens per se. However, levels of basal defense and pathogen-induced defense may not perfectly additive, given that many positive and negative feedback and cross talk mechanisms may operate in defense-related signal transduction pathways. Thus, it remains to be determined whether AtWRKY18 functions only as a positive enhancer of the basal level of plant defense or as an activator of pathogen-induced defense responses as well.
Although high levels of production of ATWRKY18 can be beneficial in plant defense responses, it also carries some costs. In the present study, we have found that transgenic plants expressing high levels of this protein exhibited stunted growth in soil. However, activation of PR gene expression and enhanced disease resistance in transgenic AtWRKY18 plants were not caused by their altered growth and morphology because these two types of phenotypes did not coincide. The deleterious effect of excessive production of AtWRKY18 on plant growth suggested that expression of the gene might require proper regulation during the activation of plant defense responses. In the present study, we have identified W box sequences in the promoter region of the AtWRKY18 gene and provided evidence that these clustered W box sequences act as negative regulatory elements. These elements may be recognized by constitutively expressed WRKY proteins and contribute to the repression of AtWRKY18 in healthy plants. Alternatively, the negative cis-acting elements may be recognized by induced WRKY proteins (possibly including ATWRKY18 itself) so that AtWRKY18 is prevented from accumulating to the point where it interferes with normal plant growth and development as demonstrated in the present study.
MATERIALS AND METHODS
Materials
[32P]dATP (>3,000 Ci mmol−1) was obtained from NEN Life Science Products (Boston); other common chemicals were purchased from Sigma (St. Louis). Arabidopsis plants were grown in a growth chamber at 22°C and 180 μEm−2 s−1 light on a photoperiod of 12 h of light and 12 h of dark. SA was dissolved in water as 100 mm stock solutions and adjusted to pH 6.5 with KOH. SA treatment was performed by spraying the plants with a 2 mm solution.
Construction of 35S-W18 and 35S-18Δ Plasmids
An EcoRI-HindIII fragment that contains the 35S promoter with double enhancers, multiple cloning sites, and 35S terminator was excised from pFF19 (Timmermans et al., 1990) and cloned into the same sites of the Arabidopsis transformation vector pOCA28 to generate pOCA30 (Du and Chen, 2000). To generate the 35S-W18 construct, the cDNA fragment that contained the full coding sequence and the 3′-untranslated region of AtWRKY18 was excised with KpnI and SalI from a cloning plasmid and subcloned into the same restriction sites of pOCA30 in the sense orientation behind the 35S promoter. To generate the 35S-W18Δ construct, the full coding sequence of AtWRKY18 was first PCR amplified with two primers (5′-TCCATGGACGGTTCTTCGTTTCTCGA-3′ and 5′-AGAA-TTCATGTTCTAGATTGCTCCATTAACC-3′), cloned into pOCA30, and completely sequenced to verify the sequence. Arabidopsis transformation was performed by the vacuum infiltration procedure (Bechtold and Pelletier, 1998). The seeds were collected from the infiltrated plants and selected in Murashige and Skoog medium containing 50 μg mL−1 kanamycin. Kanamycin-resistant plants were transferred to soil 9 d later and grown in a growth chamber.
Northern Blotting
For northern-blot analysis, total RNA (12 μg) was separated on agarose-formaldehyde gels and blotted to nylon membranes following standard procedures (Sambrook et al., 1989). Blots were hybridized with (α-32P) dATP-labeled gene-specific probes. Hybridization was performed in 1 m NaCl; 50 mm Tris/Cl, pH 7.5; 1% (w/v) SDS; 5 mm K3PO4; 100 μg mL−1 denatured salmon sperm DNA; 10% (w/v) dextran sulfate; 0.2% (w/v) bovine serum albumin; 0.2% (w/v) Ficoll 400; and 0.2% (w/v) polyvinylpyrrolidone 400 for 16 h at 65°C. The membrane was then washed for 10 min twice with 2× SSC and 1% (w/v) SDS and 10 min with 0.1× SSC and 1% (w/v) SDS at 65°C.
RT-PCR
For RT-PCR of the AtWRKY18 gene transcripts, the first cDNA was synthesized with DNase-treated total RNA (5 μg) with SuperScript RT (Life Technologies/Gibco-BRL, Rockville, MD). Quantitative PCR amplification of a base pair fragment of the AtWRKY18 gene transcripts was performed as described previously (Yu et al., 1999), using two AtWRKY18-specific primers (W18-A: 5′-actacgcaccaac-tagtcctt-3′ and W18-B: 5′-tgaattcaagcatttggacccaagtggtt-3′). The primer W18-A corresponds to a 5′-transcribed but untranslated region of AtWRKY18; therefore, RT-PCR with W18-A and W18-B as primers will amplify transcripts of only the endogenous gene but not the transgene of AtWRKY18 in the transgenic AtWRKY18 plants.
Pathogen Inoculation
Pathogen inoculations were performed by infiltration of leaves of at least six plants for each treatment with the Pseudomonas syringae pv. tomato DC3000 strain (OD600 = 0.001 in 10 mm MgCl2). Inoculated leaves were harvested 3 d after infiltration and homogenized in 10 mm MgCl2. Diluted leaf extracts were plated on King's B medium supplemented with rifampicin (100 μg mL−1) and kanamycin (25 μg mL−1) and incubated at 25°C for 2 d before counting the colony-forming units.
Determination of SA
Free and conjugated SA was extracted from Arabidopsis leaves using the same procedure described for tobacco (Nicotiana tabacum) and Arabidopsis (Bowling et al., 1994). SA separation and quantification were performed by HPLC as previously described (Bowling et al., 1994).
Production of Recombinant WRKY Proteins, Preparation of Nuclear Extracts, and EMSA
Preparation of recombinant AtWRKY18 proteins, Arabidopsis nuclear extracts, and EMSA was performed as described previously (Yu et al., 2001).
Construction of Promoter-GUS Fusion and Arabidopsis Transformation
A 3,700-bp promoter sequence of AtWRKY18 was cloned into pJL131 (Du and Chen, 2000) to generate a GUS report gene fusion. The 1,700-bp promoter fragment was generated through HindIII digestion of the 3,700-bp promoter fragment. Additional deletion constructs were generated through PCR amplification of the promoter sequences using gene-specific primers. The mutant promoter of AtWRKY18 in which the TTGAC sequences were changed to TTGAA sequences was generated by overlapping PCR. PCR-amplified fragments were all sequenced to verify the sequences. The resulting recombinant plasmids were digested with appropriate restriction enzymes and the resulting promoter-GUS fusion fragments were cloned into the Arabidopsis transformation vector pOCA28 (Du and Chen, 2000).
For the measurements of GUS activity, 4-week-old transgenic plants were infected with P. syringae pv. tomato DC3000 or sprayed with water or 2 mm SA. Leaves of the plants were collected 24 h after the treatment for determining SA- or pathogen-induced GUS activity. The leaves were homogenized in ice-cooled extraction buffer and microcentrifuged at 4°C. The GUS activity in the supernatant was measured using 4-methylumbelliferyl-β-d-glucuronide as substrate (Jefferson et al., 1987). The standard curves were prepared with 4-methylumbelliferone.
ACKNOWLEDGMENTS
We thank Dr. Allan Caplan for critically reading the manuscript and Baofang Fan for excellent technical support.
Footnotes
This work was supported in part by the National Science Foundation (grant no. MCB–9905976 to Z.C.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.001057.
LITERATURE CITED
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994;5:715–725. [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci USA. 1998;95:6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen Z. Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol Biol. 2000;42:387–396. doi: 10.1023/a:1006399311615. [DOI] [PubMed] [Google Scholar]
- Dellagi A, Helibronn J, Avrova AO, Montesano M, Palva ET, Stewart HE, Toth IK, Cooke DE, Lyon GD, Birch PR. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol Plant-Microbe Interact. 2000;13:1092–1101. doi: 10.1094/MPMI.2000.13.10.1092. [DOI] [PubMed] [Google Scholar]
- Du L, Chen Z. Identification of genes encoding novel receptor-like protein kinases as possible target genes of pathogen-induced WRKY DNA-binding proteins. Plant J. 2000;24:837–848. doi: 10.1046/j.1365-313x.2000.00923.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/s1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defense signalling: rapid gene activation by WRKY transcription factors. EMBO J. 1999;18:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2 2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H. Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet. 2000;263:30–37. doi: 10.1007/pl00008673. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Lee SH, Park HC, Bae CG, Cheong YH, Choi YJ, Han C, Lee SY, Lim CO, Cho MJ. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol Plant-Microbe Interact. 2000;13:470–474. doi: 10.1094/MPMI.2000.13.4.470. [DOI] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant-Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Lotan T, Ori N, Fluhr R. Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell. 1989;1:881–887. doi: 10.1105/tpc.1.9.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996;15:5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Timmermans MC, Maliga P, Vieira J, Messing J. The pFF plasmids: cassettes utilising CaMV sequences for expression of foreign genes in plants. J Biotechnol. 1990;14:333–344. doi: 10.1016/0168-1656(90)90117-t. [DOI] [PubMed] [Google Scholar]
- Uknes S, Dincher S, Friedrich L, Negrotto D, Williams S, Thompson-Taylor H, Potter S, Ward E, Ryals J. Regulation of pathogenesis-related protein-1a gene expression in tobacco. Plant Cell. 1993;5:159–169. doi: 10.1105/tpc.5.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang P, Fan B, Chen Z. An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with the plant defense response. Plant J. 1998;16:515–522. doi: 10.1046/j.1365-313x.1998.00311.x. [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Z, Fan B, Chen C, Chen Z. A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J. 1999;18:141–149. [Google Scholar]
- Yu D, Chen C, Chen Z. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 2001;13:1527–1540. doi: 10.1105/TPC.010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Xie Z, Chen C, Fan B, Chen Z. Expression of tobacco class II catalase gene activates the endogenous homologous gene and is associated with disease resistance in transgenic potato plants. Plant Mol Biol. 1999;39:477–488. doi: 10.1023/a:1006180708533. [DOI] [PubMed] [Google Scholar]