Abstract
HIV-1 Tat protein regulates transcription elongation by binding to the 59 nt TAR RNA stem–loop structure transcribed from the HIV-1 5′ long terminal repeat (5′-LTR). This established Tat–TAR interaction was used to investigate mRNA folding and RNA–protein interactions during early transcription elongation from the HIV-1 5′-LTR. Employing a new site-specific photo-cross-linking strategy to isolate transcription elongation complexes at early steps of elongation, we found that Tat interacts with HIV-1 transcripts before the formation of full-length TAR (TAR59). Analysis of RNA secondary structure by free energy profiling and ribonuclease digestion indicated that nascent transcripts folded into an alternative TAR RNA structure (TAR31), which requires only 31 nt to form and includes an analogous Tat-binding bulge structure. Functionally, TAR31, similar to TAR59, acts as a transcriptional terminator in vitro, and mRNA expression from TAR31-deficient HIV-1 5′-LTR mutant promoters is significantly decreased. Our results support a role for TAR31 in the control of HIV-1 mRNA transcription and we propose that this structure is important to stabilize the short early transcripts before the transcription complex commits for processive elongation. Overall, this study demonstrates that RNA folding during HIV-1 transcription is dynamic and that as the nascent RNA chain grows during transcription, it folds into a number of conformations that function to regulate gene expression. Finally, our results provide a new experimental strategy for studying mRNA conformation changes during transcription that can be applied to investigate the folding and function of nascent RNA structures transcribed from other promoters.
INTRODUCTION
RNA components of ribonucleoprotein complexes and ribozymes fold into complex three-dimensional architecture. The structural diversity of RNA includes a rich repertoire of non-standard base pairing and backbone organization and provides the basis for its biological role in macromolecular assembly and catalysis (1). RNA folding is a dynamic process governed by a number of factors, including charge, ionic strength, nucleotide composition, solvents and RNA-binding proteins. In addition, protein binding often leads to large-scale rearrangements in RNA structure and induced RNA structures contain non-standard base pairing, stacking and distortion in grooves (2). The structural diversity of mRNA suggests the potential for a relationship between varying structure and resulting functional consequences. For example, RNA folding during transcription is dynamic and RNA may test a number of conformations as the nascent chain grows. In turn, these conformations may vary in their regulatory functions, leading to changes in the rate or efficiency of transcription. Despite the undeniably important role of RNA in gene regulation, it has been difficult to acquire detailed structural information regarding how RNA folding affects gene regulation as the transcript is incrementally released from transcription complexes. Thus, RNA structure and associated function variations that occur during transcription elongation remain an open question.
In eukaryotes, RNA polymerase II (RNA Pol II) can pause, arrest, pass through terminator sequences or terminate transcription after successfully initiating RNA synthesis. The rate-limiting step in transcription elongation is the release of RNA Pol II from stalled complexes (3). In the absence of an inducer protein, RNA Pol II elongation complexes pause 20–60 nt downstream from the promoter whereas DNA- or RNA-binding activators release paused complexes by recruiting or stimulating positive-acting transcription elongation factors (4). One elegantly controlled eukaryotic transcription elongation system involves the HIV-1 Tat protein, an RNA-binding protein that enhances elongation of human RNA Pol II complexes (5). To activate transcription elongation, Tat binds to the upper stem and bulge region of trans-activation responsive (TAR) RNA, a highly structured element in nascent viral RNA that forms within the 5′ long terminal repeat (5′-LTR) region of HIV-1 during transcription (6). Wei et al. (7) identified a protein termed Cyclin T1 (CycT1) that binds Tat activation domain. CycT1 together with CDK9 forms the positive transcription elongation factor b (P-TEFb) complex (8–10). The interaction of Tat activation domain with a Ser/Thr kinase was first suggested by copurification of TAK (Tat-associated kinase) with Tat-affinity chromatography (11–15). TAK also phosphorylated the C-terminal domain (CTD) of RNA Pol II and this activity was inhibited by a nucleoside analog, DRB (13). Further analysis of transcription elongation on HIV-1 LTR revealed that TAK is a positive transcription elongation factor b, P-TEFb (8–10). Tat cooperatively binds to TAR with P-TEFb (16,17). The P-TEFb–Tat–TAR complex controls an early step in transcription elongation that results in hyperphosphorylation of the CTD of RNA Pol II and increased processivity of RNA Pol II (18). Recruitment of P-TEFb to TAR has been proposed to be both necessary and sufficient for activation of transcription elongation from the HIV-1 LTR promoter (19).
Here, we devised a new experimental strategy to investigate RNA-based control of human RNA Pol II-mediated transcription elongation and establish the structure and function of TAR during the earliest steps of transcription elongation. Elongation complexes were isolated at different stages during in vitro transcription and corresponding RNA–protein interactions were analyzed by site-specific photo-cross-linking experiments. Our results demonstrate that nascent RNA in the elongation complex folds into an alternative TAR RNA structure as soon as 31 nt are available for folding. This 31 nt TAR (TAR31) contains an inverted Tat-binding site and interacts specifically with Tat in a bulge-dependent manner. Furthermore, our studies show that TAR31 is biologically functional, acting as a termination signal and promoting HIV-1 mRNA expression.
MATERIALS AND METHODS
In vitro transcription and cross-linking
The plasmid pWT2 was derived from the p10SLT plasmid which contains the HIV-1 5′-LTR (20). Plasmid pWT2 was constructed by inserting a synthesized DNA fragment containing a triplex target sequence (5′-AAA AGA AAA GGG GGG-3′) between HindIII and NarI sites of plasmid p10SLT. Oligodeoxyribonucleotides were synthesized on an automated DNA/RNA synthesizer (ABI 392). Synthesis of psoralen and biotin containing DNA probes, triplex formation to the target sites in the plasmid DNA, photo-cross-linking and purification of RNA Pol II transcription complexes were carried out as described previously (21).
HeLa cell nuclear extracts were prepared according to the published procedures (22,23) with minor modifications (21). Pre-initiation complexes (PICs) were formed by incubating the immobilized DNA templates (200 ng) in a volume of 25 µl containing 12 µl of nuclear extract, 6 mM MgCl2 and 0.5 µg of poly(dA–dT) for 15 min at 30°C. PICs were washed with 25 µl of washing buffer A [20 mM HEPES (pH 7.9), 100 mM KCl, 20% (v/v) glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF and 6 mM MgCl2] to remove unbound proteins. PICs were walked to position U14 by incubation with 12.5 mM phosphocreatine, 20 µM three NTPs (CTP, GTP and UTP), 20 µM dATP and 10 µCi [α-32P]CTP (25 Ci/mmol, ICN; final concentration: 160 µM) for 5 min at room temperature. Transcription elongation complexes (TECs) stalled at U14 were washed with 25 µl of wash buffer B (wash buffer A containing 0.05% NP-40 and 0.015% Sarkosyl) and twice with wash buffer C (wash buffer A containing 0.05% NP-40). The TECs stalled at U14 were walked stepwise along the DNA by repeated incubation with different sets of three NTPs. Unincorporated NTPs were removed by washing the immobilized complexes with wash buffer C. To isolate RNA products from stalled TECs, 175 µl of stop solution (0.3 M Tris–HCl, pH 7.4, 0.3 M sodium acetate, 0.5% SDS and 2 mM EDTA) was added. The mixture was extracted with 200 µl of phenol/chloroform/isoamyl alcohol (50:48:2) and then with chloroform (200 µl). RNA transcripts were precipitated with ethanol and analyzed on 15% polyacrylamide–7 M urea gels. 4-Thio UTP was incorporated in the ternary complexes at U23 by stepwise walking of the elongation complex as described above. After the TECs were stalled at the desired position, they were washed three times with buffer C (described above) and then incubated with 100 ng Tat protein in buffer D (20 mM HEPES, pH 7.9, 0.5 mM DTT, 100 mM KCl, 20% glycerol and 0.2 mM EDTA) for 10 min at room temperature. The mixture was UV irradiated at 360 nm in a photochemical reactor for 15 min. The beads were separated from the mixture by magnetic particle concentrator (Dynal, Great Neck, NY) and the complexes were analyzed on a 12% polyacrylamide gel. Cross-linking yields were quantified using phosphorimaging (Molecular Dynamics).
RNA synthesis and RNA sequencing
TAR31 and TAR46 RNAs were prepared by in vitro transcription (24,25). DNA was synthesized on an Applied Biosystems ABI 392 DNA/RNA synthesizer or purchased from Integrated DNA Technologies (IDT, IA). The template strands encode the sequences for RNAs, with a T7 transcription initiation site on the 3′ end. The top strand is a short piece of DNA complementary to the T7 initiation site (5′-TAATACGACTCACTATAG-3′). The template strand of DNA was annealed to an equimolar amount of top strand DNA and transcription was carried out in transcription buffer and 4 mM NTPs at 37°C for 2–4 h. For 20 or 100 µl reactions containing 8 pmol template DNA, 40–60 U of T7 polymerase (Promega) was used. To stop transcription reactions, an equal volume of sample loading buffer was added to reactions. RNA was purified on 20% acrylamide–8 M urea denaturing gels and stored in DEPC water at −20°C.
Enzymatically transcribed RNAs were 5′ dephosphorylated by incubation with calf intestinal alkaline phophatase (Promega) for 1 h at 37°C in 50 mM Tris–HCl, pH 9.0, 1 mM MgCl2, 0.1 mM ZnCl2 and 1 mM spermidine. RNAs were purified by multiple extractions with Tris-saturated phenol and one extraction with 24:1 chloroform/isoamyl alcohol followed by ethanol precipitation. RNAs were 5′-end-labeled with 0.5 µM [γ-32P]ATP (6000 Ci/mmol) (ICN) per 100 pmol RNA by incubating with 16 U of T4 polynucleotide kinase (New England Biolabs) in 70 mM Tris–HCl, pH 7.5, 10 mM MgCl2 and 5 mM DTT. The RNAs were labeled at the 3′ end by ligation to cytidine-[5′-32P]bis-phosphate ([5′-32P]pCp) using T4 RNA ligase. Reaction mixtures (50 µl) contained 250 pmol RNA, 65 µCi [5′-32P]pCp (3000 Ci/mmol; NEN™, Boston, MA), and 40 U of T4 RNA ligase (New England Biolabs) in a buffer containing 50 mM Tris–HCl (pH 8.0), 3 mM DTT, 10 mM MgCl2, 25 mM NaCl, 50 mM ATP, 25 µg/ml BSA and 10% (v/v) dimethyl sulfoxide. After incubation at 4°C overnight, labeled RNAs were purified by phenol–chloroform extraction and ethanol precipitation. 3′- and 5′-end-labeled RNAs were gel purified on a denaturing gel, visualized by autoradiography, eluted out of the gels, and desalted on a reverse-phase cartridge.
RNA sequences were determined by base hydrolysis and nuclease digestion. Alkaline hydrolysis of RNAs was carried out in hydrolysis buffer for 8–12 min at 85°C. All ribonucleases were purchased from Pharmacia. 5′- or 3′-End-labeled RNA was incubated at room temperature with 0.05 U of 10 µl RNase T1 (40 mM Tris–HCl, pH 8.0 and 1 mM EDTA) at 50°C for 2 min, or 0.5 U of 10 µl RNase U2 (30 mM sodium citrate, pH 3.5, 1.5 mM EDTA and 0.3 µg/μl carrier tRNA) at 50°C for 5 min, 0.25 U of 10 µl RNase Bacillus cereus (Tris–HCl, pH 8.0) for 5 min. Sequencing products were resolved on 20% denaturing gels and visualized by phosphorimaging.
Structure probing of free RNA and RNA in TECs
5′-end-labeled RNA was renatured in reaction buffer and incubated at room temperature with 1 U of 10 µl RNase T1 (Tris–HCl, pH 8.0) for 5 min, 0.2–0.4 U for 10 µl RNase V1 (5 mM Tris–HCl, pH 8.1, 60 mM NaCl and 5 mM MgCl2) for 2 min, or 2.5–5 U for 10 µl nuclease S1 (5 mM MES, pH 6.3, 120 mM NaCl and 5 mM MgCl2) for 2 min. The reactions were quenched by adding sample loading buffer. Fragments were analyzed on 20% polyacrylamide–7 M urea gels.
TECs stalled at U46 were obtained by stepwise transcription. The complexes were labeled with [α-32P]UTP to obtain 3′-end-labeled U46 TECs. After washing with wash buffer C (three times), labeled complexes were re-suspended in digestion reaction buffers and incubated at room temperature with 5 U of 50 µl RNase T1 (Tris–HCl, pH 8.0) for 5 min, 0.5–1 U of 50 µl RNase V1 (5 mM Tris–HCl, pH 8.1, 60 mM NaCl and 5 mM MgCl2) for 2 min, or 0.5–2 U of 50 µl RNase B.cereus (Tris–HCl, pH 8.0) for 5 min. Reactions were quenched with 165 µl of stop solution (0.3 M Tris–HCl, pH 8.0, 0.3 M NaOAc, 2 mM EDTA and 0.05% SDS). Labeled RNAs were purified by phenol–chloroform extraction and ethanol precipitation, and analyzed on 20% polyacrylamide–7 M urea gels.
Electrophoretic mobility shift assays
RNA hairpins were transcribed in vitro and labeled as described above. GST–Tat fusion proteins (wild-type and Δ48 mutant) were expressed in Escherichia coli BL21(DE3) and purified with the B-PER GST purification kit (Pierce) following manufacturer's instructions. RNA–protein-binding reactions (20 µl) contained 50 fmol of 5′-[32P]-end labeled RNA and 1 pmol of purified GST–Tat. Complex formation was performed in TK buffer (50 mM Tris–HCl, pH 7.4, 20 mM KCl and 0.1% Triton X-100) and incubated at 4°C for 1.5 h. Complexes were separated from unbound RNA by electrophoresis in non-denaturing 8% polyacrylamide gels containing 0.1% Triton X-100. Gels were run at 4°C at 300 V for 2 h and quantified by phosphorimaging.
DNA manipulations and plasmids
All new constructs were made from vector pBC12/HIV/SEAP (renamed pwt/HIV/SEAP), which was a gift from Dr Bryan R. Cullen (26). pm1/HIV/SEAP was made through PCR amplification of a 740 bp fragment between two BglII restriction sites in pwt/HIV/SEAP. A mutant 3′ end primer was used to introduce nucleotide substitutions at positions 6 and 7 and a new base at position 7a (m1 mutations) in the HIV1 5′-LTR. To facilitate screening of mutant colonies, a new MluI restriction site was introduced flanking the BglII site on the 5′ end primer. The mutated PCR fragment was cloned into the BglII sites of pwt/HIV/SEAP. pm2/HIV/SEAP and pm3/HIV/SEAP clones were constructed via PCR amplification of a 3660 bp fragment between the two AflII restriction sites of pm1/HIV/SEAP and pwt2/HIV/SEAP, respectively. The 3′ end primer introduces three new bases and one base substitution at positions 53–56 of the wild-type HIV-1 5′-LTR. PCR fragments were cloned into AflII sites of pm1/HIV/SEAP and pwt/HIV/SEAP to obtain pm2/HIV/SEAP and pm3/HIV/SEAP, respectively. The combined mutations in pm2/HIV/SEAP allow complete base pairing of the lower stem of TAR59 and introduce a new XbaI restriction site. pm3/HIV/SEAP only has mutations introduced at positions 53–56 of TAR59. All primer sequences are available upon request.
pCMVTatHA vector expresses Tat tagged at the C-terminus with hemagglutinin (HA) from the CMV promoter and was a gift from Dr B. Matija Peterlin (27). pSV-β-galactosidase vector was purchased from Promega. Plasmid concentrations were measured at 260 nm in a Shimatzu UV-Visible 1601 spectrophotometer.
Cell culture and SEAP assays
Transcription efficiencies of wild-type and mutant HIV-1 5′-LTRs (m1, m2, m3) were compared with 293T cells in the presence or absence of Tat. Cells were co-transfected with pwt/HIV/SEAP, pm1/HIV/SEAP, pm2/HIV/SEAP, or pm3/HIV/SEAP with or without pCMVTatHA. pSV-β-galactosidase acted as a control for transfection efficiency and 350 ng of each plasmid was used for transfection. Transfection mixtures containing the indicated plasmids and 2 µl of cationic lipids (Lipofectamine 2000; Life Technologies) were incubated in 100 μl OPTIMEM I buffer at room temperature for 30 min. Transfection mixtures were added to 293T cells (500 μl) plated on poly-d-lysine coated 24-well plates at a final concentration of 5 × 105 cell/ml. After incubating for 6 h at 37°C, cells were cultured in DMEM/10% FBS at 37°C and 5% CO2. After 24 h, 20 μl of medium per sample was collected and tested for SEcreted Alkaline Phosphatase (SEAP) activity (Great EscAPe SEAP Fluorescence detection kit; Clontech). Briefly, dilution buffer was added to the collected medium to a final volume of 50 µl. Samples were heated for 30 min at 65°C to inactivate the endogenous alkaline phosphatase and, after equilibration at room temperature, 97 μl of assay buffer and 3 μl of 1 mM 4-methilumbelliferyl-phosphate (MUP) were added. After incubation at room temperature for 1 h, samples were excited at 360 nm and the emitted fluorescence was read at 445 nm on a Photon Technology International fluorescence spectrophotometer controlled by Felix software. For β-galactosidase detection, cells were lysed with 200 μl of lysis buffer (M-PER Mammalian Protein Extraction Reagent; Pierce) and incubated at room temperature for 10 min with gentle shaking. Two hundred microlitres of assay buffer (All-in-One Mammalian β-galactosidase assay kit; Pierce) were added to lysates, which were further incubated at 37°C in 5% CO2 for 30 min. Reactions were quenched by the addition of 150 μl of stop solution (Pierce). β-Galactosidase activities were quantified by measuring OD at 405 nm in a Shimatzu UV-Visible 1601 spectrophotometer. For protein quantification, 10 μl of lysate was added to 200 μl of Protein Assay (Bio-Rad) in 800 μl of H2O and the OD was measured at 600 nm. Results are representative of at least six experiments. SEAP activity was normalized for both β-galactosidase activity and protein concentration. A value of 100 was assigned to the SEAP activity associated with the wild-type HIV-1 5′-LTR in the presence of Tat. SEAP values of remaining samples were calculated relative to the wild-type.
Total RNA isolation and mRNA quantification
The 293T cells were co-transfected with pwt/HIV/SEAP, pm1/HIV/SEAP, pm2/HIV/SEAP, or pm3/HIV/SEAP with or without pCMVTatHA. pSV-β-galactosidase acted as a control for transfection efficiency and 350 ng of each plasmid was used for transfection. Two sets of identical experiments were arranged for each HIV-1 5′-LTR construct. Twenty-four hours post-transfection, the first set of samples was assayed for SEAP activity, β-galactosidase activity and protein concentration. The second set was tested for SEAP activity and RNA transcription. Total RNA was isolated using the RNeasy Kit (Qiagen). An average of 30 μg of total RNA per sample was obtained. Five micrograms of total RNA per sample were used in one-step RT/PCR to quantify SEAP mRNA levels. Nucleotides 2623–2750 were amplified from the HIV/SEAP vectors and corresponded to a non-coding insulin region that flanked the SEAP coding region. PCR amplification of nt 2103–2333 of β-galactosidase mRNA transcribed from pSV-β-galactosidase controlled for mRNA recovery and RT/PCR efficiencies. All primer sequences are available upon request.
RNA samples were treated with one unit of RQ1 RNase Free DNase (Promega) at 37°C for 30 min to remove all traces of DNA contaminants. Reactions were quenched by addition of stop solution (Promega) and heating samples at 65°C for 10 min. Each reaction contained 20 pmol of each primer, 10 μCi of [α-32P]dCTP (ICN), 1.5 μl of RT/Taq mixture in RT/Taq buffer to a final volume of 20 μl. Reverse transcription was performed at 50°C for 30 min with the denaturing step performed for 2 min at 94°C. Fifteen PCR cycles were performed at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min in a MiniCycler (MJ Research). Reactions containing 5 U of Taq (Life Technologies), but not RT, were performed as controls for DNA contamination. Reactions (15 μl) were added to 15 μl of 2× denaturing loading buffer (80% formamide, 0.01% bromophenol blue and 0.8 mM EDTA), loaded on to 6% urea gel and run at 65 W for 2 h. Amplified fragments were visualized and quantified by phosphorimaging.
RESULTS
RNA in early TECs photo-cross-links with Tat protein
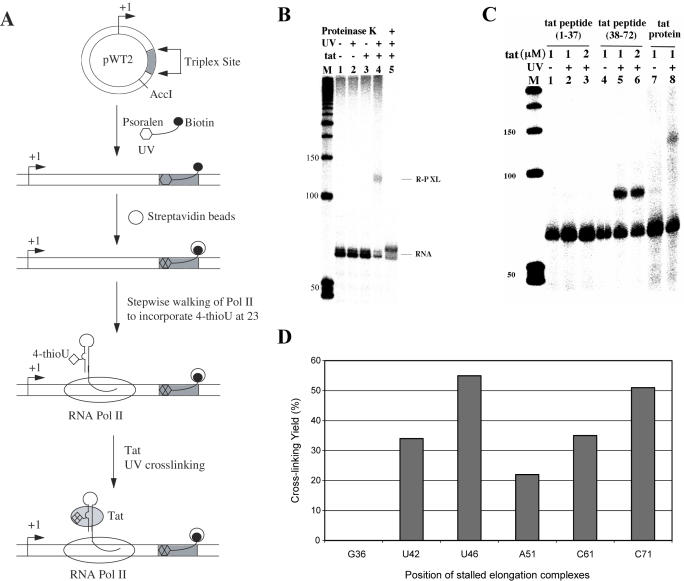
We decided to probe mRNA folding and to study RNA–protein interactions during transcription elongation at different stages. Triplex DNA structure and psoralen photochemistry were used to immobilize DNA templates and to isolate homogeneous populations of RNA Pol II ternary complexes stalled at specific sites (21,28). Figure 1A outlines our experimental strategy: (i) insertion of a target sequence for triple helix formation at a predetermined position in the DNA template; (ii) restriction enzyme digestion to release the triplex site upstream or downstream of the promoter sequences; (iii) synthesis of a third strand containing a psoralen at its 5′ end and a biotin at the 3′ end; (iv) triplex formation between the DNA template and the third strand, followed by UV irradiation (360 nm) to cross-link the complex; (v) immobilization of cross-linked DNA templates on streptavidin-conjugated magnetic beads followed by stepwise in vitro transcription to incorporate site-specific photoactive nucleosides; and (vi) incubation of stalled TECs with Tat followed by UV irradiation to form the final cross-link products.
Figure 1.
Specific Tat–TAR cross-link formation during transcription elongation. (A) Experimental design to incorporate a photoactive nucleoside into a nascent RNA chain during transcription elongation. Plasmid pWT2 harboring an inserted target sequence for triplex formation was linearized with restriction enzymes. A psoralen- and biotin-containing oligonucleotide was used to form triplex DNA. UV irradiation was then used to covalently cross-link psoralen to the template. Psoralen-cross-linked template was immobilized on streptavidin-conjugated magnetic beads and non-cross-linked DNA was washed away with buffer. Stepwise walking of RNA Pol II elongation complexes was used to incorporate the photoactive nucleoside 4-thioU into transcripts. Stalled TECs were incubated with Tat and UV irradiated to form the final cross-link product. (B) Site-specific photo-cross-linking of Tat and TAR RNA in stalled ternary complexes. Stepwise RNA Pol II walking was used to incorporate 4-thioU into TEC-associated transcripts at position 23. Tat was added to TECs stalled at C61 and UV irradiated (360 nm). The gel shows RNA from non-irradiated TECs (lane 1); UV-irradiated TECs (lane 2); TECs in the presence of 100 ng Tat but not UV irradiated (lane 3); TECs in the presence of Tat and UV irradiated (lane 4); TECs in the presence of Tat, UV irradiated, and digested with proteinase K at 37°C for 15 min (lane 5). RNA and RNA–protein cross-link are labeled as RNA and R-P XL, respectively. M is a marker lane. (C) Photo-cross-linking of TAR RNA in TECs stalled at C61 with Tat peptides. Lanes 1, 4 and 7 are non-irradiated control lanes with Tat peptides (amino acids 1–37), Tat peptide (amino acids 38–72), and full-length Tat, respectively. The gel shows RNA from TECs incubated with Tat peptide (amino acids 1–37) and UV irradiated (lanes 2 and 3); TECs incubated with Tat peptide (amino acids 38–72) and UV irradiated (lanes 5 and 6); TECs and full-length Tat and UV irradiated (lane 7). M is a marker lane (lane 8). (D) RNA–protein cross-linking between Tat and the nascent RNA chain in the TECs stalled at different positions. Elongation complexes containing 4-thioU at position 23 were stalled at G36, U42, U46, A51, C61 and C71. After incubating with Tat protein, photo-cross-linking reactions were performed and analyzed on denaturing gels as shown above. RNA–protein cross-linking yields were determined by phosphorimaging.
Pre-initiation complexes were formed on immobilized DNA templates and elongation was initiated by adding dATP, UTP, CTP and GTP. Elongation complexes were starved for ATP and stalled at U14, whereas further initiation was inhibited by sarkosyl wash as described in Materials and Methods. These stalled TECs underwent repeated incubation with different sets of three NTPs, and accordingly, were walked stepwise through transcription elongation (21,28). Since RNA Pol II can start transcription at +1 and +2 G residues in the template DNA, two transcripts were observed during walking experiments: one major product originating from the +1 start site and a minor product resulting from the +2 start site. Consistent with these observations, when transcription was initiated with GpG dinucleotide triphosphate, a single RNA product was obtained (data not shown). Viability of stalled complexes was confirmed by adding all four NTPs. This led to the production of runoff transcripts of expected lengths and indicated that TEC complexes were transcriptionally active (data not shown).
To investigate the interaction between Tat and TAR RNA, UV inducible cross-linking reagent, 4-thiouridine (4-thioU), was incorporated into RNA transcripts at position U23 during stepwise transcription. We have previously shown that that 4-thioU at U23 position in TAR RNA formed high efficiency cross-links with Tat (25). The elongation complexes carrying 4-thioU were further walked to C61 by stepwise transcription. The immobilized C61 TECs were 3′-end-labeled with [α-32P]CTP in the last step of transcription, isolated and washed thoroughly with transcription buffer. TECs containing labeled RNA were then incubated with Tat for 10 min in TK buffer, and UV irradiated at 360 nm (see Materials and Methods). Products of the photoreaction were analyzed by denaturing PAGE (12%). Irradiated RNA–protein complexes yielded a band with electrophoretic mobility less than that of RNA (Figure 1B, lane 4). This cross-linked product was observed only when RNA was irradiated in the presence of Tat (Figure 1B, compare lanes 3 and 4). Irradiation products analyzed by denaturing SDS–PAGE (15%) also showed a Tat-dependent photoproduct that had a slower electrophoretic mobility than TAR RNA (data not shown). Cross-linked products were digested with Proteinase K, which degrades cross-linked RNA–protein products but not cross-linked RNA–RNA species (25,29). Proteinase K digestion of the cross-linked products resulted in a loss of slower mobility products and a gain in free RNA (Figure 1B, lane 5), ruling out the possibility that the observed shift in mobility was due to an RNA–RNA cross-link. Finally, the cross-linked RNA–protein complex was stable at an alkaline pH (9.2), high temperatures (85°C) and under denaturing conditions (8 M urea and 2% SDS) (data not shown), suggesting that a covalent bond was formed between nascent RNA and Tat during the cross-linking reaction.
The specificity of RNA–Tat interactions was determined by performing cross-linking reactions with two different Tat peptides. TECs having 4-ThioU at position 23 and stalled at C61 were incubated with the Tat transactivation domain [Tat peptide (amino acids 1–37)] or the Tat RNA-binding domain [Tat peptide (amino acids 38–72)] and UV irradiated. RNA in the TECs did not cross-link with the transactivation domain (Figure 1C, lanes 2 and 3), but efficiently cross-linked to the RNA-binding domain to yield a slow mobility product analogous to the shifted product seen with full-length Tat (Figure 1C, compare lanes 5–6 to lane 8). These results suggested that the RNA-binding domain of Tat binds to transcripts produced from TECs stalled at C61. Competition assays further established the specificity of the cross-linking reaction between Tat and RNA from TECs stalled at C61. In these experiments, Tat and unlabeled competitor RNA were incubated with TECs stalled at C61 before performing photo-cross-linking experiments. Wild-type TAR RNA competed with TEC RNA more efficiently than bulgeless TAR RNA to inhibit formation of cross-linked products (data not shown), suggesting that TECs stalled at C61 contain a functional TAR structure that specifically interacted with Tat protein.
An alternative TAR RNA that has an inverted Tat-binding site forms during transcription elongation
To determine the 3′ end boundary of an RNA transcript that is functional for Tat binding, we incorporated 4-thioU at position 23, walked TECs to various positions, and performed photo-cross-linking experiments with full-length Tat. TECs stalled at G36, U42, U46, A51, C61 and C71 were analyzed and the quantitative yields of resulting RNA–protein cross-link products were measured. RNA from TECs stalled at G36 did not cross-link to Tat, but all others formed RNA–protein cross-links with efficiencies ranging from 20 to 55% (Figure 1D). These results indicate that RNA transcripts from TECs stalled at U42 are sufficient for Tat binding.
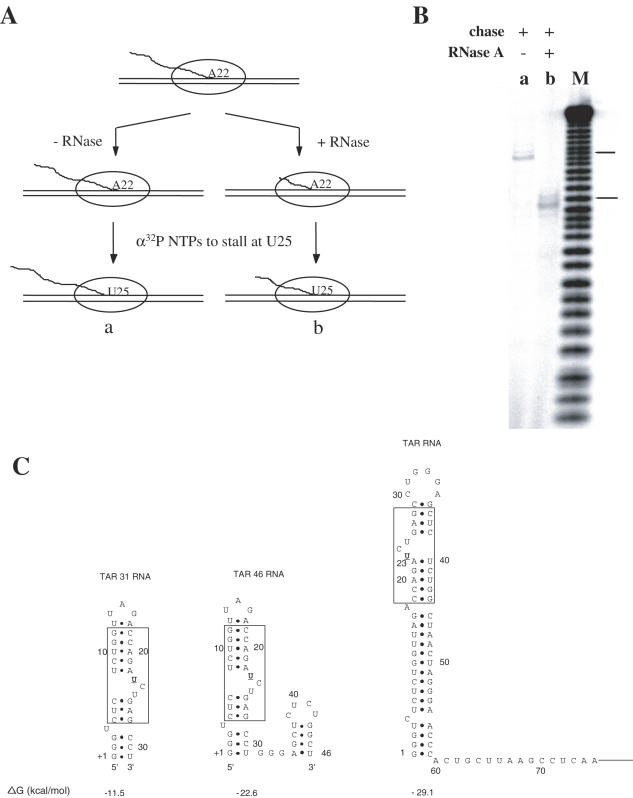
To verify the length of the transcript protected by stalled TECS, RNA Pol II was walked to position A22 and stalled TECs were divided into two fractions, with one subjected to exhaustive RNase A digestion to hydrolyze the protruding 5′ end of RNA transcripts. Both samples then were chased with NTPs to walk TECs to U25, with [α-32P]NTPs to label transcripts (Figure 2A). RNA from each sample were analyzed by PAGE and compared to an RNA marker (Figure 2B). We observe that RNase A digested approximately six nt in TECs stalled at A22, indicating that no more than 16 nt were protected by the TEC. These results were in range of the 17–19 nt protected in a previous foot printing analysis of arrested RNA Pol II elongation complexes (30). It is known that there is some heterogeneity in the length of transcript protected by different TEC and even <16 nt may be covered by the TEC if RNase A could not digest more than 6 bases owing to inefficient enzymatic activity or steric hindrance. In fact, during RNA structure probing experiments (see below and Figure 3), ribonuclease T1 cleaved at positions G32, G33 and G34 when TECs were stalled at U46, suggesting that only 12 nt of the RNA 3′ end were protected by the stalled elongation complexes.
Figure 2.
Ribonuclease footprinting of nascent RNA in RNA Pol II TECs. (A) Experimental strategy of the RNase footprinting method. TECs were stalled at position A22, subjected to exhaustive RNase A digestion, walked to U25 position and 32P-labeled. For comparison, labeled RNA from TECs stalled at U25 and not treated with RNase A was also prepared. (B) RNase footprinting experiments. The gel shows RNA from TECs stalled at U25 not treated with RNase A (lane a) and TECs stalled at U25 after RNase A digestion (lane b). Lane M is a marker lane containing an RNA hydrolysis ladder. (C) Predicted stable secondary structures of various RNA transcripts. The mfold program was used to predict secondary structures from given RNA sequences and to calculate the free energy of each structure (31). Boxed regions show common sequence and secondary structures found (including inverted) in TAR31, TAR46 and wild-type TAR.
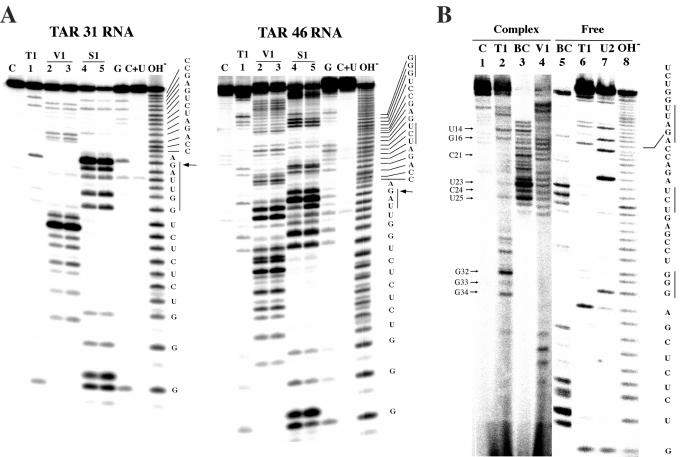
Figure 3.
Determination of alternative TAR structural elements by partial enzymatic digestion. (A) Secondary structure probing of 5′-end-labeled TAR31 and TAR46 RNA transcripts by digestion with ribonuclease T1, V1 and S1. Two concentrations of ribonuclease V1 and S1 were used. Lane C shows control RNA without digestion. Lanes G and C + U are marker lanes. Lanes labeled OH− contain an RNA alkaline hydrolysis ladder. Arrows indicate strong RNase T1 cleavage at position G16, and the vertical lines indicate loop and bulge regions uncovered by nuclease S1. (B) Structure probing of RNA in TECs stalled at U46 and comparison with free RNA. Lanes 1–4 contain 3′-end-labeled TAR46 RNA from TECs and lanes 5–8 show 3′-end-labeled free TAR46 RNA. Enzymes used for RNA digestion are shown above the lane numbers. The lane labeled BC and U2 contain RNA treated with B.cereus RNase and with RNase U2, respectively. The lane labeled OH− contains an RNA alkaline hydrolysis ladder. Single-stranded regions are marked with vertical lines. Arrows indicate strong cleavage bands.
If 12 nt were protected by TECs, only a 30 nt RNA transcript would be exposed for Tat recognition when TECs are stalled at U42. However, a 30 nt transcript cannot fold into a wild-type TAR configuration. Similarly, RNA transcripts in TECs stalled at A51 cannot adopt the wild-type TAR structure, but still cross-linked with Tat. To establish the structural attributes of these shorter transcripts, an RNA folding program (31) was used to search for RNA structures that could form at shorter lengths. Figure 2C shows an interesting stem–loop structure generated from this analysis that represents the most stable conformation of RNA transcripts containing nt 1–31 (TAR31) and 1–46 (TAR46). This stem–loop structure contained a 3 nt bulge (UCU) and sequences above and below the bulge (outlined in a box) that were identical to an inverted Tat-binding site in TAR RNA. These structural features suggested that before wild-type TAR formed, an inverted TAR structure with the capacity to bind Tat formed in association with TECs stalled at U42, U46 and A51.
To ascertain that the proposed inverted TAR RNA structure exists in early nascent transcripts, we carried out secondary structure probing experiments. TAR31 and TAR46 were synthesized by in vitro T7 transcription and 5′-end-labeled with 32P (see Materials and Methods). Secondary structure probing of 5′-end-labeled TAR31 and TAR46 RNA transcripts was performed by ribonuclease digestion (Figure 3A) and compared with RNA from the TEC stalled at U46 (Figure 3B). The RNA samples were partially digested with RNase T1, RNase V1 and nuclease S1. RNase T1 is specific for G residues in loops and single-stranded regions, RNase V1 is specific for double-stranded RNA regions, and nuclease S1 is specific for cleavage of single-stranded RNA. RNase T1 cleaved at position G16 within TAR31 and TAR46 (indicated by arrows), and nuclease S1 cleaved at positions U13, U14, A15 and G16 (highlighted by vertical lines), which was consistent with the existence of the UUAG capping loop in the structure of TAR31 and TAR46. Additional cleavage by RNase T1 at positions G32 and G34 of TAR46 (indicated by vertical lines) suggested that the predicted single-stranded region formed downstream of the inverted TAR structure. RNase V1 cleavage was observed at the 5′ and 3′ ends of TAR31 and TAR46, indicating that a double-stranded stem structure was formed from 5′ and 3′ end base pairing of the RNA transcript. Although cleavage at the UCU bulge of TAR31 was not clear in these experiments, RNase from B.cereus, which is specific for single-stranded pyrimidine residues, did cleave within the UCU region (Figure 3B). The UCU region of TAR46 was also cleaved by nuclease S1 (highlighted with vertical lines), supporting the formation of an inverted TAR structure. RNase U2 which cleaves at ApN was also used in sequencing analysis. Taken altogether, these cleavage experiments strongly support the existence of the proposed inverted TAR structure, both in isolated RNA fragments and in TEC stalled at U46.
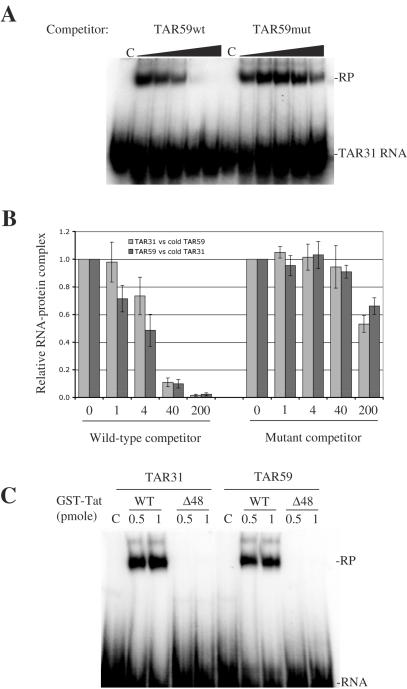
The specificity of binding of TAR31 to Tat was compared with TAR59 by electrophoretic mobility shift assay (EMSA). Short hairpin RNAs were transcribed in vitro, 5′-end-labeled with 32P, and incubated with purified GST–Tat fusion protein (see Materials and Methods). Incubation of TAR31 RNA with GST–Tat resulted in a band with reduced mobility on a native PAGE (Figure 4A). When increasing amounts of unlabeled TAR59 were added to the binding reaction, the shifted band disappeared gradually, showing that TAR59 competed with TAR31 for the binding of Tat in vitro (Figure 4A). The addition of an unlabeled mutant TAR59 hairpin that cannot form the bulge structure required for Tat binding did not affect the yield of TAR31-Tat binding. Similar results were obtained using 5′-end-labeled TAR59 with unlabeled TAR31 as a competitor. Results of competition experiments with both TAR31 and TAR59 are summarized in Figure 4B. These results show that TAR31 and TAR59 compete with each other for the binding of Tat, and in both cases the interaction is specific for the bulge structure. As a control, we used a GST-Tat Δ48 mutant, which does not contain the RNA-binding site of Tat. Incubation of TAR31 and TAR59 with this mutant protein did not result in any shifted band (Figure 4C), showing that the binding of Tat to TAR31 or TAR59 is dependent on its RNA-binding motif. These gel shift experiments confirm that the TAR31 structure makes specific interactions with Tat in a manner similar to TAR59. These interactions are both dependent on the bulge structure of TAR31 and the RNA-binding domain of Tat. However, contrary to TAR59, TAR31 did not form a stable ternary complex with Tat and Cyclin T1 (data not shown). This is an expected result because TAR31 does not contain a hexanucleotide loop which is essential for CycT1–Tat–TAR ternary complex formation.
Figure 4.
TAR31 competes with TAR59 for the binding of Tat in vitro. Binding of TAR31 and TAR59 to Tat was examined by electrophoretic mobility shift assay (EMSA). (A) Competition experiments showing the binding of 5′-end-labeled TAR31 short hairpin RNA to purified GST–Tat fusion protein in the presence of increasing amounts (0- to 200-fold molar excess) of unlabeled competitor TAR59 wild-type (TAR59wt) or bulgeless mutant (TAR59mut). The lanes labeled C are control lanes with RNA only. The RNA–protein complex is labeled RP. (B) Quantitative analysis of competition experiments with 5′-end-labeled TAR31 (light gray) and TAR59 (dark gray) in competition with unlabeled TAR59 and TAR31, respectively. Bands corresponding to RNA–Tat complex were quantified from gels and lanes without competitor were assigned an arbitrary value of 1. Values are the means ± standard error of three independent experiments. (C) Interaction between Tat and TAR31 requires the RNA-binding domain of Tat. Gel shows binding of 5′-end-labeled TAR31 and TAR59 small hairpin RNAs to wild-type GST-Tat or GST-Tat(Δ48) mutant that does not contain the RNA-binding domain.
Alternative TAR structure can induce termination of transcription elongation
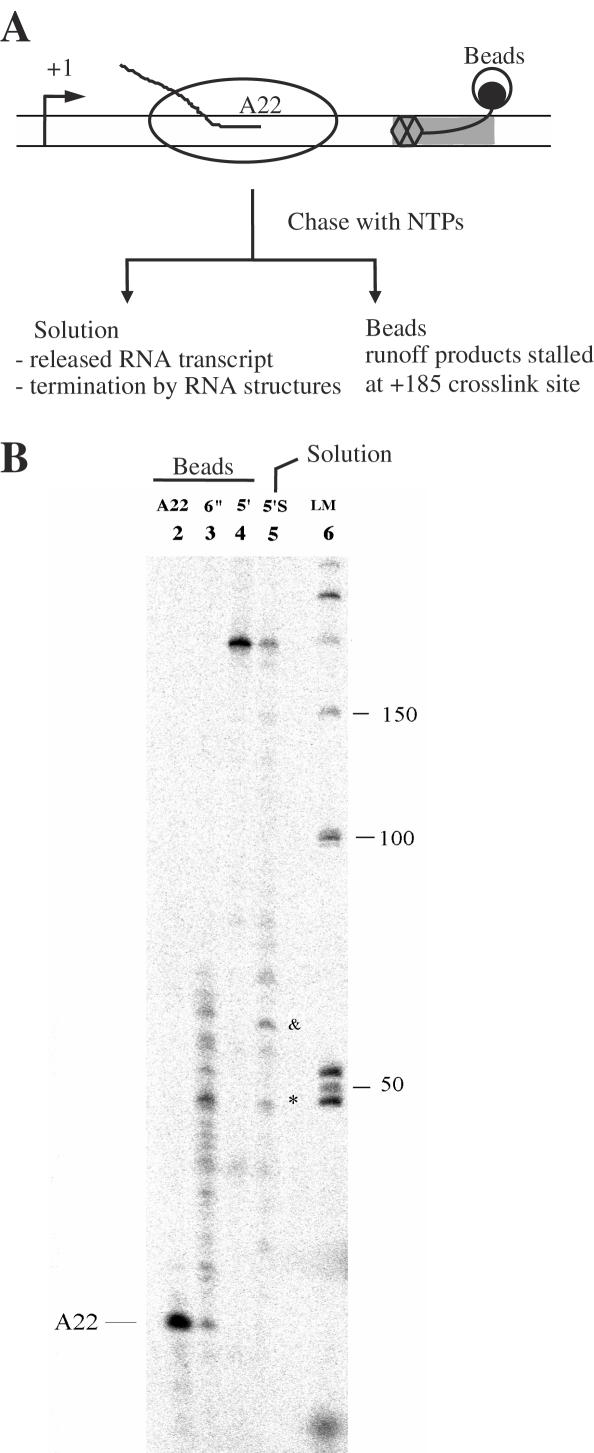
In the absence of Tat, transcription elongation has been shown to arrest after 58–61 nt are transcribed from the HIV-1 5′-LTR (32). The 58–61 nt RNA transcripts fold into the TAR RNA structure and act as a termination signal. To determine whether TAR31 was capable of terminating transcription elongation such as TAR59, we stalled TECs at position A22, chased with all four NTPs, and isolated transcripts that were associated with TECs (beads) or termination products that were released by TECs during the chase with NTPs (solution phase) (Figure 5A). Transcription reactions were monitored at 6 s and 5 min time periods (Figure 5B, lanes 3 and 4). The solution phase contained a few different termination products, the two major ones having a size of ∼46 and ∼60 nt (Figure 5B, lane 5). Quantification of band intensities in lane 5 revealed that only ∼21.8% TECs produced the runoff products and the amount of terminated transcripts containing 46 and 60 nt were 13.4 and 18.6%, respectively. The 60 nt product corresponds to TAR59 and is consistent with the capacity of full-length TAR to terminate elongation. The presence of a 46 nt RNA transcript suggests that TAR31 also acts as a transcription elongation termination signal (see Discussion).
Figure 5.
TAR31 acts as a transcription elongation termination signal. (A) Experimental outline for analyzing transcription elongation termination. A homogeneous population of TECs were stalled at A22 position and chased with unlabeled NTPs. Beads and solution phases were separated and analyzed on denaturing gels as described previously (21). Transcription reactions were monitored at 6 s and 5 min time periods. (B) Analysis of transcription products. Lane 2 shows transcripts from TECs stalled at position A22. Lanes 3 and 4 are transcripts from TECs chased with NTPs for 6 s and 5 min, respectively. Lane 5 contains transcripts released from TECs into solution during the 5 min chase with NTPs. Lane 6 is a marker lane. Two major termination signals were observed around 46 nt (*) and 60 nt (&). Quantitative analysis of band intensities in lane 5 shows that ∼21.8% TECs produced the runoff products and the amount of terminated transcripts containing 46 and 60 nt were 13.4 and 18.6%, respectively.
TAR31 is involved in the control of HIV-1 mRNA transcription, but not in Tat transactivation
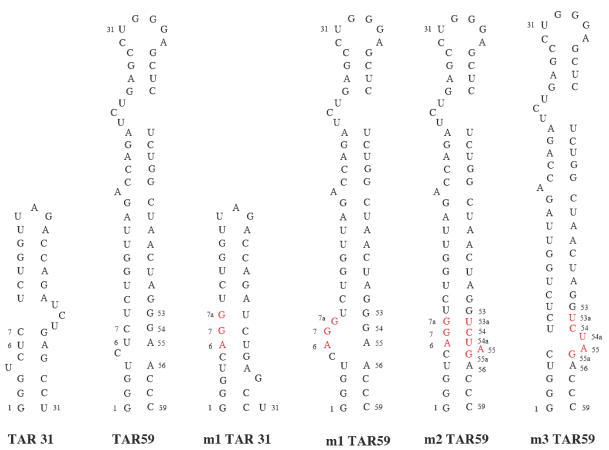
To determine whether TAR31 has a role in regulating HIV-1 5′-LTR expression, the activity of a reporter gene under TAR31-deficient HIV-1 5′-LTR promoters was tested. HIV-1 5′-LTR mutants were constructed that did not contain the TAR31-specific bulge but maintained the bulge–stem–loop structure of TAR59 (see Figure 6 and Materials and Methods). Wild-type or mutant HIV-1 5′-LTR constructs were then inserted upstream of the SEAP reporter gene. The m1 construct had mutations (m1) introduced at positions 6–7 and a nucleotide insertion (7a) within the TAR31 stem that replace the bulge structure of TAR31 with a double-stranded stem, without changing the bulge and loop region that forms the transcriptionally active portion of wild-type TAR59 (Figure 6, compare m1 TAR31 and m1 TAR59). However, the m1 mutations caused a new internal bulge to form within the lower stem structure of TAR59 (Figure 6, m1 TAR59). Although this stem region is not necessary for Tat–TAR-mediated transactivation (33), a second mutant construct was made that contained the m1 mutations and secondary compensatory mutations in the lower stem of TAR59 that included three new bases and one base substitution between positions 53–56 (Figure 6, m2 TAR59). This m2 mutant 5′-LTR retains an intact wild-type TAR59 stem structure but lacks the bulged structure of TAR31. To verify that these compensatory mutations did not affect TAR activity, a control construct was made that only had the m2 compensatory mutations in the lower stem of TAR59 and allowed formation of both the TAR31 bulge and TAR59 stem structures (Figure 6, m3 TAR59).
Figure 6.
Structure of wild-type and mutant TAR RNAs used in this study. TAR31 and TAR59 represent alternative structures of wild-type HIV-1 LTR. m1 TAR31 and m1 TAR59 represent alternative structures of a mutant LTR with two base substitutions at positions 6 and 7 and a base insertion at position 7a, preventing the formation of the TAR31 bulge without affecting the structure of TAR59 (mutated positions are shown in red). Mutant m2 contains the same mutations as mutant m1, plus three new bases and one base substitution between positions 53 and 56, which restore the wild-type lower stem structure of TAR59. Mutant m3 contains the same three new bases and one base substitution between positions 53 and 56 as mutant m2, but has wild-type nucleotides at positions 6 and 7 and does not affect the structures of TAR31 or TAR59.
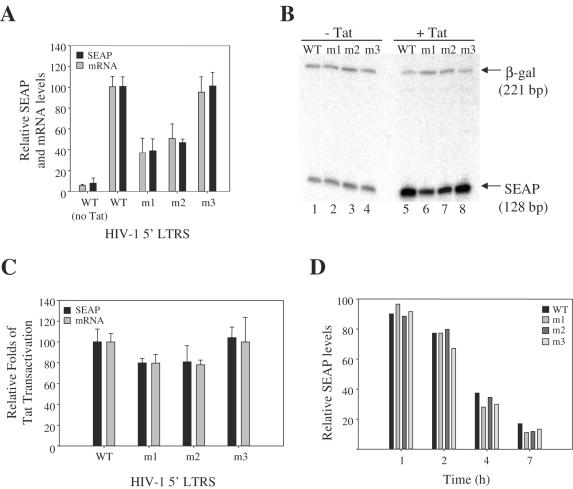
Wild-type and mutant HIV-1 5′-LTR-SEAP plasmids were transfected into 293T cells with pSV-β-galactosidase as a control for transfection efficiency. Co-transfections were performed with or without pCMV-Tat-HA, which expresses a functional Tat–hemagglutinin (HA) fusion protein. The effects of the various HIV-1 5′-LTR mutants were determined both on the levels of SEAP mRNA expressed and SEAP protein produced. Each co-transfection was performed in duplicate with SEAP, β-galactosidase activity and total protein concentration assayed in one set of experiments, and SEAP mRNA and total RNA levels measured in a mirroring set of experiments. β-Galactosidase and total protein levels were consistent among all transfections analyzed, suggesting that the HIV-1 5′-LTR mutants did not affect protein expression in general (data not shown). The presence of Tat increased the transcriptional activity from the wild-type HIV-1 promoter, resulting in 10- to 20-fold increase in SEAP protein levels. Mutants m1 and m2, which did not contain the bulged structure of TAR31, showed a 50–60% reduction in Tat-induced SEAP expression (Figure 7A). Mutant m3, which was structurally wild-type for both TAR31 and TAR59, showed SEAP levels comparable to wild-type HIV-1 promoter. Total RNA isolated from cells transfected with wild-type or mutant 5′-LTRs was subjected to [α-32P]dCTP RT–PCR to quantify SEAP and β-galactosidase mRNA levels (Figure 7A and B). β-Galactosidase mRNA was a control for total RNA extracted from each sample. In the absence of the TAR31 structure (m1 and m2 mutants), a 40–50% reduction in SEAP mRNA levels was observed that was highly correlative to the reduced SEAP expression observed with the m1 and m2 mutants (Figure 7A). The m3 mutant showed wild-type mRNA levels, indicating that the reduction in mRNA levels associated with the m1 and m2 mutants was specific to the loss of the TAR31 structure. These experiments show that the decreased SEAP expression observed when deleting the TAR31 region stems from defects at the transcriptional level and not from defects in translation.
Figure 7.
In vivo functional studies of alternative TAR. TAR31-deficient mutant HIV-1 LTRs express lower RNA levels but do not affect Tat-mediated transactivation, mRNA export or cytoplasmic mRNA degradation. (A) Levels of SEAP reporter protein and mRNA in 293T cells transfected with wild-type and mutant HIV-1 5′-LTR-SEAP constructs in the presence or absence of Tat (see text for details). SEAP values were corrected for protein concentration and β-galactosidase activity to measure transfection efficiency. Expression levels were normalized to wild-type HIV-1 5′-LTR expression levels of SEAP in the presence of Tat, which was assigned a value of 100. Each set of experiments was repeated at least six times. mRNA was isolated from cells transfected with wild-type and mutant HIV-1 5′-LTR and quantified by RT–PCR (B). mRNA levels were normalized to mRNA levels detected from the wild-type HIV-1 5′-LTR in the presence of Tat, which was assigned a value of 100. Values were obtained from at least five different sets of experiments and normalized to β-galactosidase mRNA levels to control for transfection efficiency. (B) Visualization of RNA levels transcribed from wild-type and mutant HIV-1 5′-LTRs. A 128 nt region downstream from the HIV-1 5′-LTR promoter in wild-type and mutant HIV-1 5′-LTR-SEAP plasmids and a 221 nt region of the β-galactosidase coding domain in the β-galactosidase control vector were separately reverse transcribed and amplified. PCR products were labeled with [α-32P]dCTP during the amplification step and loaded on to a 6% denaturing urea gel. Lanes 1–4 show PCR products (WT, m1, m2 and m3, respectively) from cells not expressing Tat whereas lanes 5–8 products were from cells expressing Tat. (C) Relative measure of Tat transactivation using the SEAP reporter gene assay and mRNA quantification assays. Tat transactivation was calculated as the ratio of SEAP or mRNA levels in the presence of Tat and SEAP or mRNA levels in the absence of Tat, and measured as described in Materials and Methods. (D) Rate of cytoplasmic mRNA degradation. Wild-type and mutant HIV-1 5′-LTR-SEAP vectors were transfected into 293T cells. At 24 h post-tranfection, media were replaced with fresh media containing 1 µM actinomycin D and cells were further incubated at 37°C for 1, 2, 4 or 7 h. At each indicated time, media were replaced with fresh actinomycin D-containing media; after 20 min, 20 µl aliquots were collected and tested for SEAP activity as described in Materials and Methods. After 7 h all media were removed. Cells were washed with 1× PBS and lysed for β-galactosidase detection and protein quantification as described in Materials and Methods.
TAR31 lacks a hexanucleotide loop structure and did not form a ternary complex with Tat and Cyclin T1 in vitro (data not shown), suggesting that Tat–TAR31 complexes were not involved in CycT1–Tat–TAR59-mediated transactivation. We tested this hypothesis by measuring the relative levels of Tat transactivation for wild-type and mutant 5′-LTR-SEAP constructs. Tat transactivation was established by comparing SEAP protein expression and mRNA levels in the presence and absence of Tat. The m1 and m2 mutants exhibited levels of Tat transactivation that were only slightly lower than wild-type TAR31-containing 5′-LTR promoters (Figure 7C), indicating that TAR31 did not have a role in Tat transactivation. The basal activities of the mutant 5′-LTR promoters lacking TAR31 were lower compared to TAR31-containing 5′-LTRs, which raises the possibility that TAR31-deficient mRNA transcripts were less stable than wild-type transcripts. Nuclear, non-mature TAR31-deficient mRNA may be degraded at a higher rate, or may be exported from the nucleus at a lower frequency, increasing their vulnerability to nucleases. Alternatively, cytoplasmic, mature TAR31-deficient mRNA may be targeted for degradation at a higher rate then wild-type TAR mRNA.
To address whether TAR31-deficient mRNAs had defects in export or were less stable in the cytoplasm, cells transfected with wild-type or mutant 5′-LTR promoters were treated with the general transcription inhibitor actinomycin D (1 µM) 24 h post-transfection. SEAP activity was used to evaluate the half-life of mature mRNA in the cytoplasm since we had already shown that SEAP protein levels correspond to mRNA levels. After adding actinomycin D, media were removed from cells at various time points and assayed for SEAP activity. To rule out the possibility of SEAP protein accumulation and/or degradation in the media, older media were replaced with fresh media 20 min before taking aliquots assayed for SEAP activity. SEAP activity decreased with increased exposure time to actinomycin D for all wild-type and mutant constructs tested. However, no significant differences in SEAP activity were observed for any of the constructs tested at any one particular time point after the addition of actinomycin D (Figure 7D). These results show that the rates of cytoplasmic mRNA degradation for wild-type and TAR31-deficient mRNA were the same. These results also ruled out a function for TAR31 in either mRNA export or stabilization of mature cytoplasmic mRNA. Taken together, these experiments show that TAR31 has a role in the control of HIV-1 mRNA expression during early transcription.
DISCUSSION
In this study, we identified a novel RNA structure resembling an inverted TAR that interacts specifically with Tat protein, can terminate transcription in the absence of Tat, and is important for the control of HIV-1 mRNA transcription. This analysis demonstrates that during transcription elongation, mRNA transcripts fold into different conformations to regulate gene expression. The success of the methodology established here exemplifies how in vitro transcription and cross-linking chemistry can be combined to study RNA Pol II complexes at different stages of transcription. Furthermore, our site-specific photo-cross-linking experiments provided a window for capturing RNA–protein interactions during specific steps of transcription, and Tat–TAR recognition served as a functional test to identify alternative TAR RNA structures. An RNA folding program and structural probing experiments then revealed the structural formation of the inverted TAR.
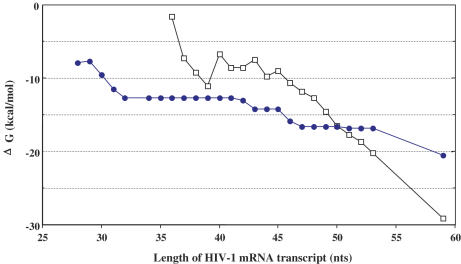
The free energy of TAR31 and wild-type TAR59 folding within RNA transcripts that varied in length was also calculated using the mfold program (31) (Figure 8). When RNA transcripts available for folding were shorter than 50 nt, RNA structures containing TAR31 had a lower free energy than TAR59, suggesting that TAR31 predominates at early stages of transcription elongation. For a 50 nt RNA transcript, the free energy of the two structures was identical and we would predict that both structures co-exist in equal ratio. When an RNA transcript grows longer than 50 nt, the free energy of TAR59 decreases dramatically, indicating that TAR59 should be most predominant at these longer lengths. The yields obtained from cross-linking experiments supported the formation of these predicted RNA structures. If TECs protect 12 nt of the RNA transcript, TECs stalled at U46 and C71 would expose RNA transcripts containing 36 and 59 nt, respectively. Thirty six free nucleotides should fold into TAR31 while 59 nt should favor TAR59 formation, and both should provide stable Tat-interacting regions. Correspondingly, the high yield of U46 and C71 cross-link products was indicative of stable formation of TAR31 and TAR59, respectively. TECs stalled at U42 should form a TAR31-like structure with a few nucleotides less than full-length TAR31. TECs halted at C61 should expose 49 nt, which most probably adopted TAR31 and TAR59 conformations since the difference in free energy values of both TAR structures was not significant (2 kcal/mol).
Figure 8.
Free energy of TAR31 and TAR59 structures in HIV-1 mRNA transcripts of different lengths. Graph shows the free energy values of TAR31 (closed circles) and TAR59 (open squares) secondary structure formation with respect to transcript length (between 28 and 59 nt). See Discussion for details.
TECs stalled at A51, which expose 39 nt transcripts, produced the lowest yields of RNA–protein cross-links. Two possibilities may explain this result. First, small differences in the free energies (1.6 kcal/mol) of the TAR31 and TAR59 conformations raises the possibility that the 39 nt transcript co-existed in both conformations. In having such a dynamic structure, 39 nt may not form a stable Tat-interacting site, explaining the low yield of cross-link products. An alternative explanation stems from an elegant study that showed the first 44 nt of HIV-1 RNA folded into an alternative structure called the HIV-1 pause hairpin (34). This hairpin structure required only 39 nt to form a stable structure. The free energy values for HIV-1 pause hairpin formation and TAR31-containing structures were −12.6 and −12.7 kcal/mol, respectively, which suggested that the pause hairpin and TAR31 co-existed when TECs were stalled at A51. The pause hairpin structure does not contain a trinucleotide bulge region and should not be recognized by Tat. Therefore, formation of this structure would decrease Tat–TAR31 cross-link efficiency. These two explanations for the lower cross-link yield with the A51 mutant are not mutually exclusive. Collectively, these analyses strongly suggest that TAR31 forms when the active center of the TEC reaches U42, wild-type TAR RNA forms after 50 nt are released, and intermediate, dynamic structures form when median length RNA becomes free from TECs. When the full-length TAR structure becomes available for Tat binding, formation of the P-TEFb–Tat–TAR ternary complex should shift the equilibrium towards the TAR59 structure and prevent the formation of alternate structures.
The Tat–TAR31 cross-link and the EMSA experiments suggest that Tat can bind to HIV-1 transcripts before TAR59 formation. The in vivo mutant HIV-1 5′-LTR analysis also indicated that the TAR31 bulge was functionally important. In the SEAP assays, HIV-1 5′-LTR mutants lacking the bulge structure of TAR31 showed half the activity of promoters containing the bulge, suggesting that TAR31 functions at the level of transcription. The decreased mRNA levels associated with TAR31-deficient transcripts could have been caused by defects in transcription initiation, Tat transactivation, mRNA transport or transcript stability. However, TAR31 did not form a detectable complex with Cyclin T1 and Tat transactivation was only slightly decreased when TAR31 was mutated. These results suggested that TAR31 was not required for assembling P-TEFb/Tat/TAR complexes, for Tat transactivation, or transcription initiation. TAR31 was also not required for mRNA export or cytoplasmic mRNA stability since wild-type and TAR31-deficient cytoplasmic mRNAs degraded at similar rates. The exact role of TAR31 remains an open question, but it is clear that it is involved during early elongation step. We propose two hypotheses for future work: (i) TAR31 structure is important to stabilize the short early transcripts before the transcription complex commits for processive elongation; and (ii) TAR 31 actively promotes transcription elongation in an as yet unknown mechanism.
In conclusion, the study presented here shows that mRNA folding during transcription can be a determinant for efficient gene expression. This study also revealed that RNA serves as a dynamic receptor for varying RNA–protein interactions. Finally, our new experimental strategy for studying mRNA conformation changes during transcription can be applied to investigate the folding and function of nascent RNA structures transcribed from other promoters.
Acknowledgments
The GST–Tat vectors were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS (NIAID, NIH) which were made available by Dr Andrew Rice. We thank Drs Bryan Cullen, John Karn, B. M. Peterlin, David Price and Andrew Rice for reagents; Donal Luse, Murali Palangat and Robert Landick for their help in establishing the stepwise transcription in our laboratory; and Tamara J. Richman for editorial assistance. We also thank Bryan Cullen, B. M. Peterlin, David Price and Stewart Shuman for helpful discussions. This work was supported in part by NIH grants (AI 43198 and AI 41404) to T.M.R. Funding to pay the Open Access publication charges for this article was provided by the NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tinoco I., Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 2.Tinoco I., Jr Physical chemistry of nucleic acids. Annu. Rev. Phys. Chem. 2002;53:1–15. doi: 10.1146/annurev.physchem.53.082001.144341. [DOI] [PubMed] [Google Scholar]
- 3.von Hippel P.H. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 4.Conaway J.W., Shilatifard A., Dvir A., Conaway R.C. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- 5.Cullen B.R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 6.Rana T.M., Jeang K.T. Biochemical and functional interactions between HIV-1 Tat protein and TAR RNA. Arch. Biochem. Biophys. 1999;365:175–185. doi: 10.1006/abbi.1999.1206. [DOI] [PubMed] [Google Scholar]
- 7.Wei P., Garber M.E., Fang S.M., Fischer W.H., Jones K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 8.Mancebo H.S., Lee G., Flygare J., Tomassini J., Luu P., Zhu Y., Peng J., Blau C., Hazuda D., Price D., et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng J., Marshall N.F., Price D.H. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M.B., Price D.H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann C.H., Carroll R.G., Wei P., Jones K.A., Rice A.P. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 1998;72:9881–9888. doi: 10.1128/jvi.72.12.9881-9888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrmann C.H., Rice A.P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann C.H., Rice A.P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X., Gold M.O., Tang D.N., Lewis D.E., Aguilar-Cordova E., Rice A.P., Herrmann C.H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc. Natl Acad. Sci. USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X., Herrmann C.H., Rice A.P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J. Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richter S., Cao H., Rana T.M. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1–Tat–TAR ternary complex formation. Biochemistry. 2002;41:6391–6397. doi: 10.1021/bi0159579. [DOI] [PubMed] [Google Scholar]
- 17.Richter S., Ping Y.H., Rana T.M. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl Acad. Sci. USA. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao S.H., Price D.H. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 2001;28:28345–28348. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 19.Bieniasz P.D., Grdina T.A., Bogerd H.P., Cullen B.R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl Acad. Sci. USA. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keen N.J., Gait M.J., Karn J. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc. Natl Acad. Sci. USA. 1996;93:2505–2510. doi: 10.1073/pnas.93.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Rana T.M. DNA damage-dependent transcriptional arrest and termination of RNA polymerase II elongation complexes in DNA template containing HIV-1 promoter. Proc. Natl Acad. Sci. USA. 1997;94:6688–6693. doi: 10.1073/pnas.94.13.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deissler H., Behn-Krappa A., Doerfler W. Purification of nuclear proteins from human HeLa cells that bind specifically to the unstable tandem repeat (CGG)n in the human FMR1 gene. J. Biol. Chem. 1996;271:4327–4334. doi: 10.1074/jbc.271.8.4327. [DOI] [PubMed] [Google Scholar]
- 24.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Rana T.M. RNA conformation in the Tat–TAR complex determined by site-specific photo-cross-linking. Biochemistry. 1996;35:6491–6499. doi: 10.1021/bi960037p. [DOI] [PubMed] [Google Scholar]
- 26.Cullen B.R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 27.Cujec T.P., Cho H., Maldonado E., Meyer J., Reinberg D., Peterlin B.M. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol. Cell. Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu Y.L., Ho C.K., Saha N., Schwer B., Shuman S., Rana T.M. Tat stimulates cotranscriptional capping of HIV mRNA. Mol. Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Wang X., Rana T.M. Protein orientation in the Tat–TAR complex determined by psoralen photocross-linking. J. Biol. Chem. 1996;271:16995–16998. doi: 10.1074/jbc.271.29.16995. [DOI] [PubMed] [Google Scholar]
- 30.Gu W., Wind M., Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl Acad. Sci. USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuker M. Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 32.Toohey M.G., Jones K.A. In vitro formation of short RNA polymerase II transcripts that terminate within the HIV-1 and HIV-2promoter-proximal downstream regions. Genes Dev. 1989;3:265–282. doi: 10.1101/gad.3.3.265. [DOI] [PubMed] [Google Scholar]
- 33.Berkhout B., Gatignol A., Rabson A.B., Jeang K.T. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- 34.Palangat M., Meier T.I., Keene R.G., Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]