Abstract
In spite of their importance in RNA metabolism, the function of DExD/H-box proteins (including DEAD-box proteins) is poorly understood at the molecular level. Here, we present recent progress achieved with the five DEAD-box proteins from Escherichia coli, which have been particularly well studied. These proteins, which have orthologues in many bacteria, participate, in particular, in specific steps of mRNA decay and ribosome assembly. In vitro, they behave as poorly processive RNA helicases, presumably because they only unwind a few base pairs at each cycle so that stable duplexes can reanneal rather than dissociate. Except for one of them (DbpA), these proteins lack RNA specificity in vitro, and specificity in vivo is likely conferred by partners that target them to defined substrates. Interestingly, at least one of them is multifunctional, presumably because it can interact with different partners. Altogether, several aspects of the information gathered with these proteins have become paradigms for our understanding of DEAD-box proteins in general.
INTRODUCTION
Putative RNA helicases of the DExD/H family belong to helicase superfamily 2 (SF2), which itself is related in sequence and in structure to helicase superfamily 1 (SF1) (1). DExD/H proteins are widely distributed in nature and they participate in a highly specific way in a variety of processes involving RNA (2–4). DEAD-box proteins constitute a major fraction of all DExD/H-box proteins. They are characterized by a core of ∼350–400 amino acids containing 9 conserved amino acid motifs (4,5). Structural studies have shown that this core folds into two ‘RecA-like domains’ separated by a flexible linker, with the conserved motifs being involved in the binding of ATP or RNA or in inter-domain contacts (3,6). In vitro, all DEAD-box proteins behave as RNA-dependent ATPases and many of them as ATP-dependent RNA helicases, contributing to the general belief that they function to rearrange RNA or ribonucleoprotein (RNP) structures (7). However, at the molecular level, their precise role is rarely understood.
Escherichia coli contains five DEAD-box proteins, and because of the powerful biochemical and genetic approaches available in this organism, it constitutes an attractive model for studying the properties of these proteins and the basis and limits of their biological specificity. Not unexpectedly then, E.coli DEAD-box proteins have been studied extensively. In particular, the role of these proteins in such important cellular processes as mRNA decay or ribosome biogenesis is more clearly understood in E.coli than in other organisms. The current state of this knowledge is presented here.
DEXD/H-BOX PROTEINS IN E.COLI
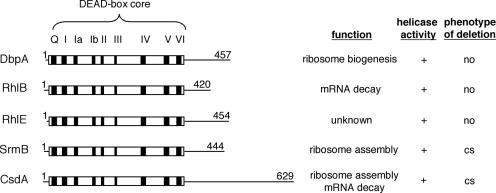
E.coli contains 5 DEAD-box genes [csdA (formerly called deaD; note that CsdA unfortunately also designates another E.coli gene encoding a cysteine sulfinate desulfinase), dbpA, rhlB, rhlE and srmB] and 13 DExH-box genes (8), among which one (hrpA) participates in RNA metabolism, and most others in DNA metabolism. Only proteins involved in RNA metabolism will be considered here. The sequence alignment of the 5 DEAD-box genes (Figure 1) shows that the common DEAD-box core (∼350 amino acids) is flanked by C-terminal extensions that differ both in size (from ∼70 to 290 amino acids) and sequence. These non-conserved extensions are thought to mediate the interactions of DEAD-box proteins with their specific partners or RNA substrates (9), as has been documented for DbpA (see below). Interestingly, the C-terminus of CsdA bears a significant similarity to that of DbpA (10), although the functional significance of this similarity is not known. Besides this similarity, the Protein Family Database of Alignment and hidden Markov models (http://www.sanger.ac.uk/software/pfam) reveal no other known motif in these C-terminal extensions. The N-terminal extensions (2–9 amino acids) also differ in sequence but they are much shorter than the C-terminal extensions and no roles have been attributed to them.
Figure 1.
Summary of structures and properties of the five DEAD-box proteins from E.coli. Regions encompassing conserved residues (i.e. helicase core) are shown as open rectangles; stripes denote the positions of the conserved motifs (Q to VI), whereas the N- and C-terminal extensions (drawn to scale) are shown as thin lines. Also indicated are the proposed functions of the proteins, their in vitro activity (+ means that the protein is able to dissociate an RNA duplex in an ATP-dependent manner), and the phenotype associated with the deletion of the gene (no, no visible growth defect; cs, cold-sensitive growth).
ENZYMATIC PROPERTIES OF THE FIVE E.COLI DEAD-BOX PROTEINS
The five E.coli DEAD-box proteins have been characterized biochemically. They all possess RNA-dependent ATPase and RNA helicase activities, i.e. they are able to dissociate short RNA duplexes in an ATP-dependent manner (11–13). Moreover, except for DbpA (see below), these ATPase and RNA helicase activities show no stringent specificity for a particular RNA in vitro, i.e. they can be detected with artificial RNA substrates. In contrast, DbpA is so far unique amongst DExD/H proteins in requiring a specific RNA motif, namely the hairpin 92 of 23S rRNA, for ATPase and helicase activities (12,14). The current model proposes that the C-terminal domain of DbpA specifically binds hairpin 92, and that in turn this binding assists the interaction of the catalytic core with nearby targets on the 23S rRNA, activating the hydrolysis of ATP and the disruption of RNA structures (12) (Figure 2A). However, the exact physiological target(s) of DbpA on 23S rRNA remain to be identified (15,16). Interestingly, the isolated C-terminal domain of the Bacillus subtilis orthologue of DbpA, YxiN, binds small RNA fragments encompassing helix 92 with the same high affinity and specificity as the full-length protein (17,18). In contrast, the isolated DEAD-box core of YxiN has a weak and non-specific RNA-binding activity that resembles that of other non-specific DEAD-box proteins. Moreover, the C-terminal domain of YxiN is able to impart RNA specificity when appended to the catalytic core of a non-specific DEAD-box protein (10). Altogether, these results indicate that the sequence-specific RNA-binding and helicase activities are separable and that some DEAD-box proteins may be functionally modular (17).
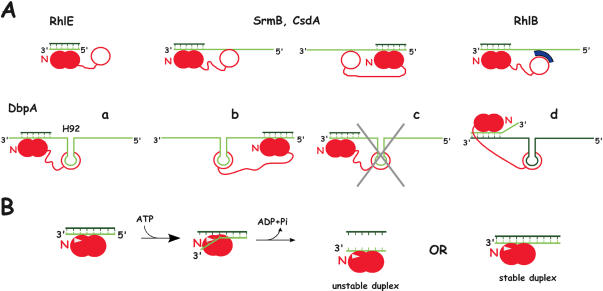
Figure 2.
Models for the unwinding activity of E.coli DEAD-box proteins. (A) Substrate recognition. The helicase is shown in red, with the core RecA-like domains and the C-terminal domain appearing as closed and open circles, respectively. The helicase core is assumed to bind to one strand of the RNA duplex, as observed with the DEAD-box protein Vasa in the presence of ATP (6); this strand is shown in light green, with the complementary strand in dark green. Moreover, the relative orientations of the core and bound RNA are assumed to be the same as in Vasa, i.e. the N-terminal domain (N) interacts with the 3′ side of the RNA. For the sake of simplicity, putative secondary binding sites (SrmB, CsdA, RhlB) are assumed to bind the same strand as the core, although this is not necessarily the case [cf DbpA, d; (23)]. (i) RhlE can unwind blunt end duplexes, and therefore its core presumably interacts directly with one strand of the duplex. (ii) SrmB and CsdA require single-stranded extensions, but these extensions can be either 5′ (left) or 3′ (right) to the duplex. We propose that in this case also the core interacts directly with one strand of the duplex, and that the extensions are used not for translocation but for binding the helicase via a secondary RNA binding site, which, by analogy with DbpA (see below), is shown here within the C-terminal domain. (iii) RhlB is inactive as a helicase unless it binds RNase E or a fragment thereof (in blue) carrying an arginine-rich region. This fragment may constitute a secondary RNA binding site that interacts with single-stranded extensions, as proposed above. The RhlB region that contacts RNase E has not been determined; it is assumed here to lie in the C-terminal domain. (iv) The DbpA C-terminal domain binds tightly to helix 92 (H92) of 23S rRNA, and the protein can unwind duplexes that are located either 3′ (a) or 5′ (b) to the helix. DbpA is unique amongst E.coli DEAD-box helicases in strictly requiring a 3′ single-stranded extension: substrates lacking such extension cannot be unwound (c). However, this extension does not need to lie on the same strand as helix 92 (d). (B) Model explaining why most DEAD-box helicases are poorly processive and do not require a single-stranded extension of definite polarity [based on (6)]. In the presence of ATP, the protein binds tightly to RNA, which can be either single- or double-stranded. However, the presence of a ‘wedge’ (white triangle) encompassing motif Ib (Figure 1) induces a kink in the bound RNA that is locally incompatible with double strandedness. Therefore, bound duplexes unwind over a few bases. The protein may preferentially bind near the duplex ends so that thermal fraying assists unwinding. After ATP hydrolysis, the partially unwound duplex is free to dissociate (‘unstable duplex’) or reanneal (‘stable duplex’), depending upon its stability. At this stage, the protein may either dissociate, or remain RNA-bound as illustrated here.
It is noteworthy that the helicase activity of CsdA, RhlE, DbpA and SrmB decreases very sharply with duplex length (12,13). A quantitative study with RhlE shows that appending only 3 extra base pairs to an 11mer duplex, i.e. increasing the unwinding energy from 18 to 24 kcal/mol, reduces the unwinding rate by as much as two orders of magnitude; meanwhile, the amount of ATP required to unwind individual duplexes increases by an even larger factor (13). Presumably, these DEAD-box proteins unwind a few base pairs during each cycle of ATP hydrolysis, as observed for several non DEAD-box helicases of the SF1 or SF2 families [e.g. with the NS3 SF2 helicase, 2–5 bp are opened per ATP hydrolysed (19)]. However, in contrast to these latter helicases, most of which are involved in DNA or RNA replication and exhibit robust, processive activity, DEAD-box proteins are apparently unable to prevent the collapse of unwound regions after each cycle of ATP hydrolysis. As a result, the final outcome—duplex reannealing or dissociation—depends dramatically upon duplex stability (Figure 2B). This reversible character of the unwinding reaction is presumably not due to the low affinity of the helicases for the conventional substrates used in unwinding assays, which would cause fast helicase dissociation; indeed, DbpA, which binds very tightly (in the 10 nM range) to RNA substrates consisting of short duplexes appended to helix 92 of 23S rRNA, is still unable to unwind these duplexes beyond a relatively modest stability threshold [i.e. 15 bp or 29 kcal/mol; (12)]. Altogether, these proteins appear poorly processive as RNA helicases, a common characteristic of DEAD-box proteins (20). A processive activity may not be physiologically relevant, however, since in natural RNA perfect helices rarely exceed 10 bp in length.
Most well-studied replicative SF1 or SF2 helicases (e.g. NS3, Rep, PcrA) require a single-stranded extension 3′ to the duplex to initiate unwinding (21), and popular models for helicase activity postulate that these proteins basically function as ATP-driven 3′→5′ translocases that travel over single-stranded nucleic acids, opening duplexes as they encounter them (21,22). The helicase activity of DbpA also requires a single-stranded extension 3′ to the duplex (23) (Figure 2A). In contrast, like most other DEAD-box proteins that have been studied in this respect, CsdA and SrmB can unwind duplexes carrying indifferently 3′ or 5′ extensions, whereas RhlE can unwind even blunt-end duplexes, again a behaviour not unique amongst DEAD-box proteins (13) (Figure 2A). Altogether, the low processivity of unwinding and the absence of requirement for single-stranded extension of definite polarity pinpoint a very different unwinding mechanism for DEAD-box proteins and for other SF2 and SF1 helicases, in spite of the relatedness of all these proteins. Indeed, it is hard to imagine that the active site of DEAD-box proteins can catalyse translocation in either 3′→5′ or 5′→3′ directions. Rather, these proteins must be able to interact directly with double-stranded RNA, though biochemical data (24) suggest that they only contact one of the two strands, presumably either of them, at a time (Figure 2A). Supporting a direct interaction with double-stranded RNA, a recent structural study on the Drosophila DEAD-box protein Vasa in the presence of ATP and oligo(U) shows that the protein binds RNA mainly via the backbone, not the bases, and therefore that it can in principle accommodate double-stranded as well as single-stranded RNA (6). However, an obstructing alpha helix (the ‘wedge’), which encompasses motif Ib and is structurally conserved amongst DEAD-box proteins, induces a kink in the RNA that would be incompatible with double-strandedness throughout the active site. Based on these and on biochemical observations, it has been proposed that DEAD-box proteins unwind double-stranded RNA not as a result of a translocation, but simply by binding RNA—whether single- or double-stranded—and then causing a reversible ATP-dependent constraint in RNA conformation that unwinds it over a few bases (6,13,24) (Figure 2B). This mechanism nicely explains the low ‘processivity’ of DEAD-box proteins as RNA helicases. However, it does not explain why DbpA requires a single-strand 3′ extension for helicase activity. Presumably, the interpretation of this odd behaviour will require structural information on DbpA itself.
Why do CsdA and SrmB require single-standed extensions (whether 3′ or 5′) for helicase activity, if these extensions are not used for translocation? Of note, these extensions must be rather long in order to trigger the ATPase and helicase activities, particularly for SrmB (25–35 nt) (13). An attractive interpretation is that CsdA and SrmB can bind RNA through additional binding site(s) besides the RNA-binding track of the helicase core. This binding would then strengthen RNA binding to the core, provided the RNA substrate is long enough to bridge the two sites (Figure 2A). In this respect, CsdA and SrmB would resemble DbpA, except that their secondary RNA-binding sites would lack sequence/structure specificity.
In contrast to the other four DEAD-box proteins, RhlB lacks detectable ATPase and RNA helicase activities on its own, but it can be stimulated by its interaction with the C-terminal half (CTH) of RNase E (11,25). In vivo, this interaction takes place within the degradosome (see below). Interestingly, studies with different RNase E fragments reveal that the stimulation of the helicase activity requires the presence of an arginine-rich region of RNase E (residues 789–820), which might contribute to RNA binding [(25); A. J. Carpousis, personal communication] and thus facilitate the interaction of the RNA substrate with the helicase core as proposed for DbpA, CsdA and SrmB (Figure 2A). RhlB is not the only example of a DEAD-box helicase stimulated by a protein partner. Indeed, the eukaryotic translation initiation factors eIF4B and eIF4H stimulate the RNA unwinding activity of eIF4A (26).
FUNCTIONS OF THE E.COLI DEAD-BOX PROTEINS
Current evidence shows that the E.coli DEAD-box proteins (and one DEAH-box protein, HrpA) participate in mRNA processing and decay, in ribosome biogenesis, and possibly in translation initiation. CsdA is unique in having been implicated in all three processes. To date, no function has been attributed to RhlE.
mRNA processing and decay
RhlB belongs to a ribonucleolytic multienzymatic complex called the RNA degradosome, which also comprises RNase E, the major endonuclease involved in mRNA decay, and polynucleotide phosphorylase (PNPase), a 3′–5′ exoribonuclease (27–29). The presence of RhlB within the degradosome and its specific activation by RNase E (see above), presumably direct RhlB to its specific targets, i.e. mRNAs. In vitro and in vivo, RhlB facilitates PNPase-mediated degradation of fragments bearing very stable secondary structures at their 3′ end, such as REP (repeated extragenic palindrome) elements (27,30,31). This activity, which requires ATP hydrolysis, is believed to reflect the RhlB-mediated unwinding of these structures that otherwise would block PNPase. RhlB also facilitates RNase E cleavage of ribosome-free, highly RNase E-sensitive mRNAs (32). Similar to PNPase, RNase E is single-strand specific, and RhlB may counteract the collapse of its cleavage sites into double-stranded structures. However, it should be stressed that RhlB is not required for activity of PNPase or RNase E in general.
In vivo, cleavage of mRNAs by RNase E (or degradosome) is generally hampered by the presence of ribosomes, presumably reflecting a competition for mRNA substrate (33,34). In contrast, a new mode of endonucleolytic cleavage, in which the ribosome plays an active role (‘killer ribosome’), has been documented recently (33). Briefly, certain nascent peptides can interact with the polypeptide exit channel, causing ribosome stalling and ultimately mRNA cleavage. Using a genetic screen to identify the factors involved, Moseley and co-workers (35) showed that the DEAH-box protein HrpA participates in the ribosome-mediated cleavage of the daa mRNA. Presumably, this helicase assists the activity of a ribosome-bound endonuclease, which may be the ribosome itself. However, the underlying mechanism remains unknown.
The cold-shock protein DEAD-box protein A, CsdA, has been implicated in the stabilization of mRNAs when overexpressed (36,37). More recently, CsdA has been shown to associate with the RNA degradosome after cold shock (38), indicating that it may play a role in mRNA decay at low temperature in vivo. Consistent with this idea, CsdA can replace RhlB in an in vitro assay for degradosome function (38). It remains to be determined whether RNase E can bind the two RNA helicases simultaneously or whether each of them is exclusive of the other. It is interesting to note, in this respect, that CsdA has been shown to bind to RNase E at a site different from RhlB (39).
In addition to CsdA, SrmB and RhlE might also participate in mRNA decay. As for CsdA, overexpression of SrmB stabilizes certain mRNAs (36). Moreover, SrmB and RhlE can also bind to RNase E, and RhlE can replace RhlB in the degradosome assay in vitro (39). Finally, these three proteins also interact in vitro with E.coli poly(A) polymerase, an enzyme which facilitates the exonucleolytic degradation of RNA fragments (40).
Ribosome biogenesis
The srmB and csdA genes were originally isolated as multicopy suppressors of mutations in ribosomal protein genes rplX (encoding L24) and rpsB (encoding S2), respectively (41,42), suggesting a role of these DEAD-box genes in ribosome biogenesis. Our laboratory showed that SrmB and CsdA are indeed involved in the assembly of the large ribosomal subunit at low temperature (43,44). The deletion of srmB or csdA leads to a severe deficit of free 50S subunits and accumulation of 40S particles corresponding to incompletely assembled large subunits. Moreover, in wild-type cells grown at low temperature, SrmB and CsdA co-sediment with pre-50S ribosomal particles, suggesting that their role in assembly is direct. Interestingly, the ribosomal protein (r-protein) content of the ΔsrmB and ΔcsdA 40S particles is different. Among those proteins missing in the ΔsrmB 40S particle is L13, an r-protein incorporated early during in vitro assembly (45), whereas only r-proteins that are incorporated late are missing from the ΔcsdA 40S particle. These results suggest that SrmB acts earlier than CsdA during assembly.
A third E.coli DEAD-box protein, DbpA, is likely involved in the biogenesis of the large ribosomal subunit. As described above, its ATPase and RNA helicase activities in vitro are specifically stimulated by hairpin 92 of the 23S rRNA, which lies at the peptidyl transferase center of the ribosome (46). No stimulation is observed with the fully assembled ribosome, suggesting that DbpA may play its role before completion of ribosome synthesis (47). Consistently, the Uhlenbeck group has isolated a dominant-negative mutant that affects assembly of the 50S subunit in vivo (L. M. Sharpe and O. C. Uhlenbeck, personal communication). However, since at 37 or 20°C the deletion of dbpA alters neither growth nor ribosome profile (O. C. Uhlenbeck, personal communication; I. Iost, unpublished data), the exact conditions under which DbpA is required for 50S biogenesis remain to be determined.
It is likely that SrmB, CsdA and DbpA assist 50S assembly by modulating RNA or RNP structures. Indeed, their unwinding activity may be required to facilitate structural transitions within the RNA and/or to allow proper binding of r-protein(s). Alternatively, these ‘RNA helicases’ may act as RNA chaperones (48) that prevent and/or resolve misfoldings. Indeed, like other large RNAs, rRNA may become trapped in incorrect structures, and presumably requires assistance to reach its active conformation (48,49). An important question regarding these RNA helicases is how they are directed to their specific targets. As noted above, the tight interaction of DbpA with hairpin 92 is presumably the basis of its specificity, and biochemical studies suggest that its physiological target is contained within a 153 nt fragment of 23S rRNA carrying helices 89–93 (47). Our in vivo data (43) suggest that SrmB might act on the structure of 23S rRNA in the vicinity of L20/L13 r-protein-binding sites. However, in contrast to DbpA, no specific RNA substrate has been found for SrmB; an attractive possibility is that SrmB interacts with an r-protein that targets it to its substrate.
The deletion of srmB or csdA leads to a cold-sensitive phenotype (Figure 1). This feature is consistent with an RNA chaperone or helicase activity, since such activities should be most significant at low temperature where RNA structures are more stable. Interestingly, at low temperature, the growth of the ΔcsdA strain is depressed more severely than that of the ΔsrmB strain, even though the defect in ribosome biogenesis appears milder. This suggests that the slow growth of the ΔcsdA strain under these conditions is due primarily to another effect, presumably a defect in mRNA decay (44).
CsdA, a multifunctional DEAD-box protein
Besides its roles in mRNA decay and 50S ribosome biogenesis, CsdA has also been proposed to play a role in the biogenesis of the small ribosomal subunit. As stated above, the csdA gene was initially identified as a multicopy suppressor of a mutation in the rpsB gene encoding r-protein S2 (42), and it was subsequently reported that CsdA overexpression restores the incorporation of S2 (and S1) into the ribosome (50). However, our laboratory has found that the composition of the 30S subunit, and in particular the amounts of r-proteins S1 and S2, was not affected by the deletion of the chromosomal csdA gene (44). It is thus possible that overexpression of CsdA relaxes its specificity so that in addition to its genuine physiological role it can fulfill other functions, e.g. in 30S assembly. Another proposed role for CsdA is to assist translation: in vitro studies suggest that it promotes translation initiation of structured mRNAs (51). Finally, CsdA has been shown to be involved in the regulation of expression of certain genes (including heat- and cold-shock proteins) at low temperature (52,53). However, whether all these potential roles of CsdA reflect direct effects remains to be demonstrated.
CsdA is not the only DEAD-box protein that is involved in several different biological processes (9). An emerging idea is that DEAD-box proteins are simply motor proteins that are directed to their specific targets (and possibly regulated) by their partners, as documented for DbpA and RhlB whose activity is determined by helix 92 and by RNase E, respectively. Those DEAD-box proteins that are able to interact with several partners could then play multiple roles: CsdA may be an example of this situation.
ARE THE E.COLI RNA HELICASES ESSENTIAL AND ARE THEY INTERCHANGEABLE IN VIVO?
In contrast to the yeast Saccharomyces cerevisiae where most of the DEAD-box proteins are essential, no member of this family is essential to E.coli under standard laboratory conditions at 37°C. Indeed, rhlB (31), rhlE (I. Iost, unpublished data) and dbpA (O. C. Uhlenbeck, personal communication; I. Iost, unpublished data) can be individually deleted without a noticeable effect on growth, whereas the deletion of srmB and csdA leads to a growth defect at low temperature only (Figure 1). Yet, these genes have been conserved during evolution (see below). It is possible that their function is required only under specific, yet unidentified physiological conditions, or that one RNA helicase can substitute for the absence of another. To test this latter possibility, we combined some DEAD-box gene deletions with the idea that the double mutants might show a synthetic lethal phenotype. Among the 10 possible combinations, 6 were tested (ΔcsdAΔsrmB, ΔcsdAΔrhlE, ΔcsdAΔdbpA, ΔrhlEΔdbpA, ΔrhlEΔsrmB and ΔsrmBΔdbpA), but in no case was the growth of double mutants slower than that of single mutants (growth was monitored on rich medium plates at low temperatures; I. Iost, unpublished data). In addition, we have shown that overexpression of SrmB does not suppress the cold sensitivity of the ΔcsdA mutant and vice versa (44). As for proteins involved in mRNA decay, it has been shown that RhlB, RhlE and CsdA are interchangeable in the degradation of a structured mRNA fragment in vitro (38,39) but whether this is true in vivo remains to be shown. As the only clear example of in vivo interchangeability, we have found that overexpression of RhlE corrects the cold-sensitive growth of the ΔcsdA strain (I. Iost, unpublished data). It is thus possible that RhlE can replace the function of CsdA in mRNA decay. Interestingly, RhlE was identified as a component of the cold-adapted Pseudomonas syringae degradosome (54). Except for the case of RhlE that has to be examined further, we believe that the E.coli DEAD-box proteins are not generally interchangeable in vivo. This property, which is presumably shared by most members of the family, may be due to their different C-terminal extensions.
PHYLOGENETIC CONSERVATION OF THE FIVE E.COLI DEAD-BOX PROTEINS IN EUBACTERIA
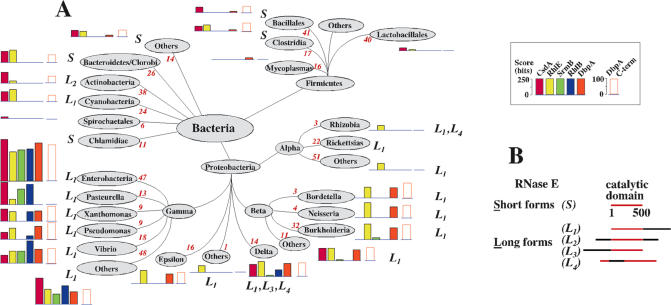
Another way of assessing the physiological importance of the five E.coli DEAD-box proteins is to examine their phylogenetic conservation. To this end, we used Blast analysis (Blast-P, with default settings) to search for possible orthologues of these proteins in annotated microbial genomes (543 eubacterial sequences at the time of analysis; www.ncbi.nih.gov). The probes consisted of the whole sequence of the five proteins, and homology was estimated from BLAST scores, which primarily reflect the number of matches between the candidate sequence and the probe used. When the probes were assayed against one another, scores ranging from 182 to 275 bits were observed, owing to the presence in each probe of the 9 conserved DEAD-box motifs. Therefore, to identify possible orthologues of a given E.coli DEAD-box protein in other bacteria, we only considered homology scores >275 bits, i.e. we only retained candidates whose homology to the probe was higher that the homology between any two E.coli DEAD-box proteins. In Figure 3A, this threshold value has been subtracted from the observed scores to highlight the contribution of motifs specific to individual proteins.
Figure 3.
(A) Distribution of putative orthologues of the E.coli DEAD-box proteins over the bacterial phylogenic tree, as revealed by BLAST analysis. The whole sequence of the proteins (coloured rectangles) or, for DbpA, the C-terminal extension (open rectangles; see Figure 1 and text for details) have been used as probes. The tree was adapted from a web page of The National Center for Biotechnology Information, National Library of Medicine, USA (www.ncbi.nlm.nih.gov). For a given DEAD-box protein (see inset for the colour code) and for each phylum, class or order, the height of the histogram is proportional to the highest score observed for a representative of this phylum, class or order. The scores obtained with the whole sequence probes have been corrected to take into account the homology that exists between unrelated DEAD-box proteins (see text). Red figures (italics) correspond to the number of sequences in the databank. Note that eventually bacteria distant from E.coli contain other DEAD-box helicases whose sequence are unrelated to those of E.coli and therefore have not been detected here. The mention ‘S’ or ‘L’ following the histogram refers to RNase E, the genuine partner of RhlB in E.coli. ‘S’ (Short) means that all identified RNase E orthologues consist of the catalytic region only, whereas ‘L’ (Long) means that at least one RNase E representative carries an extension of >100 residues with no homology to the catalytic core of E.coli RNase E (first 500 residues). Such extension could act as a scaffold for the assembly of the degradosome. The subscript (1–4) refers to the position of the extension with respect to the catalytic core (see B). (B) Schematic drawing showing the different configurations of RNase E observed in the phylogenetic tree.
Using these criterias, RhlB and SrmB had the narrowest phylogenetic representation among the five E.coli DEAD-box proteins: they occur almost exclusively in gamma-proteobacteria, with less significant hits in beta- and delta-proteobacteria (Figure 3A). Moreover, within gamma-proteobacteria, they are less conserved in Xantomonadales and Pseudomonadales. The early separation of these orders from other gamma-proteobacteria presumably explains this divergence. Indeed, Xantomonadales are distant from other gamma-proteobacteria from the viewpoint of their 16S rRNA sequence (55) and of other elements of their translational apparatus; the latter is also true for Pseudomonadales (56,57).
CsdA and RhlE have a broader phylogenetic distribution than SrmB and RhlB. Putative CsdA orthologues with relatively high scores were found not only in Proteobacteria (classes beta- gamma- and delta-), but also in distant phyla such as Cyanobacteria, Actinobacteria, Bacteroides/Chlorobia and Firmicutes (Figure 3A). RhlE also showed broad distribution, but with several unusual features. As expected, its highest scores occurred in enterobacteria, as for other E.coli DEAD-box proteins. However, with very few exceptions, mainly from Alteromonadales and Oceanospirillales ('others gammaproteobacteria' in Figure 3A) the next most significant hits were found not in other gamma-proteobacteria but in beta- (Bulkhoderia, Neisseria), delta- and epsilon-proteobacteria and also in the distant phylum Cyanobacteria. An intriguing possibility is that the enterobacterial gene was transferred horizontally from one of these distant bacteria after enterobacteria diverged from other gamma-proteobacteria. Another unique feature of RhlE is its multiple occurrence (up to six) in some organisms, particularly beta-proteobacteria, and, within gamma-proteobacteria, in Alteromonadales and Vibrionaceae. As a consequence, the average number of RhlE-like proteins in organisms where they could be detected exceeded 2 (511 hits per 249 organisms), whereas for other DEAD-box proteins this figure was close to 1. Since no role has been attributed to RhlE (see above), the significance of these multiple occurrences remains elusive.
DbpA-like proteins were readily identified in all classes of proteobacteria except alpha-proteobacteria. However, in other phyla, only hits of borderline significance were observed. In particular, YxiN, the B.subtilis DEAD-box helicase that is functionally equivalent to DbpA (see above) was not detected. In an effort to increase the sensitivity of the detection, we decided to use the C-terminal extension of DbpA, which carries the helix 92 binding site, as an alternative probe for BLAST analysis. The same library of annotated genomes was probed with the last 119 amino acids of the DbpA protein. Only candidates annotated as DEAD-box or SF2 helicase and showing scores ≥30 bits (corresponding to a probability of random occurrence of ∼0.01 per microbial genome) were retained. Under these conditions, the probe was specific: with the E.coli genome, it produced no hits with DEAD-box proteins other than DbpA. With proteobacteria, the phylogenetic pattern produced with this probe was qualitatively very similar to that obtained with the full-length probe (Figure 3A). However, scores were smaller, revealing that besides the C-terminal region, the helicase core of DbpA also contain motifs that contribute to the identity of the protein. Beyond proteobacteria, the C-terminal probe became more efficient than the full-length probe for detecting DbpA-like proteins (Figure 3A). This observation suggests that, in distant phyla, DbpA-specific motifs are less conserved within the helicase core than within the C-terminal extension. Interestingly, this behaviour seems unique to DbpA; with other E.coli DEAD-box proteins, the phylogenetic distribution produced with either the full-length or C-terminal probes were more similar (not illustrated). Of note, provided the C-terminal probe was used, the phylogenetic distribution of DbpA-like proteins was as broad as for CsdA or RhlE (Figure 3A).
It should be stressed that, according to the above analysis, many bacterial orders or species showed no evidence for orthologues of individual E.coli DEAD-box helicases. There may be two reasons for this situation. First, as noted above, these proteins are dispensable for E.coli in rich medium at 37°C and they presumably only confer an advantage under special conditions (e.g. at low temperature). Hence, they may not be needed in organisms experiencing a constant, high temperature lifestyle: this may explain, for example, the loss of DbpA from the Pasteurellales, all of which are parasites or symbionts of mammals. Second, proteins functionally equivalent to E.coli DEAD-box helicases may in fact be more widespread than suggested above, because what is important for function is not the conservation of the protein sequence per se but that of interactions with specific partners. This point is best illustrated with RhlB. Although this protein is essentially confined to gamma-proteobacteria, its molecular partner, RNase E, is much more widely distributed. Some orders or phyla only contain the catalytic domain (‘short’ form) of RNase E, but in many other cases this domain carries non-catalytic extensions that may serve as degradosome scaffolds and therefore are potential RhlB partners (‘long’ forms) (58,59) (Figure 3A and B). Of note, all bacterial orders or phyla having putative RhlB orthologues also contain ‘long’ RNase E forms with which these orthologues might interact; conversely, orders or phyla containing only ‘short’ RNase E forms, or lacking RNase E altogether, also lack recognizable RhlB (Figure 3A). More intriguing is the situation where long forms of RNase E can be identified whereas the RhlB partner cannot. RNase E has been purified and characterized in two of these cases. In the gamma-proteobacterium P.syringae, in which we detected no RhlB (C-terminal probe) or only a RhlB with very low score (full-length probe), RNase E was reported to bind a DEAD-box protein resembling RhlE (see above). Similarly, whereas we detected no RhlB-like proteins in alpha-proteobacteria, biochemical studies on the alpha-proteobacterium Rhodobacter capsulatus reveal the existence of a degradosome with two uncharacterized DEAD-box proteins (60). It is likely that, in both cases, these RNase E-associated DEAD-box helicases are true functional equivalent of RhlB from E.coli, even if they do not resemble it.
CONCLUSION
Whereas in most cases the general processes involving the DEAD-box proteins from E.coli or S.cerevisiae have been identified, the precise molecular role of these proteins remains obscure. A major goal for the future is to identify their physiological targets and the way they find them. In particular, the factors that interact with these proteins must be identified. A considerable advantage of E.coli compared to other organisms is the possibility of reconstituting in vitro some reactions that involve RNA helicases, such as ribosome assembly (45) or exonucleolytic mRNA degradation (30). These systems provide a basis for assessing the role of the RNA helicases in specific processes.
Whereas the above BLAST searches show that E.coli DEAD-box genes have orthologues in many other bacteria, none were found in eucaryotic organisms such as the yeast S.cerevisiae. Yet, some helicases from yeast may be functionally related to those of E.coli. Thus, the yeast exosome, a key player in RNA degradation and processing whose multisubunit catalytic core is related to bacterial PNPase, contains putative RNA helicases (61). Whether these proteins play exactly the same role as RhlB in the degradosome remains to be seen. As for ribosome biogenesis, it involves as many as 15 DEAD-box helicases in yeast. Presumably, some of them are required for reactions that have no counterpart in E.coli, such as the dissociation of the small nucleolar RNAs from pre-rRNA or the nuclear export of the ribosomes (62,63). Determining whether others play the same role as SrmB, CsdA or DbpA in ribosome assembly or r-protein incorporation, will be an interesting issue for future work.
Acknowledgments
We are much indebted to F. Loussala for his help in BLAST searches and analysis and to Drs C. Condon and A. J. Carpousis for critical reading of the manuscript. We thank Prof. O. C. Uhlenbeck for communication of unpublished data. This work has been funded by CNRS, ENS and by grants from the Agence Nationale de la Recherche and from the Ministère de l'Education Nationale, de la Recherche, et de la Technologie (ACI ‘DRAB’) to M.D. Funding to pay the Open Access publication charges for this article was provided by CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gorbalenya A.E., Koonin E.V. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 2.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 3.Rocak S., Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nature Rev. Mol. Cell. Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 4.Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 6.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 7.Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 8.Perutka J., Wang W., Goerlitz D., Lambowitz A.M. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J. Mol. Biol. 2004;336:421–439. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Silverman E., Edwalds-Gilbert G., Lin R.J. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 10.Kossen K., Karginov F.V., Uhlenbeck O.C. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 11.Vanzo N.F., Li Y.S., Py B., Blum E., Higgins C.F., Raynal L.C., Krisch H.M., Carpousis A.J. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bizebard T., Ferlenghi I., Iost I., Dreyfus M. Studies on three E.coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 14.Nicol S.M., Fuller-Pace F.V. The ‘DEAD box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polach K.J., Uhlenbeck O.C. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 16.Karginov F.V., Uhlenbeck O.C. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karginov F.V., Caruthers J.M., Hu Y., McKay D.B., Uhlenbeck O.C. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 18.Wang S., Hu Y., Overgaard M.T., Karginov F.V., Uhlenbeck O.C., McKay D.B. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumont S., Cheng W., Serebrov V., Beran R.K., Tinoco I., Jr, Pyle A.M., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers G.W., Jr, , Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 21.Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 22.Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diges C.M., Uhlenbeck O.C. Escherichia coli DbpA is a 3′→5′ RNA Helicase. Biochemistry. 2005;44:7903–7911. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- 24.Rogers G.W., Jr, Lima W.F., Merrick W.C. Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 25.Vanzo N.F. Toulouse, France: Université Paul Sabatier de Toulouse; 1999. Le dégradosome d'ARN: étude structurale et fonctionnelle d'un complexe impliqué dans la dégradation de l'ARN chez Escherichia coli. [Google Scholar]
- 26.Rogers G.W., Jr, , Richter N.J., Lima W.F., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 27.Py B., Higgins C.F., Krisch H.M., Carpousis A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 28.Miczak A., Kaberdin V.R., Wei C.-L., Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpousis A.J., Vanzo N.F., Raynal L.C. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- 30.Coburn G.A., Miao X., Briant D.J., Mackie G.A. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khemici V., Carpousis A.J. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- 32.Khemici V., Poljak L., Toesca I., Carpousis A.J. Evidence in vivo that the DEAD-box RNA helicase RhlB facilitates the degradation of ribosome-free mRNA by RNase E. Proc. Natl Acad. Sci. USA. 2005;102:6913–6918. doi: 10.1073/pnas.0501129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deana A., Belasco J.G. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- 34.Dreyfus M., Joyce S. Translation Mechanisms. Georgetown, TX, USA: Landes Bioscience/Eureka.com; 2003. The interplay between translation and mRNA decay in prokaryotes: a discussion on current paradigms; pp. 165–183. [Google Scholar]
- 35.Koo J.T., Choe J., Moseley S.L. HrpA, a DEAH-box RNA helicase, is involved in mRNA processing of a fimbrial operon in Escherichia coli. Mol. Microbiol. 2004;52:1813–1826. doi: 10.1111/j.1365-2958.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- 36.Iost I., Dreyfus M. mRNAs can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 37.Brandi A., Spurio R., Gualerzi C.O., Pon C.L. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 1999;18:1653–1659. doi: 10.1093/emboj/18.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prud'homme-Genereux A., Beran R.K., Iost I., Ramey C.S., Mackie G.A., Simons R.W. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol. Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 39.Khemici V., Toesca I., Poljak L., Vanzo N.F., Carpousis A.J. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 40.Raynal L.C., Carpousis A.J. Poly(A) polymerase I of Escherichia coli: characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- 41.Nishi K., Morel-Deville F., Hershey J.W.B., Leighton T., Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- 42.Toone W.M., Rudd K.E., Friesen J.D. deaD, a Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charollais J., Pflieger D., Vinh J., Dreyfus M., Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 44.Charollais J., Dreyfus M., Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nierhaus K.H. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 46.Tsu C.A., Kossen K., Uhlenbeck O.C. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsu C.A., Uhlenbeck O.C. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 48.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder R., Barta A., Semrad K. Strategies for RNA folding and assembly. Nature Rev. Mol. Cell. Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 50.Moll I., Grill S., Grundling A., Blasi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol. 2002;44:1387–1396. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- 51.Lu J., Aoki H., Ganoza M.C. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int. J. Biochem. Cell Biol. 1999;31:215–229. doi: 10.1016/s1357-2725(98)00142-3. [DOI] [PubMed] [Google Scholar]
- 52.Jones P.G., Mitta M., Kim Y., Jiang W., Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc. Natl Acad. Sci. USA. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka K., Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 2001;183:2808–2816. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purusharth R.I., Klein F., Sulthana S., Jager S., Jagannadham M.V., Evguenieva-Hackenberg E., Ray M.K., Klug G. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 2005;280:14572–14578. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- 55.Olsen G.J., Woese C.R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tchufistova L.S., Komarova A.V., Boni I.V. A key role for the mRNA leader structure in translational control of ribosomal protein S1 synthesis in gamma-proteobacteria. Nucleic Acids Res. 2003;31:6996–7002. doi: 10.1093/nar/gkg883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asai T., Zaporojets D., Squires C., Squires C.L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Condon C., Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K., Cohen S.N. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- 60.Jager S., Fuhrmann O., Heck C., Hebermehl M., Schiltz E., Rauhut R., Klug G. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 2001;29:4581–4588. doi: 10.1093/nar/29.22.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Symmons M.F., Williams M.G., Luisi B.F., Jones G.H., Carpousis A.J. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 62.El Hage A., Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 63.Kos M., Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]