Abstract
DNA helicases are required for virtually every aspect of DNA metabolism, including replication, repair, recombination and transcription. A comprehensive description of these essential biochemical processes requires detailed understanding of helicase mechanisms. These enzymes are ubiquitous, having been identified in viruses, prokaryotes and eukaryotes. Disease states, such as xeroderma pigmentosum, Cockayne's syndrome, Bloom's syndrome and Werner's syndrome, have been linked to defects in specific genes coding for DNA helicases. Helicases have been placed into different subfamilies based on sequence comparison. The largest subgroups are termed superfamily 1 and superfamily 2. A proposed mechanism for helicases in these classes has been described in terms of an ‘inchworm model’. The inchworm model includes conformational changes driven by ATP binding and hydrolysis that allow unidirectional translocation along DNA. A monomeric form of the enzyme is proposed to have two DNA-binding sites that enable sequential steps of DNA binding and release. Significant differences exist between helicases in important aspects of the models such as the oligomerization state of the enzyme with some helicases functioning as monomers, some as dimers and others as higher-order oligomers.
INTRODUCTION
Helicase mechanisms
DNA and RNA helicases are a ubiquitous yet diverse group of enzymes present in viruses, bacteria and eukaryotes (1–5). They are defined by their ability to catalyze the unwinding of duplex nucleic acids. Helicases convert chemical energy of nucleoside triphosphate (NTP) hydrolysis to the mechanical energy necessary to transiently separate the strands of duplex nucleic acids. Helicases provide the single-stranded DNA (ssDNA) intermediates necessary for replication, recombination and repair. Different helicases can be distinguished by co-factor utilization, substrate preference, directionality of unwinding, processivity and effects of other proteins on their activity. There are six known human diseases caused by defective helicases. The clinical abnormalities in these diseases are quite diverse, suggesting that different processes involving DNA manipulation are defective (6). Some helicases are encoded by viruses and are targets for antiviral drug development such as herpes simplex virus and hepatitis C virus (7,8). The prominent role of both human and viral helicases in human disease, coupled with the central importance of these proteins in the most basic aspects of nucleic acid metabolism, makes this class of enzymes an attractive target for study.
Substantial progress has been made in the past five years by a number of research groups studying helicase mechanisms. Much of the biochemical knowledge of DNA helicases comes from the study of bacterial and phage enzymes. Two recent review articles from Delagoutte and von Hippel provide excellent summaries of many current models for helicase function (2,3). Helicases can function as monomers, dimers or higher-order oligomers (9–11), and translocate along ssDNA with different directional biases (12–17). There are enormous differences in processivity among helicases, and the mechanistic and kinetic details of translocation and unwinding also vary considerably. RecBCD helicase, for example, utilizes two motors bound to each strand of the DNA duplex to create a rapid and highly processive helicase (18,19), which has been visualized at the single molecule level (20,21). Hexameric helicases appear to exclude one strand of the duplex from the central channel of the hexamer to unwind double-stranded DNA (dsDNA), although they can accommodate both strands and function as molecular DNA pumps in other cases (4,22–24). A dimeric inchworm mechanism has been proposed for UvrD and Rep helicases (10,25). PcrA helicase functions efficiently as a monomer during translocation on ssDNA (12,26). T4 Dda and HCV NS3 helicases are functional as monomers but more processive when acting cooperatively (27–29). Some helicases have been found to function as molecular motors that can displace proteins in their paths (30–33).
SF1 AND SF2 HELICASES CAN FUNCTION AS MONOMERS, DIMERS OR LARGER OLIGOMERS
Many SF1 and SF2 helicases are proposed to function via ‘inchworm’ mechanisms that require coordinated alternate binding of nucleic acid at two different sites within the functional unit of the helicase. This can be accomplished by a monomer containing two binding sites or by a dimer with a single binding site per subunit. Both variations of this mechanism have been supported by structural and biochemical studies to account for the activities of different helicase enzymes.
The most thorough structural characterization of an SF1 helicase is for PcrA, which is essential for replication in Bacillus subtilis and Staphylococcus aureus. Based on X-ray crystallographic, biochemical and biophysical studies, a detailed description of the mechanism for PcrA has been proposed (5,9,26,34,35). The mechanism describes specific conformational changes within the DNA-binding site that are coupled to ATP binding and hydrolysis. DNA passes through a complex active site by 1 bp for each ATP hydrolyzed. Disruption of the duplex occurs as the ssDNA is pulled unidirectionally through the enzyme. Comparison of a ‘substrate’ complex structure (ADPNP-bound) and a ‘product’ complex structure (sulfate-bound) reveals conformational changes associated with ATP hydrolysis consistent with an inchworm mechanism for translocation (9). Nucleotide binding results in closure of the binding cleft and repositioning of DNA-binding domains in a manner that increases the affinity of the enzyme for duplex DNA. Translocation is proposed to occur via alternate binding of DNA at two different sites within the PcrA monomer coordinated by ATP binding and hydrolysis.
The dimeric inchworm model for Escherichia coli UvrD and Rep helicases is based on extensive structural and biochemical studies including single molecule experiments (10,25). Rep helicase has been crystallized in two different conformations bound to ssDNA (36). As with PcrA, these conformational differences suggest that nucleotide binding and hydrolysis cyclically alter the nucleic acid binding properties of the enzyme in order to regulate translocation. However, dimer formation is required for Rep activity in vitro (37), with nucleotide co-factors regulating cooperativity of nucleic acid binding to the two sites within each dimer (38). Biochemical and single molecule studies of UvrD have demonstrated that dimer formation is also critical for helicase activity of this enzyme (10). Monomeric UvrD binds specifically to ssDNA/dsDNA junctions, but association of a second monomer with the protein–DNA complex is necessary for unwinding to occur. The low stability of the dimeric complex is proposed to account for the low processivity of UvrD helicase.
Some helicases are active as monomers but exhibit enhanced unwinding activity under conditions in which multiple monomers are able to function cooperatively (27–29). The unwinding mechanisms of these helicases are not as well characterized as those of the strictly monomeric or dimeric helicases. In the case of Dda, cooperativity is presumed to be associated with transient interactions between monomers bound to the same nucleic acid substrate molecule and traveling in the same direction on DNA despite the absence of any observed quaternary structure (27,28). Chemical cross-linking and gel filtration chromatography indicate that Dda does not form an oligomeric species in solution (39). Unwinding activity of wild-type Dda was not significantly reduced by the addition of an ATPase-deficient mutant, indicating no dominant-negative effect (39). Unwinding by Dda under pre-steady-state conditions exhibited biphasic kinetics with the amplitude of the first phase equivalent to the enzyme concentration (40). Taken together, these results indicate that oligomerization of Dda is not required for DNA unwinding.
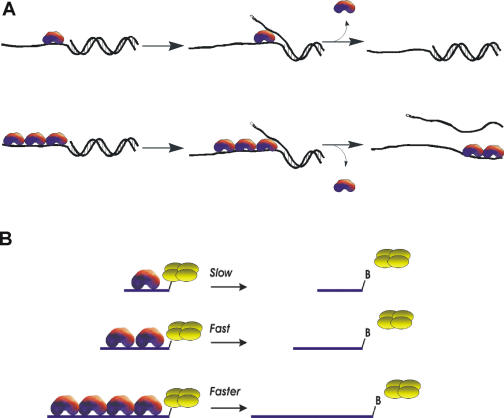
However, Dda exhibits greater activity for unwinding partial-duplex substrates that are of sufficient length to allow binding of multiple monomers (Figure 1) (28). Multiple molecules of Dda can unwind more substrate under single-turnover conditions because of the enhanced likelihood that a molecule will complete the unwinding process before dissociating from the substrate. Streptavidin displacement activity also increases as substrate length increases (27). These results indicate that the binding of adjacent molecules along the DNA strand contributes to the increased unwinding or displacement activity of Dda via transient protein–protein interactions (Figure 1A) or through functional cooperativity (Figure 1B). The authors have proposed a variation of the inchworm model for helicase activity by Dda termed the ‘cooperative inchworm’ model. The essence of this model is that helicase monomers can function independently during translocation, but when the monomers encounter a ‘challenge', such as duplex DNA or a protein block, multiple monomers enhance the likelihood of overcoming the challenge. As the challenge increases in difficulty, such as with the displacement of streptavidin, then the importance of cooperativity in enzyme activity increases.
Figure 1.
Cooperative inchworm model for activities exhibited by Dda. (A) Increased activity is observed for DNA unwinding, bypass of DNA lesions and for the displacement of DNA-binding proteins. Enhanced unwinding can occur due to increased enzymatic activity, due to protein–protein interactions or simply due to increased probability for unwinding. When monomeric Dda dissociates from the substrate, reannealing can occur if insufficient base pairs are unwound. When multiple Dda molecules are bound, unwinding can continue after dissociation of one of the monomers. (B) Multiple molecules assembled along the ssDNA function together to increase the rate of displacement of streptavidin from biotin-labeled oligonucleotides.
Hepatitis C virus NS3 helicase exhibits similar functional cooperativity characteristics. Levin and Patel (41) demonstrated that both ATPase and unwinding activities of NS3 are dependent on protein concentration and that an ATPase-deficient mutant has a dominant-negative effect on unwinding by wild-type NS3. The authors proposed an unwinding mechanism involving formation of a transient dimeric species. Tackett et al. (42) showed that efficient unwinding by NS3 requires binding of multiple monomers per nucleic acid substrate molecule. Mackintosh et al. (43) crystallized two NS3 helicase monomers with a weak protein interface bound to a single oligonucleotide. Mutation of the protein–protein interface resulted in reduced replication of HCV subgenomic replicons in vivo. However, the mutations at the interface did not dramatically influence helicase activity in vitro.
INTERACTION OF HELICASES WITH NUCLEIC ACID
Most helicases require an ssDNA overhang adjacent to duplex DNA in order to initiate unwinding. The strand containing the overhang is referred to as the loading strand whereas the complementary strand is referred to as the displaced strand. The specifics of the interactions of particular helicase enzymes with the loading and/or displaced strands vary considerably depending on the unwinding mechanism of each helicase.
PcrA helicase interacts with both single-stranded and duplex regions of the unwinding substrate (9). Mutation of the duplex interaction domains impairs unwinding activity but does not affect translocation on ssDNA (44). In contrast, deletion of a homologous domain from Rep helicase does not impair unwinding activity (45), suggesting that the sort of duplex interaction required for unwinding by PcrA may not be critical to the Rep helicase unwinding mechanism.
Several studies have examined the effects of modified nucleic acid substrates on unwinding by helicase enzymes. Replacing the displaced strand with a peptide nucleic acid (PNA) mimic has no effect on unwinding by Dda helicase. Dda was able to unwind DNA–PNA substrates at rates similar to those observed for DNA–DNA substrates (46), indicating that the rate-limiting step for unwinding of short oligonucleotides by Dda is insensitive to the chemical nature of the displaced strand and the thermal stability of oligonucleotide substrates. DNA unwinding by Dda can occur simply as a consequence of translocation along the loading strand. The effects of modification of the loading strand of DNA on unwinding by Dda helicase have also been studied using a 5′-DNA–PNA–DNA-3′ chimera. A single molecule of Dda is not able to translocate efficiently through the DNA lesion. However, multiple Dda molecules can bypass the lesion with no reduction in unwinding rate compared to that observed for a DNA substrate (47). This implies that the bypass of the lesion is not due to an increase in the number of chances for bypass, but instead is due to a change in the activity of the lead molecule of helicase, consistent with the cooperative inchworm model.
Strand specificity for the SF2 RNA helicase NPH-II has been investigated by introducing specific chemical modifications into each strand of the RNA substrate. NPH-II required physical continuity of the phosphodiester linkages, suggesting that this enzyme tracks along the RNA backbone (48). The effects of inserting polyglycol linkers into unwinding substrates have also been investigated. Rep helicase is able to initiate unwinding of a DNA substrate despite the presence of polyglycol lesions in the 3′-overhang region of the loading strand (49). NS3 helicase is able to translocate through polyglycol lesions in the duplex region of the loading strand of an RNA substrate (50), indicating that translocation by these enzymes does not require continuous specific interaction with the unwinding substrate. Vaccinia helicase NPH-II, in contrast, cannot translocate past polyglycol discontinuities (50), suggesting a translocation mechanism significantly different from that of Rep and NS3. Specificity for RNA unwinding by some SF2 helicases has been investigated. NPH-II appears to recognize the ribose moieties to distinguish DNA from RNA (51).
One of the major mechanistic issues related to helicase activity is the kinetic and physical step size of the enzyme. The physical step size is the number of base pairs unwound during a single ATP hydrolysis event whereas the kinetic step size is the number of bases unwound per rate-limiting kinetic cycle. The kinetic step size and the physical step size may not be equivalent and depend on the specific mechanism utilized. This area of investigation has been pioneered by the Lohman laboratory (52,53), and kinetic step sizes of 4–5 bp have been determined for the UvrD (52) and RecBCD helicases (54). A recent study from the Bujalowski laboratory has determined the kinetic step size of a hexameric helicase, DnaB, to be 1 bp (55). These investigators made the observation that several base pairs near the end of a duplex can melt spontaneously owing to thermal instability and the presence of the enzyme bound to the DNA.
Recent studies of T4 Dda and HCV NS3 helicases indicate that translocation by monomeric helicases may proceed in a series of non-uniform steps punctuated by pauses of varying lengths (56–58). Most recently, a model incorporating Brownian motion has been proposed for the NS3 helicase domain and the gene 4 helicase from bacteriophage T7 (59,60). This proposed mechanism does not require two intrinsic binding sites for nucleic acid within the active site of a helicase, unlike the mechano-chemical mechanism described by the inchworm model.
HELICASES DISPLACE NUCLEIC ACID BINDING PROTEINS
Helicases and other enzymes that move along DNA or RNA are likely to encounter proteins that are bound to the nucleic acid. The resulting protein–protein collision can lead to dissociation of the helicase from the DNA, displacement of the DNA-binding protein or temporary stalling of the motor. The biological relevance of such protein–protein interactions is gaining attention of researchers in this field. Enzymes containing helicase motifs have been shown to remove proteins bound to DNA (30,31) and RNA (32,33). Chromatin remodeling proteins share regions of homology to helicases (61). Chromatin remodelers disrupt the interaction between DNA and histones but do not necessarily unwind dsDNA. A number of models have been proposed for how remodeling can occur including nucleosome sliding, nucleosome dissociation or histone replacement with a variant histone. Examples of chromatin remodelers are the SWI2/SNF2 class of enzymes (62). One helicase-like protein that is involved in DNA repair is the Mfd protein (63). When RNA polymerase is stalled at a site of DNA damage, Mfd is thought to ‘push’ the polymerase to correctly position the enzyme past the site of damage. Alternatively, Mfd can completely displace RNAP from the DNA, thereby allowing DNA repair to occur.
Kaplan and O'Donnell (64) showed that DnaB can displace dsDNA-binding proteins during translocation on dsDNA. However, specific interactions with other proteins can impede DnaB. For example, DnaB is stopped upon encountering the replication terminator protein Tus (65). The ability of DnaB to dislodge proteins from DNA was proposed to facilitate branch migration in DNA recombination or DNA repair. In Saccharomyces cerevisiae, termination of replication forks is controlled in a sequence-specific fashion by the interaction of the Fob1p replication terminator protein (66). Stalled replication forks can be released by the Rrm3p helicase (termed a ‘sweepase’). The action of Rrm3p is modulated by other proteins such as Tof1p–Csm3p complex (67). Hence, a complex series of protein collisions controls the number of replication forks that can proceed through normal termination sites in this organism.
Two putative yeast helicases, Sub2 and Prp28, reportedly disrupt RNA–protein complexes during spliceosomal assembly (68,69). Structural reorganization of ribonucleoprotein complexes was catalyzed in vitro by the activity of the RNA helicase NPH-II (33). The ability of RNA helicases to remove proteins from dsRNA in the absence of RNA unwinding has also been demonstrated previously (32). It is likely that many functions of RNA helicases await discovery.
The ability of helicases to displace proteins bound to DNA was used to investigate the mechanism of Dda helicase. Dda was found to catalyze dissociation of streptavidin from the 3′ end of a biotinylated oligonucleotide, but not from the 5′ end (15). These results indicated that Dda travels with a strong directional bias on ssDNA and illustrated that helicases generate force during translocation (Figure 1B). Other helicases such as NS3 were later shown to translocate in the opposite direction as Dda, but were also capable of displacing streptavidin (16). It was initially surprising to determine that helicases can displace such tightly bound proteins as streptavidin from biotinylated DNA. However, there exists a relationship between force production and disruption of normal equilibria such that even a small force can lead to a substantial change in the equilibrium binding constant for a protein and its ligand (70). In this case, the protein was streptavidin and the ligand was biotin. However, the same principle applies to proteins binding to DNA. Dda can displace proteins from DNA including the E.coli lac repressor (71) and the E.coli Ter protein (72). However, not all protein–DNA complexes are displaced by Dda. Dda was unable to dislodge a GAL4–DNA complex, even under conditions which should favor binding of more than one molecule of Dda to the substrate (73). Therefore, some specific protein–DNA complexes are able to sequester Dda in a manner that appears to trap Dda and reduce the ATP hydrolysis activity of the enzyme. It is possible that the GAL4–DNA complex adopts a unique structure that perturbs Dda's interaction with DNA. The mechanism for protein displacement by Dda helicase was investigated by designing a substrate containing a DNA-binding site for the E.coli trp repressor (74). The monomeric form of Dda was insufficient to displace the E.coli trp repressor from dsDNA under single-turnover conditions. When the substrate was designed to allow more than one Dda helicase to bind, trp repressor was readily displaced. These results indicate that multiple Dda molecules act to displace DNA-binding proteins in a manner that correlates with previously reported DNA unwinding activity and streptavidin displacement activity.
SUMMARY
In summary, helicases in the superfamily 1 and superfamily 2 classes have been proposed to function in an inchworm fashion. The fundamental aspects of this mechanism can be accommodated by monomeric forms of these enzymes; however, some helicases function as dimers and there is still debate regarding the role of specific subdomains in the overall mechanism for DNA unwinding. The activity of some of these helicases increases through functional cooperativity when multiple molecules assemble along ssDNA. Future work in this area will focus on the interaction of helicases with other DNA-binding proteins, such as single-stranded binding proteins, polymerases and recombinases. It is likely that these interactions will modulate helicase activity significantly, as has already been shown for PcrA and Rep helicases (75,76).
Acknowledgments
We thank members of the Raney laboratory for helpful discussions and comments. This work was funded by NIH grant R01 GM59400 and NIH grant R01 AI060563 (K.D.R.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 2.Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 3.Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 4.Patel S.S., Picha K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 5.Soultanas P., Wigley D.B. Unwinding the ‘Gordian knot’ of helicase action. Trends Biochem. Sci. 2001;26:47–54. doi: 10.1016/s0968-0004(00)01734-5. [DOI] [PubMed] [Google Scholar]
- 6.Ellis N.A. DNA helicases in inherited human disorders. Curr. Opin. Genet. Dev. 1997;7:354–363. doi: 10.1016/s0959-437x(97)80149-9. [DOI] [PubMed] [Google Scholar]
- 7.De Francesco R., Rice C.M. New therapies on the horizon for hepatitis C: are we close? Clin. Liver. Dis. 2003;7:211–242, xi. doi: 10.1016/s1089-3261(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 8.Kleymann G. New antiviral drugs that target herpes virus helicase primase enzymes. Herpes. 2003;10:46–52. [PubMed] [Google Scholar]
- 9.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 10.Maluf N.K., Fischer C.J., Lohman T.M. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 11.Bujalowski W., Klonowska M.M., Jezewska M.J. Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 12.Dillingham M.S., Wigley D.B., Webb M.R. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 13.Fischer C.J., Maluf N.K., Lohman T.M. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004;344:1287–1309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Kim D.E., Narayan M., Patel S.S. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 15.Morris P.D., Raney K.D. DNA helicases displace streptavidin from biotin-labeled oligonucleotides. Biochemistry. 1999;38:5164–5171. doi: 10.1021/bi9822269. [DOI] [PubMed] [Google Scholar]
- 16.Morris P.D., Byrd A.K., Tackett A.J., Cameron C.E., Tanega P., Ott R., Fanning E., Raney K.D. Hepatitis C virus NS3 and simian virus 40 T antigen helicases displace streptavidin from 5′-biotinylated oligonucleotides but not from 3′- biotinylated oligonucleotides: evidence for directional bias in translocation on single-stranded DNA. Biochemistry. 2002;41:2372–2378. doi: 10.1021/bi012058b. [DOI] [PubMed] [Google Scholar]
- 17.Young M.C., Schultz D.E., Ring D., von Hippel P.H. Kinetic parameters of the translocation of bacteriophage T4 gene 41 protein helicase on single-stranded DNA. J. Mol. Biol. 1994;235:1447–1458. doi: 10.1006/jmbi.1994.1100. [DOI] [PubMed] [Google Scholar]
- 18.Dillingham M.S., Spies M., Kowalczykowski S.C. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 19.Taylor A.F., Smith G.R. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 20.Bianco P.R., Brewer L.R., Corzett M., Balhorn R., Yeh Y., Kowalczykowski S.C., Baskin R.J. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 21.Spies M., Bianco P.R., Dillingham M.S., Handa N., Baskin R.J., Kowalczykowski S.C. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 22.Bujalowski W., Jezewska M.J. Interactions of Escherichia coli primary replicative helicase DnaB protein with single-stranded DNA. The nucleic acid does not wrap around the protein hexamer. Biochemistry. 1995;34:8513–8519. doi: 10.1021/bi00027a001. [DOI] [PubMed] [Google Scholar]
- 23.Egelman E.H., Yu X., Wild R., Hingorani M.M., Patel S.S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jezewska M.J., Surendran R., Bujalowska D., Bujalowska W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? J. Biol. Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 25.Ha T., Rasnik I., Cheng W., Babcock H.P., Gauss G.H., Lohman T.M., Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 26.Dillingham M.S., Wigley D.B., Webb M.R. Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- 27.Byrd A.K., Raney K.D. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nature Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 28.Byrd A.K., Raney K.D. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 29.Levin M.K., Wang Y.H., Patel S.S. The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 2004;279:26005–26012. doi: 10.1074/jbc.M403257200. [DOI] [PubMed] [Google Scholar]
- 30.Flores M.J., Sanchez N., Michel B. A fork-clearing role for UvrD. Mol. Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 31.Macris M.A., Sung P. Multifaceted role of the Saccharomyces cerevisiae Srs2 helicase in homologous recombination regulation. Biochem. Soc. Trans. 2005;33:1447–1450. doi: 10.1042/BST0331447. [DOI] [PubMed] [Google Scholar]
- 32.Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 33.Jankowsky E., Gross C.H., Shuman S., Pyle A.M. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 34.Dillingham M.S., Soultanas P., Wigley D.B. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dillingham M.S., Soultanas P., Wiley P., Webb M.R., Wigley D.B. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl Acad. Sci. USA. 2001;98:8381–8387. doi: 10.1073/pnas.131009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korolev S., Hsieh J., Gauss G.H., Lohman T.M., Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 37.Cheng W., Hsieh J., Brendza K.M., Lohman T.M. E.coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 2001;310:327–350. doi: 10.1006/jmbi.2001.4758. [DOI] [PubMed] [Google Scholar]
- 38.Bjornson K.P., Wong I., Lohman T.M. ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E.coli Rep helicase: implications for ATP-driven helicase translocation. J. Mol. Biol. 1996;263:411–422. doi: 10.1006/jmbi.1996.0585. [DOI] [PubMed] [Google Scholar]
- 39.Morris P.D., Tackett A.J., Babb K., Nanduri B., Chick C., Scott J., Raney K.D. Evidence for a functional monomeric form of the bacteriophage T4 DdA helicase. Dda does not form stable oligomeric structures. J. Biol. Chem. 2001;276:19691–19698. doi: 10.1074/jbc.M010928200. [DOI] [PubMed] [Google Scholar]
- 40.Nanduri B., Byrd A.K., Eoff R.L., Tackett A.J., Raney K.D. Pre-steady-state DNA unwinding by bacteriophage T4 Dda helicase reveals a monomeric molecular motor. Proc. Natl Acad. Sci. USA. 2002;99:14722–14727. doi: 10.1073/pnas.232401899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin M.K., Patel S.S. The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 42.Tackett A.J., Chen Y., Cameron C.E., Raney K.D. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J. Biol. Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 43.Mackintosh S.G., Lu J.Z., Jordan J.B., Harrison M.K., Sikora B., Sharma S.D., Cameron C.E., Raney K.D., Sakon J. Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus. J. Biol. Chem. 2006;281:3528–3535. doi: 10.1074/jbc.M512100200. [DOI] [PubMed] [Google Scholar]
- 44.Soultanas P., Dillingham M.S., Wiley P., Webb M.R., Wigley D.B. Uncoupling DNA translocation and helicase activity in PcrA: direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng W., Brendza K.M., Gauss G.H., Korolev S., Waksman G., Lohman T.M. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc. Natl Acad. Sci. USA. 2002;99:16006–16011. doi: 10.1073/pnas.242479399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tackett A.J., Morris P.D., Dennis R., Goodwin T.E., Raney K.D. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 47.Eoff R.L., Spurling T.L., Raney K.D. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- 48.Kawaoka J., Jankowsky E., Pyle A.M. Backbone tracking by the SF2 helicase NPH-II. Nature Struct. Mol. Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 49.Amaratunga M., Lohman T.M. Escherichia coli Rep helicase unwinds DNA by an active mechanism. Biochemistry. 1993;32:6815–6820. doi: 10.1021/bi00078a003. [DOI] [PubMed] [Google Scholar]
- 50.Beran R.K., Bruno M.M., Bowers H.A., Jankowsky E., Pyle A.M. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 51.Kawaoka J., Pyle A.M. Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–649. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ali J.A., Lohman T.M. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 53.Lucius A.L., Maluf N.K., Fischer C.J., Lohman T.M. General methods for analysis of sequential ‘n-step’ kinetic mechanisms: application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 2003;85:2224–2239. doi: 10.1016/s0006-3495(03)74648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucius A.L., Vindigni A., Gregorian R., Ali J.A., Taylor A.F., Smith G.R., Lohman T.M. DNA unwinding step-size of E.coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J. Mol. Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 55.Galletto R., Jezewska M.J., Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 56.Eoff R.L., Raney K.D. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nature Struct. Mol. Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 57.Serebrov V., Pyle A.M. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 58.Dumont S., Cheng W., Serebrov V., Beran R.K., Tinoco I., Jr, Pyle A.M., Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levin M.K., Gurjar M., Patel S.S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nature Struct. Mol. Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 60.Stano N.M., Jeong Y.J., Donmez I., Tummalapalli P., Levin M.K., Patel S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lusser A., Kadonaga J.T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 62.Mohrmann L., Verrijzer C.P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Park J.S., Marr M.T., Roberts J.W. E.coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell. 2002;109:757–767. doi: 10.1016/s0092-8674(02)00769-9. [DOI] [PubMed] [Google Scholar]
- 64.Kaplan D.L., O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 65.Mulugu S., Potnis A., Shamsuzzaman, Taylor J., Alexander K., Bastia D. Mechanism of termination of DNA replication of Escherichia coli involves helicase–contrahelicase interaction. Proc. Natl Acad. Sci. USA. 2001;98:9569–9574. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohanty B.K., Bastia D. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 2004;279:1932–1941. doi: 10.1074/jbc.M309078200. [DOI] [PubMed] [Google Scholar]
- 67.Mohanty B.K., Bairwa N.K., Bastia D. The Tof1p–Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J.Y., Stands L., Staley J.P., Jackups R.R., Jr, Latus L.J., Chang T.H. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 69.Kistler A.L., Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Evans E. Probing the relation between force—lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 71.Yancey-Wrona J.E., Matson S.W. Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res. 1992;20:6713–6721. doi: 10.1093/nar/20.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bedrosian C.L., Bastia D. Escherichia coli replication terminator protein impedes simian virus 40 (SV40) DNA replication fork movement and SV40 large tumor antigen helicase activity in vitro at a prokaryotic terminus sequence. Proc. Natl Acad. Sci. USA. 1991;88:2618–2622. doi: 10.1073/pnas.88.7.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maine I.P., Kodadek T. Inhibition of the DNA unwinding and ATP hydrolysis activities of the bacteriophage T4 DDA helicase by a sequence specific DNA–protein complex. Biochem. Biophys. Res. Commun. 1994;198:1070–1077. doi: 10.1006/bbrc.1994.1152. [DOI] [PubMed] [Google Scholar]
- 74.Byrd A.K., Raney K.D. Displacement of a DNA binding protein by Dda helicase. Nucleic Acids Res. 2006;34:3020–3029. doi: 10.1093/nar/gkl369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scott J.F., Eisenberg S., Bertsch L.L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc. Natl Acad. Sci. USA. 1977;74:193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petit M.A., Dervyn E., Rose M., Entian K.D., McGovern S., Ehrlich S.D., Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]