Abstract
Bacteriophage T7 helicase (T7 gene 4 helicase-primase) is a prototypical member of the ring-shaped family of helicases, whose structure and biochemical mechanisms have been studied in detail. T7 helicase assembles into a homohexameric ring that binds single-stranded DNA in its central channel. Using RecA-type nucleotide binding and sensing motifs, T7 helicase binds and hydrolyzes several NTPs, among which dTTP supports optimal protein assembly, DNA binding and unwinding activities. During translocation along single stranded DNA, the subunits of the ring go through dTTP hydrolysis cycles one at a time, and this probably occurs also during DNA unwinding. Interestingly, the unwinding speed of T7 helicase is an order of magnitude slower than its translocation rate along single stranded DNA. The slow unwinding rate is greatly stimulated when DNA synthesis by T7 DNA polymerase is coupled to DNA unwinding. Using the T7 helicase as an example, we highlight critical findings and discuss possible mechanisms of helicase action.
INTRODUCTION
The stable form of DNA in the cell is double stranded whose complementary strands exist as annealed to each other to ensure stability of the stored genetic information. In order to use the information encoded in the DNA sequence, the individual strands need to be opened. Helicases are the enzymes that catalyze this strand separation reaction using the energy of NTP binding and hydrolysis (1–3). The fundamental nature of this reaction and the requirement for helicases in cell metabolism and propagation make helicases essential components of the cellular proteome. Indeed mutations in many helicases lead to human diseases such as cancer and premature aging (4,5).
Although separating the strands of a duplex DNA for replication, repair and recombination is the most common reaction catalyzed by helicases, these proteins also act during transcription, translation, splicing and in various processes that require structural rearrangement of nucleic acids or nucleic acid–protein complexes (1–3,6). This is probably the reason why mutations in helicases lead to pathologically unrelated diseases. As diverse as their physiological roles may be, many of the activities of helicases can be explained by the ability of helicases to translocate along nucleic acids as nucleic acid motors. Some helicases have the ability to translocate along long stretches of nucleic acid whereas others operate within only a few bases.
Advances in understanding the role of helicases come from identification of new proteins with helicase functions, their characterization, correlation of their mutations with diseases and obtaining molecular level details via structural, biochemical and biophysical methods. In this review, we focus on the mechanism of a ring-shaped helicase from bacteriophage T7. We outline the common structural themes elucidated by various groups and discuss our interpretation of the biochemical findings aiming to construct a working model of how DNA unwinding is catalyzed by this member of the ring-shaped class of helicases.
STRUCTURAL INSIGHTS
Quaternary organization
Bacteriophage T7 codes for most proteins that it requires for the replication of its genome (7). T7 gp4 gene transcript codes for two proteins: gp4A, the full-length product of gp4, has primase and helicase activities and gp4B, a truncated protein that results from a translational initiation at the methionine codon 64 in the gp4 coding sequence, has only helicase activity (8–11). The helicase activity is indistinguishable in the two proteins (12,13) and hence they are generally referred to as T7 helicase. In the presence of dTTP, T7 helicase assembles into a ring-shaped hexamer with a ring diameter of 13 nm and a central channel diameter of 2.5 nm according to electron microscopy studies (14). These dimensions, although suitable for holding ssDNA in the ring center, can also accommodate dsDNA (15). Crystal structures reveal that the nucleotide cofactors bind at the interfaces between the monomers (16,17). In addition to this small region of interaction, each subunit makes most of its contacts with a neighboring subunit by intercalating one of its helices into a helix bundle of the neighboring subunit (16,17).
The ring-shaped T7 helicase displays dramatic structural flexibility and the hexamer can deviate from a 6-fold symmetrical structure (17). The subunits of the ring can adopt different relative conformations depending on their nucleotide ligation states (17). T7 helicase ring adopts a more compact conformation when it binds DNA and dTTP (18). A total of seven structures for three different helicase fragments have been solved (16,17,19). The structure of gp4D, a helicase domain fragment, was solved as a trimer and modeled as a ring-shaped hexamer (17). The 56 kDa gp4B protein (G317V, K318M) crystallized as a ring-shaped heptamer (19) and a shorter helicase domain fragment crystallized as a helical filament (16). This flexibility, together with the tendency of the ring subunits to form asymmetric structures makes ring opening a feasible mechanism for self-assembling around DNA. Indeed, biochemical studies indicate that the preformed helicase ring binds DNA via a ring opening mechanism (20). Whether the break in the helicase ring persists after the ring assembles around the DNA and how it affects its processivity of translocation and unwinding are not known.
The finding that T7 helicase can exist as a heptameric ring raises interesting questions regarding its biological significance. It has been suggested that the heptameric helicase with its enlarged central channel might be the form that translocates along duplex DNA whereas the hexameric form can only translocate along ssDNA (19). A recent study has shown that the fraction of heptameric rings is higher in the presence of dTDP, and hexamers are still the dominant species in the presence of dTTP (21). Furthermore, the heptameric species does not bind efficiently to DNA, even though the extra subunit might have a distint role in the assembly of the helicase ring around DNA.
Monomer fold
Helicases posses a domain topology that is analogous to that of the RecA protein (22–24). To bind a nucleotide, all helicases employ the Walker A (P-loop) and B motifs in addition to a variable number of other signature sequences. In each T7 subunit, two amino acid residues (R504, Y535) are involved in stacking the nucleotide base and about five (S314, G317, K318, S319, T320) from the Walker A motif contribute to the stabilization or coordination of the charges on the nucleoside phosphate groups (Figure 1A). D424 from the Walker B motif acts as the major stabilizer of the Mg2+ ion. It has been proposed that one residue (E343) serves as the initiator of hydrolysis, another one (H465) is a critical component of the connection bridge between DNA and nucleotide binding sites (Figure 1B) and arginine finger (R522) is involved in coordinating the inter-subunit cooperativity (16,17,25). Among these, the critical roles of R522, K318, D424 and E343 have been corroborated via site directed mutagenesis (25–28).
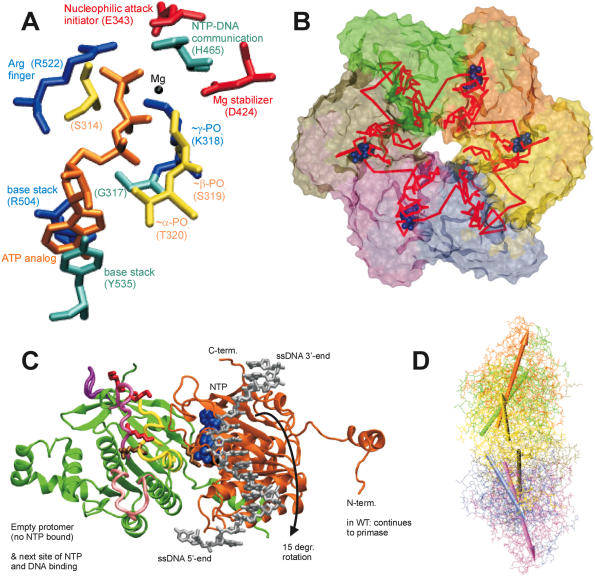
Figure 1.
(A) Critical residues coordinating the bound nucleotide. Amino acids around the nucleotide binding site of T7 helicase are shown with brief notes on their possible functions. The nucleotide is in orange, Mg(II) in black, Arg and Lys in blue, Asp and Glu in red, Ser and Thr in yellow, His, Tyr and Gly in cyan. Among the included residues, 314, 317, 318, 319 and 320 are a part of the Walker A loop, whereas residue 424 is a part of the Walker B motif. All the residues are from the B chain in the strucutre 1e0j (17), except R522 which is from the C chain. (B) Conformational change transmission network in the structural model of the ring-shaped hexameric T7 helicase. All six subunits of the T7 helicase domain are illustrated by a surface representation and the red lines show one of the possible conformational change transmission paths with an undefined (and un-implied) directionality and sequence. The red network simply traces the inter-subunit sensor R522, the hypothetical DNA binding loops (424–439, 464–475, 503–513), the residue that is likely to establish intra-domain nucleotide ligation state communication (H465), a residue proposed to initiate the nucleophilic attack (E343), residues that disrupt helicase functioning when mutated (S345, E348, S496, R487, G488, S345, G451) (27) and eight other amino acids that are critical in coordinating or stabilizing the bound cofactor (S314, G317, K318, S319, T320, D424, R504, Y535) (17). The order of residues is as follows: 522, 451, 496, 488, 487, (475–464), (439–424), (503–413), 314, (317–320), 343, 345, 348, 535, next522. Coloring of the subunits A–F is as follows: yellow, orange, green, tan, mauve, iceblue. (C) Two helicase protomers with a possible delineation of single stranded DNA. Two subunits of the hexameric helicase are viewed from the central channel, their C-termini facing upward. The DNA is oriented with its 5′ end facing down in the figure. DNA is initially bound to the right subunit, which is bound to NTP and thus is in a high affinity state for DNA. The difference in orientations between unbound and NTP docked conformations is a rotation of 15°, which might correspond to the power stroke (17). The directionality of the rotation upon NTP docking (shown with an arrow) agrees with the proposed orientation of DNA. The probability of DNA being caught by the left subunit that supplies the Arginine finger is increased both because of its likelihood of binding to NTP in the next step and because of its vicinity to the center of the ring [5 å closer than the other neighbor (17)]. We took the single DNA strand from NDB code bd0070 and manually positioned it on a dimer from PDB code 1e0j (17). We used VMD and its associated routines (62,63) for visualization and POV-Ray (http://www.povray.org/) for rendering. The ligand-free subunit is in green (chain C in the structure), the NTP bound one is in orange (chain B). The DNA is shown in silver and the nucleotide in blue. The putative DNA binding loops [424–439 in pink (lowermost), 464–475 in mauve (middle), 503–513 in purple (uppermost loop)] (17), the motif H4 (480–500 in yellow) and the mutationally tested residues on them are also indicated (K467, K471, K473 in red R487, R522 in tan) on the C subunit (green subunit). (D) Relative orientations of helicase monomers. To get an idea for the inherent symmetry in a hexameric ring, we can look at the orientations of the six monomers with respect to a common reference, such as the line passing through the central channel through bound DNA. If for all subunits, a chosen line within the monomer that connects one of its residues close to the inner channel to another one distal to the inner channel, is equally perpendicular to the line of DNA, then we would conclude that the monomers are symmetrically arranged with respect to the above chosen definition. When we apply this type of an analysis to T7 hexamer (17), we see that the monomers are not arranged symmetrically (17). For instance the upper part of the figure has A, B and C subunits in yellow, orange and green. A is closest to the screen and C is the distal one. As seen, there is an apparent rotation (∼15°) between successive units (17). To relate to the mechanism shown in (C) [the DNA in (D) has its 5′ end towards the right side of the figure], we can use two simple approaches: As the DNA is initially bound to subunit B, upon NTP hydrolysis the orientation of subunit B would become similar to that of subunit A (because the electron density in the crystal structure was lesser for subunit A nucleotide than for subunit B nucleotide, thus A is more like an NDP state whereas B is more like an NTP state) (17), as a result the B subunit rotation would be in a direction to extrude DNA towards the 5′end (right side). As a second approach to interpret the figure, we can consider the biochemical finding that the NTP docking is the power stroke step (32): in this case after an empty subunit (such as C) receives DNA, it would move it as soon as the nucleotide docks and as the subunit C is already in the maximally rotated state, the only way it can rotate will be in the direction to reset its orientation to that of monomer B (since C is binding NTP as it is doing this, it is already natural for it to assume the state of the NTP bound monomer). In both cases the direction of extruding DNA is toward the right, ie. the 5′ end of DNA, thus making the helicase movement 5′–3′. The relative arrangement of the upper trimer and the lower trimer is not significant, as the lower one was just modeled upon the upper one via symmetry assumptions. The coloring is the same as in (B), arrows having the same colors as the monomers that they represent.
T7 helicase hydrolyses several NTPs, but it is puzzling that only dTTP supports unwinding (29). What is the basis for discrimination of dTTP from other nucleotides (12)? It seems that the 2′-OH group of ribose might play a role since ribonucleotides are especially disfavored (UTP versus dTTP, ATP versus dATP). However, the reasons for the inhibitory role of the 2′-OH group and the base specificity are not clear.
A structure of the ring helicase with DNA bound remains an important challenge as the mode of binding to DNA holds the key to the mechanism that helicase employs to translocate and to unwind DNA. Although studies are consistent with DNA binding in the central channel of the ring, the exact residues that bind DNA are not known. Mutagenesis and biochemical studies have identified regions and residues that are likely candidates for interactions with the DNA. Mutation of R487 or the three Lysines K467, K471, and K473 were shown to compromise DNA binding (27,30). Additionally sequence and structure analysis indicates that motif H4 and the three specific loops (424–439, 464–475, 503–513) that face the central channel of the ring might be involved in DNA binding (17) (Figure 1C, green subunit).
MECHANISTIC FINDINGS
Nucleotide binding and hydrolysis
T7 helicase hexamer has a total of six binding sites for nucleotides. Cooperative interactions among the protomers are observable as the active site for binding and hydrolysis of nucleotides resides near a cleft between subunits. Biochemical studies indicate that both in the absence and in the presence of DNA, a hexamer is able to bind up to four dTTPs (31). The <6 nt binding observed is attributed to negative cooperativity also observed in other hexameric helicases (29,32–35). Pre-steady state kinetic analysis has shown that T7 helicase hydrolyzes ∼4 dTTP/hexamer (32). Having a catalytic site number greater than three decreases the probability of some sites being non-catalytic at all times, as might have been in analogy to F1 ATPase (36,37). Therefore, it has been proposed that the NTPase cycles propagate through all six helicase subunits. The slow step during each cycle of dTTP hydrolysis is the release of Pi at saturating dTTP concentration and nucleotide binding at low concentrations of dTTP (32,38). DNA binding stimulates the dTTPase rate by increasing both the dTTP hydrolysis and the Pi release rates (32). A closer look at a single NTPase cycle reveals that the act of translocating DNA is accomplished at the dTTP docking step, when the initial binding of the nucleotide is followed by a conformational change in the helicase subunit that results in stronger binding of dTTP (32). Thus, the energy from the dTTP hydrolysis cycles that is used to move the DNA comes from the dTTP binding and product release steps as opposed to coming directly from the hydrolysis step, parallel to the observation in another hexameric helicase (39).
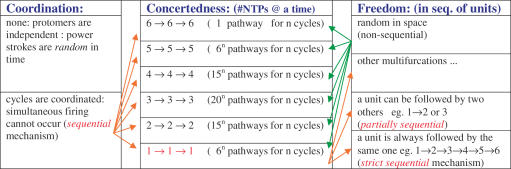
The number of conceivable mechanisms for completing a cycle of NTP hydrolysis is large if the spatial progression and temporal regulation of six subunits is considered (Figure 2). Kinetic and modeling studies have determined the number of subunits involved in dTTP hydrolysis and the cooperative nature of hydrolysis within the subunits of the ring. A stochastic mechanism of dTTP hydrolysis or random firing by the ring subunits was eliminated as a possible mechanism owing to mutant poisoning experiments (30,32). Experimental data as well as computational modeling support a partially or strictly sequential order for the dTTP hydrolysis cycles (30,32). As the dTTPase cycles propagate through each subunit, it was proposed that the DNA from one subunit is transferred to the next while the remaining subunits recharge their dTTP states and prepare for their turn.
Figure 2.
Possible NTPase mechanisms for ring-shaped helicases. Three factors define the nature of the NTPase mechanism employed by the ring subunits (32) and these are arranged in the three columns. (i) Coordination defines the deterministic nature of subunit firing: If firings are random in time, a single firing event by a subunit can be followed by co-firing by two subunits or more, thus there is no regulation on the number of units that hydrolyze NTP and employ the power stroke to the DNA at the same time. On the other hand, if the cycles are fully coordinated, a firing unit of a certain number of protomers has to wait until the preceding unit of the same size finishes firing. (ii) Concertedness or the number of subunits that hydrolyze NTP at a time (all-concerted versus paired versus 1 by 1): This is simply the number of subunits that hydrolyze NTP at the same time. (iii) Freedom or the degeneracy of permutations in the firing sequence. A firing by a certain number of subunits is followed by firing by any other similarly sized unit if firings are random in space, thus firing by first unit can be followed by any of the available units of the same size. On the contrary, if the firings are non-random, they would have a defined sequentiality: a certain unit will be more likely to be followed by another certain unit. The simplest mechanism that agrees with the data on T7 helicase is a partially or strictly sequential mode with a firing unit size of one monomer (32).
Of interest is the spatial arrangement of subunits that are following each other in the sequential mechanism (Figure 2). Structures of the various conformational states of a ring-shaped helicase in the process of hydrolyzing NTP or translocating nucleic acid are yet to be characterized. One can postulate that the directional movement of the nucleic acid is achieved by the polar binding of the helicase to the nucleic acid and by the directionality of the helicase domain motions that might act as power strokes. In fact, one of the structures of T7 helicase solved provides clues to these domain motions (17). Binding of NTP causes a 15° rotation of that subunit (Figure 1D). Thus among the six protomers some will be rotated relative to each other while others will rotate in the opposite direction to maintain a closed ring conformation. Structural analysis indicates that the direction of rotation when binding NTP from an unbound state is towards the N-terminal part of the ring (when viewed from inside of the channel) (17). Together with the observation that the NTP-state has a higher affinity for the DNA (31) and that NTP docking is the power stroke step (32), this implies that the 5′ end of the DNA strand should be facing the N-terminal side of the channel so as to be extruded from that side. This is consistent with the 5′→3′ directionality of helicase translocation and that the N-terminal primase site trails at the 5′ end of the DNA (14). According to this model, DNA would be passed from an NTP bound subunit to the neighboring one that supplies the Arginine finger (17). Thus, a hand off between two neighboring subunits is the most probable sequence (Figure 1C and D).
Translocation along DNA
Although there is no consensus on a detailed unwinding mechanism of helicases, unidirectional translocation of the helicase along ssDNA must play a fundamental role in the unwinding process of most helicases. Comparisons between translocation parameters, especially the rate, and those of unwinding provide important clues to the mechanisms used by helicases to separate complementary strands. As important as it is, there is no simple way to measure translocation rate and processivity. The reason is that helicases can bind to any place on a nucleic acid and some distribution function needs to be assumed. Also a helicase can behave differently at the ends of DNA, which increases the complexity of analyses. Also, possible secondary structures of ssDNA need to be kept in mind to avoid any complications.
T7 helicase is capable of traveling along ssDNA with a speed comparable with its unwinding rate in vivo, and it was found that it moves about three bases per dTTP hydrolyzed (40). Its processivity (probability of making a step forward as opposed to dissociating from the DNA) is virtually 1. The unwinding parameters of T7 helicase on dsDNA, on the other hand, are far below their optimal values (41). T7 helicase unwinds about 10 times slower, and it has a processivity that can go down to 0.9 depending on the sequence properties of the DNA. The reason for a slower rate of unwinding duplex DNA as opposed to translocation along ssDNA probably lies in the tendency of the DNA to stay double stranded. Closing of the complementary strands can act as an opposing force to slow down the helicase, which is consistent with the observation that the rate and processivity defects are enhanced with increasing GC content in the DNA (I. Donmez, Y. J. Jeong and S. S. Patel, unpublished data).
Models of nucleic acid unwinding
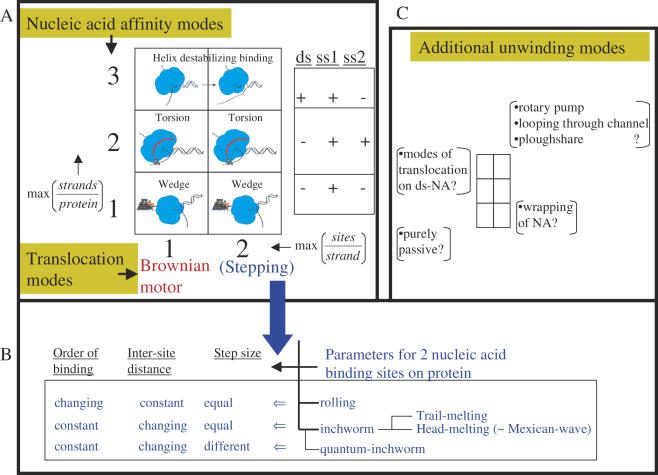
Having a distinct strand separation activity requires the helicase to interact not only with the strand that it is translocating along, but also with the displaced strand or perhaps with the duplex DNA near the junction. High resolution structural information is not available for any of the DNA binding modes of T7 helicase, and biochemical studies have not detected strong binding to both strands of the DNA or to dsDNA. The strand exclusion model or wedge model is most consistent with available data and it postulates that the ring interacts mainly with the one strand of DNA, and as it translocates along that strand, it excludes the complementary strand from its inner channel (Figure 3A). The force production from directional translocation along the ssDNA can bring about the separation of the complementary DNA strands without invoking an elaborate base pair melting mechanism. Even though this is the simplest unwinding model to account for our observations, further structural and biophysical studies are needed to establish the translocation and DNA unwinding mechanisms. There are many other helicase translocation and unwinding mechanisms that have been proposed in the literature (Figure 4 and its legend). Whether helicases with similar structures will employ similar mechanisms remains to be determined.
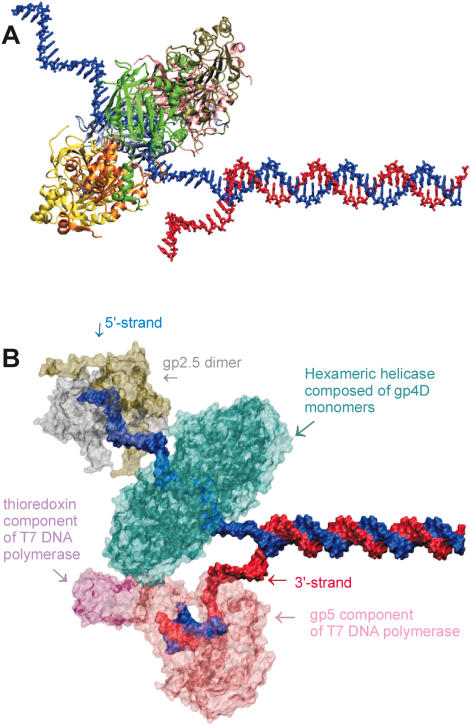
Figure 3.
(A) Strand exclusion model: T7 helicase ring (4D domain) positioned on the 5′ strand of DNA. An illustration of the wedge mechanism of unwinding: one of the strands of DNA is threaded through the central channel of the ring while the other one is left outside. (B) Components of the T7 replisome assembled on fork DNA. A possible assembly of T7 DNA polymerase (gp5 complexed with Escherichia coli thioredoxin), T7 single stranded binding protein (gp2.5, active as a dimer), and T7 helicase 4D domains as a hexameric ring. The excluded molecules are the primase domain of helicase and the second DNA polymerase on the lagging strand. We used X3DNA (64) to model DNA structure of a random DNA sequence, Swiss PDB Viewer (65) for manually rearranging and docking of protein and DNA structures, VMD and its associated routines (62,63) for representations and POV-Ray (http://www.povray.org/) for rendering the structures. DNAP is from 1t7p (66), gp2.5 is from 1je5 (67) and helicase the gp4D fragments from 1e0j (17). DNAP is in pink/mauve, gp2.5 dimer in silver/tan, gp4D hexamer in cyan, 5′ strand in blue and 3′ strand in red.
Figure 4.
Possible mechanisms of helicase translocation and nucleic acid unwinding. Nucleic acid unwinding is considered to be a combination of two processes: unidirectional translocation (achieved by the protein via polar binding to DNA) and strand separation (a function of transient sets of nucleic acid affinity modes of the protein). The six-part box in (A) illustrates the possible unwinding modes resulting from the various nucleic acid binding modes (y-axis) and translocation modes (x-axis). The second box on the right in (A) shows an experimental scheme that parallels the three nucleic acid binding modes of the y-axis. For example, if the only type of binding between the protein and DNA is one protein site bound to one strand of the DNA, then the stoichiometry max (strands/protein) would be 1. If the maximal stoichiometry is two strands per protein the value would be 2, and if one protein can bind to both duplex and single stranded nucleic acid, the y-axis value would be 3 (= 2 + 1). The nucleic acid affinity modes outlined on the y-axis define the unwinding mechanisms depicted with pictures within the boxes. The wedge unwinding mode results from the helicase interacting exclusively with one strand, a torsional component of unwinding would necessitate interaction with an additional strand of nucleic acid, and direct destabilization of the nucleic acid junction would require interaction with the duplex region at the junction in addition to the single strand on which the helicase is translocating. The x-axis defines the translocation mechanisms, and the two numerical values it can take correspond to the maximum observable number of nucleic acid sites that the helicase can transiently and simultaneously bind to during translocation. If there is only one possible nucleic acid binding site, the helicase may move unidirectionally along the nucleic acid using a Brownian motor (BM) or ratchet type mechanism (68,69). If the helicase binds the nucleic acid via two sites, it can employ stepping modes of translocation, such as rolling or inchworm. The stepping translocation modes are further elaborated in (B). In the rolling mechanism, the two helicase sites that bind successive regions of the nucleic acid alternate their affinity while retaining the distance between them but not their relative order. In other words, one helicase site is ahead in one step, but the other one moves in front during the next step. In the inchworm mechanism, the helicase polarity is maintained during translocation; thus, the same side of the protein faces the same end (3′ or 5′) of the nucleic acid at all times. To move, the distance between the two protein sites and the binding affinity periodically changes. A widely accepted mechanism combines the inchworm translocation mode with the duplex destabilization mode. In this mechanism, the two contact sites, protein–dsDNA and protein–ssDNA, can be in one of two relative orders. If the dsDNA binding site is ahead of the other, the mechanism might be referred to as head-melting, or trail-melting in the other case. The mode of head-melting can be likened to a Mexican-wave movement along the DNA (70,71). In the case where these two sites take steps of different sizes, the mechanism was given the name quantum inchworm (72). Further modifications of inchworm mechanism taking into account requirements for the nature of the displaced strand or the predominant mode of interaction with the loading strand (i.e. hydrophobic or electrostatic) have also been proposed (73–75). We display all six combinations as physically plausible, even though not all have been proposed previously. The most popular mechanisms are shown by the coordinates (1,1), (1,2) and (2,3).In addition to the above modes, several other types of unwinding mechanisms have been proposed as illustrated in (C). For example a purely passive mechanism is probably employed by the chemical formaldehyde (76), ssRNA wrapping might be used by Rho, dsDNA translocation might be used by DnaB during branch migration (15), ploughshare is proposed for MCM proteins (77), and looping through channel is proposed for SV40-large T antigen (78). In addition, the torsional model can work with a duplex DNA in the central channel in addition to the mentioned mode of binding two single strands separately. Classifying most of these specific mechanisms requires extra dimensions in the 2 × 3 graph of part (A).
DEPENDENCE OF T7 HELICASE UNWINDING RATE ON ASSISTING PROTEINS
T7 helicase moves at its maximal speed along ssDNA, but slows down considerably when it is unwinding a duplex DNA (41). DNA synthesis by T7 DNA polymerase at the replication fork junction accelerates the unwinding rate of T7 helicase, indicating functional coupling of the two motor proteins (42,43). A part of the reason for the observed enhanced activity of T7 helicase is likely to be the shift of the ssDNA-dsDNA equilibrium by the DNA synthesis activity of the polymerase (44). However, additional factors must be involved based on the result that the observed acceleration of the helicase rate is DNA polymerase rate-dictated. One factor could be that the DNA polymerase prevents back-slippage of the helicase. Evidence for helicase slippage comes from the identification of a mutant that has a higher NTP hydrolysis efficiency but a slower rate, which points to an evolutionary compromise between efficiency and rate (45). If the efficiency can be sacrificed in the wild-type T7 helicase, then slippage would be a major cause for observing suboptimal rates. The presence of the DNA polymerase behind the helicase would prevent helicase slippage contributing to the enhancement of the thermodynamic efficiency and the rate of unwinding. Alternatively, helicase rate enhancement may be due to pushing of the helicase by the DNA polymerase, provided that helicase has a conformation enabling it to slide (diffuse via non-specific interactions) along DNA. If architecturally the two proteins are moving together on corresponding bases of the complementary strands rather than one being strictly behind the other, then the DNA polymerase may be simply providing ssDNA to the helicase by sterically shifting the dsDNA–ssDNA equilibrium at the junction, owing to its limited (and non-self-sufficient) strand separation activity. Determining the exact mechanism and all the factors involved in the observed helicase rate stimulation will require further structural and biochemical studies.
Events are more complicated in vivo (Figure 3B), as there are potentially two DNA polymerases that need to work with the helicase. Efficient replication of the genome requires the leading and lagging strand syntheses to be coordinated, which is achieved by the looping of the lagging strand so as to enable the parallel progression of the two polymerases (46). Single strand binding proteins (either T7 gp2.5 or host SSB) are known to be critical for coupling of these two processes (47). Maintenance of simultaneous progression of synthesis of the two strands also requires a mechanism for recycling the lagging strand DNA polymerase (47,48). Lagging strand is being synthesized discontinuously and Okazaki fragments of an average size of 3000 bases are synthesized upon encountering the primase recognition sites at proper distances starting with the sequence GTC (49,50). Coordination could be mediated by the physical interactions between the different proteins. T7 DNA polymerase's thioredoxin binding domain has affinity for both helicase and gp2.5, whereas helicase and gp2.5 seem to interact via their acidic C-terminal residues (51–56). A recent report brought evidence for the way switches are made onto new Okazaki fragments (57). It seems that the entire replication machinery stalls as the primase synthesizes the necessary RNA primer and resumes upon primer completion. Apart from synthesizing tetraribonuclotide primers and unwinding DNA, T7 helicase also mediates strand exchange and homologous recombination with the help of gp2.5 (58,59). This separate functionality is likely to be important for replication of bacteriophage T7, which is probably done via a recombinational mechanism (60,61).
PERSPECTIVE
The replication system of bacteriophage T7 is one of the simplest for studying the action of a helicase during replication and in particular for studying the mechanisms of translocation and base pair separation by a ring-shaped helicase. The ability to assemble T7 replication proteins and to reconstitute efficient DNA replication in vitro without any accessory factors and hydrolyzable substrates enable studies to address questions that are more difficult to investigate in other systems. Future studies are envisioned that involve determination of the helicase ring structures in different nucleic acid and nucleotide ligation states; determining the coupling nature of the NTPase reaction and the helicase reaction; understanding the cooperativity between DNA polymerase and the helicase; determining the architecture of the complex on the DNA and understanding the effects of single-stranded DNA binding proteins on the activities of the helicase and the replisome.
Acknowledgments
The authors thank the previous and present members of the Patel laboratory that contributed to the studies of helicases and T7 replication proteins and all the members for critical discussions. The authors acknowledge the support of NIH (GM55310). Funding to pay the Open Access publication charges for this article was provided by NIH grant GM55310.
Conflict of interest statement. None declared.
REFERENCES
- 1.Matson S.W., Bean D.W., George J.W. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 2.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 3.Patel S.S., Picha K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 4.van Brabant A.J., Stan R., Ellis N.A. DNA helicases, genomic instability, and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2000;1:409–459. doi: 10.1146/annurev.genom.1.1.409. [DOI] [PubMed] [Google Scholar]
- 5.Mohaghegh P., Hickson I.D. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- 6.Patel S.S., Donmez I. Mechanisms of helicases. J. Biol. Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 7.Dunn J.J., Studier F.W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 8.Lechner R.L., Richardson C.C. A preformed, topologically stable replication fork. Characterization of leading strand DNA synthesis catalyzed by T7 DNA polymerase and T7 gene 4 protein. J. Biol. Chem. 1983;258:11185–11196. [PubMed] [Google Scholar]
- 9.Matson S.W., Richardson C.C. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J. Biol. Chem. 1983;258:14009–14016. [PubMed] [Google Scholar]
- 10.Matson S.W., Tabor S., Richardson C.C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J. Biol. Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 11.Nakai H., Richardson C.C. Leading and lagging strand synthesis at the replication fork of bacteriophage T7. Distinct properties of T7 gene 4 protein as a helicase and primase. J. Biol. Chem. 1988;263:9818–9830. [PubMed] [Google Scholar]
- 12.Bernstein J.A., Richardson C.C. Purification of the 56-kDa component of the bacteriophage T7 primase/helicase and characterization of its nucleoside 5′-triphosphatase activity. J. Biol. Chem. 1988;263:14891–14899. [PubMed] [Google Scholar]
- 13.Bernstein J.A., Richardson C.C. Characterization of the helicase and primase activities of the 63-kDa component of the bacteriophage T7 gene 4 protein. J. Biol. Chem. 1989;264:13066–13073. [PubMed] [Google Scholar]
- 14.Egelman E.H., Yu X., Wild R., Hingorani M.M., Patel S.S. Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA. 1995;92:3869–3873. doi: 10.1073/pnas.92.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan D.L., O'Donnell M. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 16.Sawaya M.R., Guo S., Tabor S., Richardson C.C., Ellenberger T. Crystal structure of the helicase domain from the replicative helicase–primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 17.Singleton M.R., Sawaya M.R., Ellenberger T., Wigley D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 18.Yong Y., Romano L.J. Nucleotide and DNA-induced conformational changes in the bacteriophage T7 gene 4 protein. J. Biol. Chem. 1995;270:24509–24517. doi: 10.1074/jbc.270.41.24509. [DOI] [PubMed] [Google Scholar]
- 19.Toth E.A., Li Y., Sawaya M.R., Cheng Y., Ellenberger T. The crystal structure of the bifunctional primase–helicase of bacteriophage T7. Mol. Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 20.Ahnert P., Picha K.M., Patel S.S. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase–primase protein. EMBO J. 2000;19:3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crampton D.J., Ohi M., Qimron U., Walz T., Richardson C.C. Oligomeric States of Bacteriophage T7 Gene 4 Primase/Helicase. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.05.037. 360, 667–677. [DOI] [PubMed] [Google Scholar]
- 22.Gorbalenya A.E., Koonin E.V. Helicases—Amino-Acid-Sequence Comparisons and Structure-Function-Relationships. Curr. Opin. Struc. Biol. 1993;3:419–429. [Google Scholar]
- 23.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 24.Story R.M., Steitz T.A. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 25.Crampton D.J., Guo S., Johnson D.E., Richardson C.C. The arginine finger of bacteriophage T7 gene 4 helicase: role in energy coupling. Proc. Natl Acad. Sci. USA. 2004;101:4373–4378. doi: 10.1073/pnas.0400968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel S.S., Hingorani M.M., Ng W.M. The K318A mutant of bacteriophage T7 DNA primase–helicase protein is deficient in helicase but not primase activity and inhibits primase–helicase protein wild-type activities by heterooligomer formation. Biochemistry. 1994;33:7857–7868. doi: 10.1021/bi00191a013. [DOI] [PubMed] [Google Scholar]
- 27.Washington M.T., Rosenberg A.H., Griffin K., Studier F.W., Patel S.S. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J. Biol. Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 28.Mendelman L.V., Notarnicola S.M., Richardson C.C. Evidence for distinct primase and helicase domains in the 63-kDa gene 4 protein of bacteriophage T7. Characterization of nucleotide binding site mutant. J. Biol. Chem. 1993;268:27208–27213. [PubMed] [Google Scholar]
- 29.Hingorani M.M., Patel S.S. Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry. 1996;35:2218–2228. doi: 10.1021/bi9521497. [DOI] [PubMed] [Google Scholar]
- 30.Crampton D.J., Mukherjee S., Richardson C.C. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 31.Picha K.M., Patel S.S. Bacteriophage T7 DNA helicase binds dTTP, forms hexamers, and binds DNA in the absence of Mg2+. The presence of dTTP is sufficient for hexamer formation and DNA binding. J. Biol. Chem. 1998;273:27315–27319. doi: 10.1074/jbc.273.42.27315. [DOI] [PubMed] [Google Scholar]
- 32.Liao J.C., Jeong Y.J., Kim D.E., Patel S.S., Oster G. Mechanochemistry of t7 DNA helicase. J. Mol. Biol. 2005;350:452–475. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 33.Bujalowski W., Klonowska M.M. Negative cooperativity in the binding of nucleotides to Escherichia coli replicative helicase DnaB protein. Interactions with fluorescent nucleotide analogs. Biochemistry. 1993;32:5888–5900. doi: 10.1021/bi00073a023. [DOI] [PubMed] [Google Scholar]
- 34.Jezewska M.J., Kim U.S., Bujalowski W. Interactions of Escherichia coli primary replicative helicase DnaB protein with nucleotide cofactors. Biophys. J. 1996;71:2075–2086. doi: 10.1016/S0006-3495(96)79406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jezewska M.J., Lucius A.L., Bujalowski W. Binding of six nucleotide cofactors to the hexameric helicase RepA protein of plasmid RSF1010. 1. Direct evidence of cooperative interactions between the nucleotide-binding sites of a hexameric helicase. Biochemistry. 2005;44:3865–3876. doi: 10.1021/bi048037+. [DOI] [PubMed] [Google Scholar]
- 36.Boyer P.D. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 37.Abrahams J.P., Leslie A.G., Lutter R., Walker J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 38.Jeong Y.J., Kim D.E., Patel S.S. Kinetic pathway of dTTP hydrolysis by hexameric T7 helicase-primase in the absence of DNA. J. Biol. Chem. 2002;277:43778–43784. doi: 10.1074/jbc.M208634200. [DOI] [PubMed] [Google Scholar]
- 39.Rajendran S., Jezewska M.J., Bujalowski W. Multiple-step kinetic mechanism of DNA-independent ATP binding and hydrolysis by Escherichia coli replicative helicase DnaB protein: quantitative analysis using the rapid quench-flow method. J. Mol. Biol. 2000;303:773–795. doi: 10.1006/jmbi.2000.4124. [DOI] [PubMed] [Google Scholar]
- 40.Kim D.E., Narayan M., Patel S.S. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002;321:807–819. doi: 10.1016/s0022-2836(02)00733-7. [DOI] [PubMed] [Google Scholar]
- 41.Jeong Y.J., Levin M.K., Patel S.S. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl Acad. Sci. USA. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delagoutte E., von Hippel P.H. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001;40:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 43.Stano N.M., Jeong Y.J., Donmez I., Tummalapalli P., Levin M.K., Patel S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Hippel P.H., Delagoutte E. A general model for nucleic acid helicases and their ‘coupling’ within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 45.Washington M.T., Patel S.S. Increased DNA unwinding efficiency of bacteriophage T7 DNA helicase mutant protein 4A′/E348K. J. Biol. Chem. 1998;273:7880–7887. doi: 10.1074/jbc.273.14.7880. [DOI] [PubMed] [Google Scholar]
- 46.Park K., Debyser Z., Tabor S., Richardson C.C., Griffith J.D. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J. Biol. Chem. 1998;273:5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 47.Lee J., Chastain P.D., 2nd, Kusakabe T., Griffith J.D., Richardson C.C. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol. Cell. 1998;1:1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 48.Debyser Z., Tabor S., Richardson C.C. Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell. 1994;77:157–166. doi: 10.1016/0092-8674(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 49.Lee S.J., Richardson C.C. Essential lysine residues in the RNA polymerase domain of the gene 4 primase-helicase of bacteriophage T7. J. Biol. Chem. 2001;276:49419–49426. doi: 10.1074/jbc.M108443200. [DOI] [PubMed] [Google Scholar]
- 50.Lee J., Chastain P.D., II, Griffith J.D., Richardson C.C. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J. Mol. Biol. 2002;316:19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 51.Hamdan S.M., Marintcheva B., Cook T., Lee S.J., Tabor S., Richardson C.C. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl Acad. Sci. USA. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong D., Richardson C.C. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein–protein interactions. J. Biol. Chem. 1998;273:6556–6564. doi: 10.1074/jbc.273.11.6556. [DOI] [PubMed] [Google Scholar]
- 53.Notarnicola S.M., Mulcahy H.L., Lee J., Richardson C.C. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 54.Kim Y.T., Tabor S., Churchich J.E., Richardson C.C. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J. Biol. Chem. 1992;267:15032–15040. [PubMed] [Google Scholar]
- 55.Kim Y.T., Richardson C.C. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein–protein interactions. J. Biol. Chem. 1994;269:5270–5278. [PubMed] [Google Scholar]
- 56.He Z.G., Rezende L.F., Willcox S., Griffith J.D., Richardson C.C. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J. Biol. Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 57.Lee J.B., Hite R.K., Hamdan S.M., Xie X.S., Richardson C.C., van Oijen A.M. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 58.Kong D., Griffith J.D., Richardson C.C. Gene 4 helicase of bacteriophage T7 mediates strand transfer through pyrimidine dimers, mismatches, and nonhomologous regions. Proc. Natl Acad. Sci. USA. 1997;94:2987–2992. doi: 10.1073/pnas.94.7.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong D., Richardson C.C. Single-stranded DNA binding protein and DNA helicase of bacteriophage T7 mediate homologous DNA strand exchange. EMBO J. 1996;15:2010–2019. [PMC free article] [PubMed] [Google Scholar]
- 60.Sadowski P.D., Vetter D. Genetic recombination of bacteriophage T7 DNA in vitro. Proc. Natl Acad. Sci. USA. 1976;73:692–696. doi: 10.1073/pnas.73.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Powling A., Knippers R. Recombination of bacteriophage T7 in vivo. Mol. Gen. Genet. 1976;149:63–71. doi: 10.1007/BF00275961. [DOI] [PubMed] [Google Scholar]
- 62.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. , 27–38. [DOI] [PubMed] [Google Scholar]
- 63.Varshney A., F.P.B., Wright W.V. Linearly scalable computation of smooth molecular surfaces. IEEE Comput. Graph. Appl. 1994;14:19–25. [Google Scholar]
- 64.Lu X.J., Olson W.K. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 66.Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 67.Hollis T., Stattel J.M., Walther D.S., Richardson C.C., Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl Acad. Sci. USA. 2001;98:9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levin M.K., Gurjar M., Patel S.S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nature Struct. Mol. Biol. 2005;12:429–435. doi: 10.1038/nsmb920. [DOI] [PubMed] [Google Scholar]
- 69.Astumian R.D. Thermodynamics and kinetics of a Brownian motor. Science. 1997;276:917–922. doi: 10.1126/science.276.5314.917. [DOI] [PubMed] [Google Scholar]
- 70.Soultanas P., Wigley D.B. DNA helicases: ‘inching forward’. Curr. Opin. Struct. Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 71.Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 72.Bianco P.R., Kowalczykowski S.C. Translocation step size and mechanism of the RecBC DNA helicase. Nature. 2000;405:368–372. doi: 10.1038/35012652. [DOI] [PubMed] [Google Scholar]
- 73.Beran R.K., Bruno M.M., Bowers H.A., Jankowsky E., Pyle A.M. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 74.Eoff R.L., Spurling T.L., Raney K.D. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- 75.Kawaoka J., Jankowsky E., Pyle A.M. Backbone tracking by the SF2 helicase NPH-II. Nature Struct. Mol. Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 76.Delagoutte E., von Hippel P.H. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi T.S., Wigley D.B., Walter J.C. Pumps, paradoxes and ploughshares: mechanism of the MCM2-7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 78.Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J.A., Fanning E., Jochimiak A., Szakonyi G., Chen X.S. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003;423:512–518. doi: 10.1038/nature01691. [DOI] [PubMed] [Google Scholar]