Abstract
The Pif1p family of DNA helicases is conserved from yeast to humans. To date, four members of this family have been analyzed in some detail by in vitro and in vivo assays: the two baker's yeast helicases, ScPif1p and Rrm3p, the fission yeast Pfh1p and the human enzyme hPif1p. In vitro, these enzymes are 5′ to 3′ DNA helicase and show little processivity. In vivo, ScPif1p, Rrm3p and probably Pfh1p, function in both the nucleus at specific genomic loci and in mitochondria, where they are needed for the stable maintenance of the genome as accessory helicases to the replication machinery. Interestingly, they act on common DNA substrates but appear to have largely non-overlapping cellular functions, ranging from Okazaki fragment processing, telomerase inhibition, to helping the replication fork progress through non-nucleosomal protein–DNA complexes. For example, both ScPif1p and Rrm3p affect the replication of telomeres, but in a different way: Pif1p inhibits telomerase-mediated telomere elongation by directly removing telomerase from a DNA end, whereas Rrm3p facilitates replication through telomeric DNA. Here we review the current knowledge on the Pif1-like helicases, as a first step towards understanding the basis of their functional specialization and mechanism of action.
INTRODUCTION
Helicases were originally recognized as enzymes that can unwind double-stranded nucleic acids. This property is still useful to characterize helicases biochemically, but several helicases are now known to be able to perform other biochemical transactions with nucleic acids. For example, some helicases, like the yeast Srs2p (1,2) or the phage T4 Dda helicase (3) are able to remove proteins from single-stranded DNA. To perform diverse tasks, helicases use different strategies to recognize and load onto their substrates, to move directionally on a nucleic acid strand and to displace what is attached or annealed to the nucleic acid onto which they load. These biochemical processes are best studied in vitro. However, identifying the relevant physiological substrates for a given helicase is a challenge that can only be accomplished through in vivo studies.
Studies of WRN and BLM helicases have highlighted the association of human homologs of RecQ helicases with cancer predisposition and/or premature aging (4,5). Since helicase genes whose mutation causes human disease invariably have counterparts in model organisms that are amenable to genetic analysis, such as the budding yeast Saccharomyces cerevisiae, model organisms are particularly useful to obtain the requisite in vitro and in vivo data that are needed to understand the functions of these helicases and which can provide insights into the role(s) of their human counterparts.
This review focuses on one family of helicases whose prototype is the budding yeast ScPif1p. The Pif1p helicase family, which belongs to the SF1 superfamily of helicases, is conerved from yeast to humans (6). So far, four members of this family have been studied in some detail: the two budding yeast members, ScPif1p and Rrm3p, the fission yeast Schizosaccharomyces pombe member Pfh1p and the human counterpart of the family, hPif1p. Although no pathology has been reported in association with a human Pif1p deficiency, recent studies on Pif1 helicases in yeast model systems indicate that these enzymes play important roles in the maintenance of genomic integrity. Here we review the in vivo and in vitro data relevant to our current understanding of their functions and mechanism of action.
EVOLUTIONARY CONSERVATION OF PIF1 HELICASES
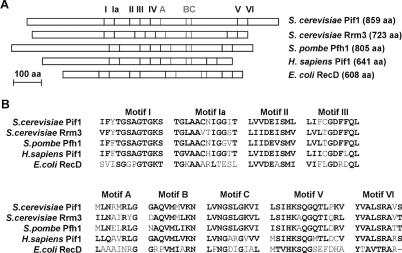
Most helicases from various organisms have been classified into two superfamilies, superfamily I (SFI) and II (SFII), based on the conservation of seven helicase motifs (I, Ia, II, III, IV, V and VI) (7). Pif1 helicases are members of the SFI superfamily of DNA helicases and have been found in the entire eukaryotic phylum, from yeast to human (6). Amino acid sequences of Pif1 helicases share a high degree of conservation (>60%) over the region of 300–500 amino acids encompassing the seven helicase motifs that are the hallmark of the SFI superfamily, but show little similarity in size and sequence within the remaining N- and C-terminal regions of the proteins (Figure 1). It has been argued that Pif1 helicases share a significant sequence homology with the Escherichia coli helicase RecD in the region containing the seven SFI helicase signature motifs (8,9). Interestingly, this homology includes not only the seven helicase motifs, but also three additional motifs of unknown functions, named motifs A, B and C, which cluster between the helicase motifs IV and V and are unique to RecD and Pif1 helicases (Figure 1). Therefore, it has been proposed that the Pif1-like helicase belong to the RecD subfamily of DNA helicases (8,9). As discussed below, biochemical studies have shown that RecD and Pif1 helicases are ssDNA dependent ATPases and 5′–3′ helicases (10,11), which further supports the evolutionary link between RecD and Pif1 helicases. RecD is a subunit of the RecBCD complex, a bipolar DNA helicase and nuclease that is essential for DNA repair and homologous recombination in E.coli (12). Interestingly, Rrm3p, ScPif1p and Pfh1p have been shown to interact genetically with DNA2, which encodes in budding and in fission yeast a helicase/endonuclease involved in Okazaki fragment processing (13–15). It is tempting to speculate that Pif1 helicases can participate in a functional module containing helicases of different polarities and endonuclease activity that cooperate in a similar fashion that the RecBCD complex. In this article, the baker's yeast Pif1p is referred to as ScPif1p and the human protein as hPif1p.
Figure 1.
(A) Schematic structures of Pif1-like helicases and E.coli RecD helicase. The seven SFI conserved helicase motifs are represented with black bars, and the three additional motifs conserved between RecD and Pif1 helicases with grey bars. (B) Alignments of conserved motifs of Pif1-like helicases and E.coli RecD helicase. Amino acids are shown by single letter code and highly conserved residues are shown in bold.
IN VITRO PROPERTIES OF PIF1-LIKE HELICASES
Owing to the difficulty of purifying them, limited in vitro studies of Pif1-like helicases have been achieved so far. Indeed, these helicases are prone to aggregation and are poorly soluble. Four members of the family have been tediously purified and studied in vitro: ScPif1p, Pfh1p and N-terminal truncated versions of Rrm3p and hPif1p. Like RecD (10,11), the four helicases exhibit ssDNA stimulated ATPase activity and 5′–3′ helicase activity on synthetic DNA substrates containing a 5′ single-stranded DNA region, and none of the Pif1 helicases studied so far is able to unwind blunt DNA substrates (9,16–19). It has also been shown that ScPif1p and hPif1p are unable to bind RNA (9,20), but they are able to unwind RNA/DNA hybrids when the loading strand is made of DNA (9,20). Indeed, ScPif1p has a preference for unwinding RNA/DNA hybrids or forked DNA substrates [(16), J.-B. Boulé and V. A. Zakian, manuscript in preparation). The physiological relevance of these properties will be discussed below, although additional in vitro work will be necessary to assess more extensively the enzymatic characteristics of these helicases.
IN VIVO FUNCTIONS OF PIF-1 LIKE HELICASES
Budding yeast Pif1p (ScPif1p)
Mitochondrial DNA maintenance
The PIF1 gene (Petite Integration Frequency) was first isolated as a non-essential gene involved in the maintenance of mitochondrial DNA (mtDNA) (21). ScPif1p is needed for high-frequency recombination between wild type and certain defective (rho-) mitochondrial genomes (21). In the absence of ScPif1p, yeasts lose their mtDNA, especially at high temperatures, and are defective in mtDNA repair. The helicase activity of ScPif1p is required for its role in maintenance of mtDNA as like a pif1Δ strain, pif1-K264A cells lose mtDNA (J.-Q. Zhou and V. A. Zakian, unpublished data). Recently, a recombination-independent role of ScPif1p in mitochondria was discovered in which the helicase cooperates with base excision repair to resist spontaneous oxidative mtDNA damage (22). In the context of mtDNA damage, ScPif1p may inhibit replication progression to allow time for repair (23). Although the mechanism by which ScPif1p participates in the maintenance of mtDNA is unknown, it has been shown that mitochondrial genomes that contain AT-rich palindromes show increased PIF1-dependent recombination, leading to the proposal that Pif1p recognizes and resolves particular DNA structures in mitochondrial DNA (24).
Inhibition of telomerase-mediated telomere addition
The PIF1 gene was re-isolated in a screen to identify genes that affect telomeres (25), suggesting that the enzyme has a nuclear function in addition to its role in mitochondria. Analysis of the 5′ region of the PIF1 ORF reveals two in frame AUG codons separated by 40 codons: translation from the first AUG generates a protein with a mitochondrial localization signal, while the protein translated from the second AUG lacks this signal and localizes in the nucleus. Indeed, two isoforms of ScPif1p can be detected by western blotting, the mitochondrial isoform running slightly faster than the nuclear isoform, owing to the loss of the mitochondrial localization peptide by proteolysis upon mitochondrial import (17). When the first AUG in the PIF1 ORF is mutated (pif1-m1 mutant), cells express only the nuclear (larger) isoform (17) and display mitochondrial defects but have wild type length telomeres (25). Mutation of the second AUG (pif1-m2 mutant) generates only the mitochondrial (smaller) isoform (17); pif1-m2 cells have wild type mitochondrial function but defects in telomere maintenance (25).
Cells lacking nuclear ScPif1p have longer telomeres than wild-type cells (25), and this effect on telomere length is telomerase dependent (17). Conversely, ScPif1p over-expression results in telomere shortening (17). The molecular mechanism of ScPif1p action on telomerase has been well-documented by a combination of in vitro and in vivo approaches. Using recombinant Pif1p and partially purified yeast telomerase, it was found that in vitro, ScPif1p reduces the processivity of yeast telomerase (20). In contrast, catalytically inactive ScPif1p-K264A, in which the invariant lysine in the Walker A box is mutated to alanine, does not affect telomerase action. Since ScPif1p-K264A binds as well as wt ScPif1p to single-stranded DNA, Pif1p does not inhibit telomerase by competing with it for binding to the DNA substrate but rather acts catalytically. This in vitro effect on telomerase processivity is sufficient to explain why telomeres are shorter when ScPif1p is over-expressed and longer in its absence. These in vitro data also provide insight into the mechanism by which ScPif1p limits the processivity of telomerase: ScPif1p releases Est2p, the catalytic reverse transcriptase telomerase subunit, from DNA (20).
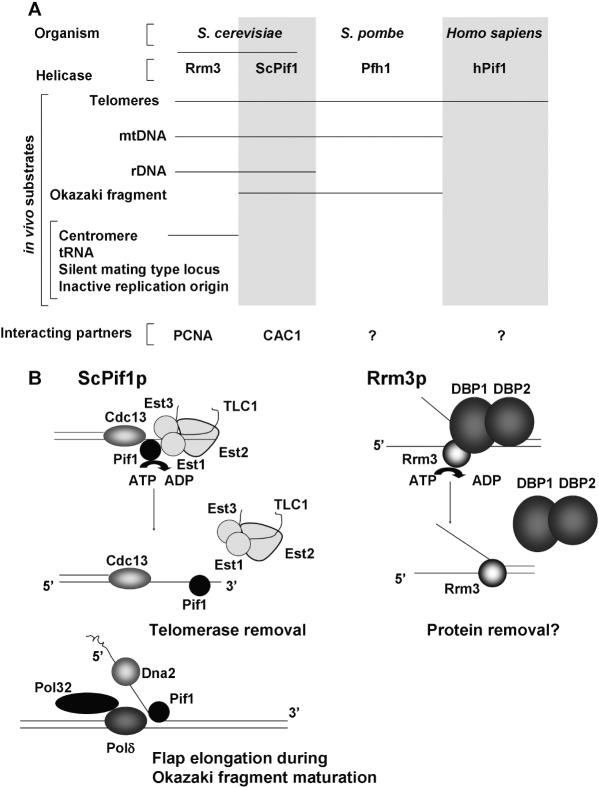
Using a chromatin immuno-precipitation assay (26), it was shown that ScPif1p also releases telomerase from telomeres in vivo (20). In pif1Δ cells, more Est1p is bound to telomeres [Est1p is an essential component of telomerase that is thought to function in both recruitment and activation (27,28)]. When wt ScPif1p is over-expressed, the amount of telomere-bound Est1p and Est2p decreases. As with the in vitro studies, over-expression of Pif1p-K264A does not affect Est1p or Est2p telomere binding. These effects are specific for telomerase components as the telomere binding protein Cdc13p is not affected in the same way by depletion or over-expression of Pif1p. The preference of ScPif1p for RNA/DNA unwinding (J.-B. Boulé and V. A. Zakian , manuscript in preparation) raises the possibility that Pif1p removes telomerase from DNA by unwinding the hybrid formed between it and telomerase RNA. Alternatively or in addition, ScPif1p might be able to remove the catalytic subunit Est2p directly (Figure 2).
Figure 2.
(A) Genomic loci which maintenance is affected by Pif1-like helicases. (B) Left panel: mechanistic models explaining Pif1p action at telomeres and during Okazaki fragment processing, based on available genetic and biochemical evidences (15,20). Right panel: model of how Rrm3p could help replication fork progression through a protein–DNA complex (DBP: DNA Binding Protein).
Inhibition of de novo telomere addition at double strand breaks
The most striking telomere phenotype of ScPif1p deficient cells is the fate of induced or spontaneous double strand breaks. In wild-type cells, double strand breaks are almost always repaired by homologous recombination and only rarely by telomere addition. However, in pif1Δ or pif1-m2 cells, the rate of telomere addition to spontaneous or induced double strand breaks increases ∼200- to almost 1000-fold (25,29,30). Since this increase only occurs in cells expressing telomerase (30), Pif1p inhibits the ability of telomerase to add a telomere de novo.
Cells have the ability to distinguish telomeres from double strand breaks. Whereas even a single double strand break elicits a DNA damage response (31), normal telomeres are not recognized as damaged DNA. It is tempting to speculate that ScPif1p helps the cell distinguish telomeres from DNA damage. However, ScPif1p inhibits both telomerase-mediated lengthening of telomeres (17,20,25) and telomerase-mediated telomere addition to double strand breaks (25,29,30). Telomerase is recruited to yeast telomeres in G1 phase as a result of a specific interaction between telomerase RNA and a Ku subunit (32). Whether ScPif1p is able to displace telomerase bound to telomeres in G1, where it is probably not base-paired to single-stranded DNA, is currently unknown. In addition, there is a Ku-independent pathway for telomerase recruitment in late S/G2 phase that is lost in a cdc13-2 mutant (26). ScPif1p acts on this Cdc13p-dependent pathway as telomeres are longer in mutants that have only this second pathway for telomerase recruitment when they also lack ScPif1p (L. R. Vega, J. Phillips and V. A. Zakian, manuscript in preparation). Both the Ku and the Cdc13p-dependent pathways also act during recruitment of telomerase to double strand breaks (33,34). Mutating either of the two telomerase recruitment pathways in pif1 mutant cells reduces the increase in telomere addition that characterizes this strain (30). Thus, ScPif1p appears to inhibit telomere addition by both the Ku and the Cdc13p-dependent pathways. Taken together, these data do not support a role for ScPif1p in distinguishing telomeres from double strand breaks, but rather support a model in which ScPif1p indiscriminately limits telomerase action both at telomeres and double strand breaks.
Other loci affected by Pif1p
In addition to its effects on telomeres, nuclear ScPif1p is required for efficient fork arrest at a site called the replication fork barrier (RFB) within the ribosomal DNA (rDNA) (35). Another recent genetic study also suggests that Pif1p has a non-telomeric role during replication of nuclear DNA (15). This study found that deletion of PIF1 rescues the slow growth phenotype of dna2 mutant cells and even the lethality of DNA2 deletion. The Dna2p helicase/endonuclease acts to process long flaps that arise during Okazaki fragment maturation when excessive strand displacement by the lagging strand DNA polymerase polδ occurs (36). The genetic interactions between ScPif1p and Dna2p suggest that ScPif1p also participates in the formation of long flaps. The dna2Δ pif1Δ mutants retain some defects in DNA replication and repair, but these defects are further suppressed by deletion of the POL32 gene, a polδ subunit which when mutated leads to decreased strand displacement by polδ (15). Although additional work is needed to identify unambiguously the substrate for ScPif1p in this process, these results are consistent with a role for ScPif1p in 5′ flap extension during Okazaki fragment processing (Figure 2). Interestingly, as described below, a similar role during Okazaki fragment processing has been proposed for fission yeast Pfh1p, suggesting at least a partial functional conservation between ScPif1p and its ortholog in fission yeast.
Budding yeast Rrm3p
The RRM3 gene was originally discovered as an inhibitor of recombination between rDNA repeats (37). RRM3, like PIF1, is a non-essential gene, and rrm3Δ pif1Δ cells are also viable (35).
Role of Rrm3p in the replication of non-nucleosomal genomic loci
There is considerable information on the in vivo roles of Rrm3p. Using two-dimensional (2D) gel electrophoresis, it has been shown that Rrm3p is needed for timely fork progression through ∼1400 chromosomal sites, including rDNA genes, tRNA genes, centromeres, inactive replication origins, the silent mating type loci, telomeres and subtelomeric regions (35,38,39). In the absence of Rrm3p, increased replication pausing at these sites results in DNA breakage and recombination. A consequence of the increased fork breakage in rrm3 cells is the hyperphosphorylation and activation of the intra-S-phase checkpoint protein kinase Rad53p (39) and the need for DNA repair and fork restart activities for viability (8,40). All the effects of Rrm3p require its helicase and/or translocase activity, since a catalytically inactive Rrm3p has the same effects on DNA replication, breakage and recombination as deleting Rrm3p.
The mechanism by which Rrm3p facilitates fork progression through these discrete chromosomal sites is poorly understood. Interestingly, the diverse loci affected by Rrm3p have in common their assembly into stable, non-nucleosomal protein–DNA complexes. Removing these protein–DNA complexes by mutating their binding sites (39) or by deleting a factor that binds the site (41) alleviates the Rrm3p dependent pauses. These data support a model in which Rrm3p promotes fork movement past non-nucleosomal protein–DNA complexes, perhaps by removing protein complexes from DNA. The possibility that Rrm3p acts by removing protein complexes suggests an interesting parallel with ScPif1p, which removes telomerase from DNA [(20), Figure 2]. Alternatively, Rrm3p, like ScPif1p, may preferentially unwind RNA–DNA hybrids, suggesting that its effects on fork progression might involve effects on RNA primers. So far, full-length Rrm3p has not been purified. Rather, in vitro work has been done on an amino truncated form of the protein (18). Since the amino terminus of Rrm3p is required for its in vivo role in DNA replication (42), a mechanistic understanding of Rrm3p will await a better in vitro assay to test the different models. Taken together these observations suggest that Rrm3p is involved in genome replication as an accessory helicase that helps fork progression through obstacles such as bound proteins or maybe particular DNA structures. It was shown that Rrm3p interacts directly with PCNA by yeast two-hybrids and pull-down assays (43). In addition to its well-described role as a polymerase processivity factor, PCNA serves as a docking protein for additional factors (44). It is possible, as suggested by Schmidt et al. (43), that during general replication, replicative proteins occupy the interaction site on PCNA, but when the replication fork reaches an obstacle and stalls, these protein disengage, allowing accessory proteins like Rrm3p to dock and help resume fork progression. However, since Rrm3p moves with the replication fork through both Rrm3p-dependent and independent sites, Rrm3p is unlikely to be recruited to stalled forks although its activity may be triggered by fork arrest (A. Azvolinsky, S. Dunaway, J. Z. Torres, J. B. Bessler and V. A. Zakian, manuscript in preparation).
Mitochondrial DNA maintenance
Like ScPif1p, Rrm3p's functions do not appear to be nuclear limited. Based on sequence analysis, we predicted that Rrm3p encodes a mitochondrial targeting sequence (45). A recent analysis of the yeast mitochondrial proteome indicates that Rrm3p localizes to mitochondria (46). In addition, deletion of Rrm3p in a pif1Δ strain partially suppresses the accelerated loss of mtDNA that occurs in the absence of ScPif1p, suggesting that Rrm3p, like ScPif1p, affects mtDNA replication, recombination or repair (47). Again, ScPif1p and Rrm3p affect the same DNA substrate but in different ways.
Fission yeast pfh1p
The Schizosaccharomyces pombe pfh1+ (PIF1-homolog-1) gene encodes an 805 amino acid protein that is ∼60% similar to both Pif1p and Rrm3p over a ∼450 amino acid region that contains the seven helicase motifs (6,45,48). Like multi-cellular eukaryotes, S.pombe encodes only a single Pif1 family helicase that has roughly equal similarity to the two baker's yeast proteins. Thus, its sequence does not predict if Pfh1p functions like ScPif1p or Rrm3p. In contrast to baker's yeast Pif1p and Rrm3p, Pfh1p is essential, and its helicase activity is required for its essential function(s) (45). Although cells depleted of Pfh1p go through S phase, they arrest with a terminal phenotype consistent with a defect in a post-initiation step in DNA replication.
Based on the functions of ScPif1p and Rrm3p, replication of mtDNA, rDNA and telomeres were examined in cells lacking Pfh1p. Although no evident defect in the abundance or structure of rDNA or mtDNA was found, telomeres are modestly shorter in the absence of Pfh1p. Since pfh1+ is still essential in cells with circular chromosomes, maintenance of telomeric DNA cannot be the sole essential function of the Pfh1p helicase. Although Pfh1p depleted cells have no apparent defects in mtDNA, Pfh1p is predicted to be targeted to mitochondria (45). Indeed, subcellular localization experiments indicate that there are two forms of Pfh1p, one localized to nuclei and one to mitochondria (S. Pinter and V. A. Zakian, unpublished data).
Role of Pfh1p in Okazaki fragment maturation
The best indications so far of Pfh1p's functions come from dissection of its genetic interactions. For example, a search for suppressors of the temperature sensitive cdc24-M38 mutant allele identified cold sensitive alleles of pfh1+ (48). Cdc24p, although not required for bulk DNA synthesis, plays an important role in chromosome replication since compromising its function leads to arrest with incompletely replicated chromosomes (49,50). The cdc24+ gene interacts with genes encoding subunits of the DNA polymerase δ holoenzyme, as well as with the dna2+ gene which encodes a nuclease-helicase required for Okazaki fragment processing. Interestingly, mutant alleles of pfh1 can also suppress the temperature-sensitive phenotype of the dna2-C2 allele, indicating that when the function of Dna2p or Cdc24p is compromised, inactivation of Pfh1p can restore viability (13). These data suggest that Pfh1p, in concert with Cdc24p and Dna2p, is involved in Okazaki fragment processing, although the mechanism by which this is accomplished is unclear. Since Pfh1p is a 5′ to 3′ helicase, it has been suggested that Pfh1p is involved in the generation of flaps from the 5′ end of the Okazaki fragments together with the displacement activity of polymerase δ (13), similar to what has been recently proposed for ScPif1p in budding yeast (15).
Human Pif1p (hPIF)
The human homolog of ScPif1p was first identified by homology search (17) and a full length cDNA was eventually isolated from a HEK 293 cells cDNA library [(9), M. Mateyak and V. A. Zakian, manuscript submitted]. An amino truncated version of the protein was expressed in E.coli and purified, and as expected, hPif1p displays 5′–3′ helicase activity in vitro (9). When hPif1p is stably overexpressed in the human HT1080 fibrosarcoma cell line, telomeres shorten by 0.5 to ∼1.5 kb after 55 population doublings and this effect requires hPif1p helicase activity (9). In vitro, similar to what was observed in the yeast system (20), hPif1p is able to limit human telomerase processivity. Although we do not see an effect of hPIF over-expression on telomere length, we find that hPIF and TERT (the catalytic subunit of human telomerase) are associated in vivo (M. Mateyak and V. A. Zakian, manuscript submitted). More studies are needed to assess how these results are relevant to telomerase regulation in vivo, but these data are a first indication that lessons learned from studies in model organisms provide paradigms to determine the roles of hPif1p in the cell.
CONCLUSIONS AND FUTURE DIRECTIONS
Despite their sequence similarity, it is clear from in vivo studies that Pif1 helicases do not have identical functions. Indeed, in the case of the two baker's yeast proteins, their functions appear to be non-overlapping, even though they affect the same DNA targets. Therefore, one possibility is that the Pif1 family sequence similarity reflects their acting at common DNA targets (Figure 2). The three fungal enzymes are predicted to have both mitochondrial and nuclear forms, and all three proteins affect telomeres. However, the three proteins do not affect telomeres and mtDNA in the same way, and the effects of Rrm3p, ScPif1p and Pfh1p on chromosomal replication are not limited to telomeres.
The basis for the recruitment of Pif1-like helicase to common DNA targets is unclear. One can imagine that genomic loci like rDNA, tRNA genes, telomeres or mitochondrial DNA form particular DNA structures upon replication that need to be resolved by structure-specific helicases for replication to progress efficiently. Another possibility is that the sequence similarity reflects common mechanism(s) of action. The four Pif1 family members that have been characterized are all rather non-processive 5′–3′ DNA helicases. In addition, ScPif1p has the more unusual properties of preferentially unwinding RNA/DNA hybrids and of displacing the telomerase catalytic subunit from DNA (20). The role of Rrm3p in moving replication past protein–DNA complexes suggests that it too might be able to displace proteins from DNA (39,41). In addition, the involvement of Pif1 helicases in Okazaki fragment processing suggests that they could function in 5′ flap extension, possibly in combination with other helicases. It is possible that Pif1 family helicases share some or all these mechanistic properties. These alternative mechanistic models are summarized in Figure 2.
The fact that Pif1 family members have different cellular function is not surprising. Although these proteins have >30% similarity in all pairwise combinations over an ∼400 amino acid regions that contains the helicase motifs (6,17), their N- and C-terminal regions are not similar in sequence or even size. For Rrm3p, the 249 N-terminus is essential for its in vivo role in promoting DNA replication and also acts as a negative regulator of protein abundance (42). In addition, the Rrm3p amino terminus contains a putative PCNA interaction motif and interacts with PCNA by two-hybrid criteria (43). Thus, the N- and C-terminal regions of Pif1 family helicases may promote protein–protein interactions important to recruit the helicase to specific sites of action. These motifs might contribute to or even dictate the cellular functions of the helicases (51–53). In addition, the helicase domains of Pif1 family members are similar but not identical, and these domains are also likely to contribute to the in vivo specificity of the helicases. For example, a hybrid protein having the N-terminus and C-terminus of Rrm3p fused to the helicase domain of Pif1p does not provide Rrm3p function, even though the fusion protein is stably expressed (42). The core helicase might contain determinants of substrate specificity or preference as well as requirements for loading and strand recognition, thereby specifying most of the biochemical properties that can be studied in vitro. One clear objective is to develop in vitro studies aimed at determining the substrate preferences of these helicases and their interacting partners. These types of analysis will allow us to link the numerous genetic observations made on the Pif1 helicases to their biochemical properties and to understand how, at the mechanistic level, these conserved helicases operate in the cell.
Acknowledgments
This work was supported by NIH grants GM26938 and GM43265. JBB was supported in part by a fellowship from the association de la Recherche contre le Cancer (ARC) and in part from a fellowship from the NJ Commission on Cancer research. Funding to pay the Open Access publication charges for this article was provided by the NIH grants mentioned above.
Conflict of interest statement. None declared.
REFERENCES
- 1.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein, Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 2.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 3.Byrd A.K., Raney K.D. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 4.Mankouri H.W., Hickson I.D. Understanding the roles of RecQ helicases in the maintenance of genome integrity and suppression of tumorigenesis. Biochem. Soc. Trans. 2004;32:957–958. doi: 10.1042/BST0320957. [DOI] [PubMed] [Google Scholar]
- 5.Cheok C.F., Bachrati C.Z., Chan K.L., Ralf C., Wu L., Hickson I.D. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 2005;33:1456–1459. doi: 10.1042/BST0331456. [DOI] [PubMed] [Google Scholar]
- 6.Bessler J.B., Torres J.Z., Zakian V.A. The Pif1p subfamily of helicases: region specific DNA helicases. Trends Cell Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 7.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt K.H., Kolodner R.D. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D.-H., Zhou B., Huang Y., Xu L.-X., Zhou J.-Q. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor A.F., Smith G.R. RecBCD enzyme is a DNA helicase with fast and slow motors from opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 11.Dillingham M.S., Spies M., Kowalczykowski S.C. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 12.Smith G.R. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- 13.Ryu G.H., Tanaka H., Kim D.H., Kim J.H., Bae S.H., Kwon Y.N., Rhee J.S., MacNeill S.A., Y.S. Seo Y.S. Genetic and biochemical analysis of Pfh1p DNA helicase function in fission yeast. Nucleic Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Budd M.E., Tong A.H.Y., Polaczec P., Peng X., Boone C., Campbell J.L. PLoS Gen. 2004;1:634–650. doi: 10.1371/journal.pgen.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Budd M.E., Reis C.C., Smith S., Myung K., Campbell J.L. Evidence suggesting that Pif1 helicase functions in DNA replication with the DNA2 helicase/nuclease and DNA polymerase δ. Mol. Cell Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahaye A., Leterme S., Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 17.Zhou J.-Q., Monson E.M., Teng S.-C., Schulz V.P., Zakian V.A. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 18.Ivessa A.S., Zhou J.-Q., Schulz V.P., Monson E.M., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J.-Q., Qi H., Schulz V., Mateyak M., Monson E., Zakian V.A. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol. Biol. Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulé J.B., Vega L.R., Zakian V.A. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 21.Foury F., Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Rourke T.W., Doudican N.A., Mackereth M.D., Doetsch P.W., Shadel G.S. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doudican N.A., Song B., Shadel G.S., Doetsch P.W. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foury F., van Dyck E. A PIF-dependent recombigenic signal in the mitochondrial DNA of yeast. EMBO J. 1985;4:3525–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz V.P., Zakian V.A. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 26.Taggart A.K.P., Teng S.C., Zakian V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- 27.Taggart A.K., Zakian V.A. Telomerase: What are the EST proteins doing? Curr. Opin. Cell Biol. 2003;15:275–280. doi: 10.1016/s0955-0674(03)00040-1. [DOI] [PubMed] [Google Scholar]
- 28.Lundblad V. Telomere replication: an Est fest. Curr. Biol. 2003;13:R439–R441. doi: 10.1016/s0960-9822(03)00365-8. [DOI] [PubMed] [Google Scholar]
- 29.Mangahas J.L., Alexander M.K., Sandell L.L., Zakian V.A. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myung K., Chen C., Kolodner R.D. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 31.Sandell L.L., Zakian V.A. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 32.Fisher T.S., Taggart A.K.P., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- 33.Stellwagen A.E., Haimberger Z.W., Veatch J.R., Gottschling D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bianchi A., Negrini S., Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Ivessa A.S., Zhou J.-Q., Zakian V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 36.Garg P., Burgers P.M.J. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 37.Keil R.L., McWilliams A.D. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in. Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivessa A.S., Zhou J.-Q., Schulz V.P., Monson E.M., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 40.Torres J.Z., Schnakenberg S.L., Zakian V.A. The Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra S phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004;24:3198–3212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres J.Z., Bessler J.B., Zakian V.A. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bessler J.B., Zakian V.A. The amino terminus of the Saccharomyces cerevisiae DNA helicase Rrm3p modulates protein function altering replication and checkpoint activity. Genetics. 2004;168:1205–1218. doi: 10.1534/genetics.104.028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt K.H., Derry K.L., Kolodner R.D. Saccharomyces cerevisiae RRM3, a 5′ to 3′ DNA helicase, physically interacts with proliferating cell nuclear antigen. J. Biol. Chem. 2002;277:45331–45337. doi: 10.1074/jbc.M207263200. [DOI] [PubMed] [Google Scholar]
- 44.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J.-Q., Qi H., Schulz V., Mateyak M., Monson E., Zakian V.A. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 sub-family of DNA helicases. Mol. Biol. Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prokisch H., Scharfe C., Camp D.G., 2nd, Xiao W., David L., Andreoli C., Monroe M.E., Moore R.J., Gritsenko M.A., Kozany C., et al. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004;2:e160. doi: 10.1371/journal.pbio.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Rourke T.W., Doudican N.A., Zang H., Eaton J.S., Doetsch P.W., Shadel G.S. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene. 2005;354:86–92. doi: 10.1016/j.gene.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka H., Ryu G.H., Seo Y.S., Tanaka K., Okayama H., MacNeill S.A., Yuasa Y. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002;30:4728–4739. doi: 10.1093/nar/gkf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nasmyth K., Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharmyces pombe. Mol. Gen. Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka H., Tanaka K., Murakami H., Okayama H. Fission yeast cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol. 1999;19:1038–1048. doi: 10.1128/mcb.19.2.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider S., Schwer B. Functional domains of the yeast splicing factor Prp22p. J. Biol. Chem. 2001;276:21184–91. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 53.Ziegelin G., Niedenzu T., Lurz R., Saenger W., Lanka E. Hexameric RSF1010 helicase RepA: the structural and functional importance of single amino acid residues. Nucleic Acids Res. 2003;31:5917–29. doi: 10.1093/nar/gkg790. [DOI] [PMC free article] [PubMed] [Google Scholar]