Abstract
Homologous recombination (HR) is a ubiquitous cellular pathway that mediates transfer of genetic information between homologous or near homologous (homeologous) DNA sequences. During meiosis it ensures proper chromosome segregation in the first division. Moreover, HR is critical for the tolerance and repair of DNA damage, as well as in the recovery of stalled and broken replication forks. Together these functions preserve genomic stability and assure high fidelity transmission of the genetic material in the mitotic and meiotic cell divisions. This review will focus on the Rad54 protein, a member of the Snf2-family of SF2 helicases, which translocates on dsDNA but does not display strand displacement activity typical for a helicase. A wealth of genetic, cytological, biochemical and structural data suggests that Rad54 is a core factor of HR, possibly acting at multiple stages during HR in concert with the central homologous pairing protein Rad51.
INTRODUCTION
Homologous recombination (HR) is a high fidelity and template-dependent DNA repair pathway found in all organisms studied. HR serves in the non-mutagenic tolerance of DNA damage, in the repair of complex DNA damage, such as single-stranded DNA (ssDNA) gaps, double-stranded DNA breaks (DSBs) and interstrand crosslinks, as well as in the recovery of stalled and collapsed replication forks (1,2). Historically prominent is the role of HR during prophase of the first meiotic division, where it contributes to high fidelity segregation of the homologs and to the generation of genetic diversity among the meiotic products.
RAD54 is a core constituent of the RAD52 epistasis group that encodes the proteins that are essential for HR in eukaryotes. Rad54 protein is a member of the Snf2-family of SF2 helicases that contains many prominent chromatin-remodeling proteins including Snf2, ISWI and others. This group of proteins shares a common core that includes seven motifs proposed to identify helicases (3). However, rather than operating like DNA helicases, which are capable of separating the strands of duplex DNA, the Snf2-related proteins are viewed as motor proteins that translocate on duplex DNA and remodel specific protein–duplex DNA complexes (4). The particular functions of these proteins appear to involve specific protein interactions mediated by domains outside the core motor domain. The budding yeast Saccharomyces cerevisiae genome encodes 17 Snf2-related proteins (Table 1). Interestingly, at least seven of them, Rad54, Rdh54/Tid1, Rad5, Rad16, Rad26/CS-B, as well as the Ino80 and Swr1 complex, have specific functions during DNA repair.
Table 1.
Snf2-family members functioning in DNA repair
| S.cerevisiae | Human | Repair pathway |
|---|---|---|
| Rad54 | Rad54 | HR |
| Tid1/Rdh54 | Rad54B? | HR (meiosis), adaptation from checkpoint arrest |
| Rad26 | CS-B | Transcription-coupled repair |
| Rad5 | ? | Post-replication repair |
| Rad16 | ? | Genome-wide nucleotide excision repair |
| Ino80 | ? | Chromatin remodeling at DSB |
| Swr1 | SRCAP | Chromatin remodeling at DSB |
Previous reviews provide excellent overall outlines of HR and the RAD52 group proteins (1,5–8), as well as detailed discussions of the Snf2-related chromatin remodeling factors (9–11). In this review, we focus on the Rad54 protein. Versatile like the proverbial Swiss Army knife, Rad54 has been postulated to function at multiple stages during HR. Biochemical analyses of complex in vitro recombination assays led to a number of mutually non-exclusive models, as reviewed previously (12). We will discuss results from genetic, biochemical and cytological experiments, as well as insights from the recently accomplished determinations of the Rad54 protein structure to highlight the mechanistic models for the function of Rad54 during HR.
THE RAD52 EPISTASIS GROUP AND HR
Overview
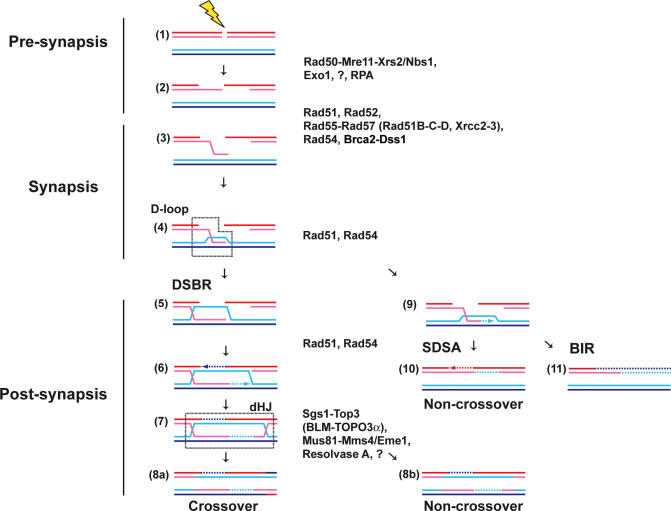
HR can be divided conceptually into three stages (Figure 1). First, in pre-synapsis a recombination-proficient DNA substrate (tailed DSB or gap) is generated either by specific enzymatic action or as a consequence of genotoxic stress (e.g. replication problems). Second, synapsis generates a physical connection (D-loop) between the recombinogenic substrate and an intact homologous duplex DNA template leading to the formation of heteroduplex (or hybrid) DNA. Third, in post-synapsis contiguous DNA strands are restored by priming DNA synthesis from the invading 3′ end on the template DNA and resolving the ensuing junction intermediates. These basic features of HR are often studied and mostly schematized in the context of initiation by a DSB (Figure 1), but initiation from a ssDNA gap is highly relevant in the context of spontaneous DNA damage and in the recovery of stalled replication forks. HR comprises a number of interrelated pathways that share basic mechanistic aspects (1). The original double-strand break repair model (DSBR; Figure 1, left) involves a double-Holliday junction intermediate, whose resolution leads to crossover and non-crossover outcomes (13). Later work identified an asymmetry between the two ends of the DSB, leading to the synthesis-dependent strand annealing model (SDSA; Figure 1, middle), where the invading strand retreats after DNA synthesis and anneals with the second end [reviewed in (1)]. In addition, break-induced replication (BIR; Figure 1, right) was proposed to copy an entire chromosome arm by a replication fork assembled at the D-loop, skipping the involvement of the second end of the DSB (14).
Figure 1.
Pathways of DSBR by HR. HR can be conceptually divided into three stages: Pre-synapsis (1,2), synapsis (3,4) and post-synapsis (5–11). The proteins identified to function at the individual stages are listed, alternative human nomenclature is listed in brackets. / indicates alternative nomenclature in different organisms (Xrs2/Nbs1: S.cerevisiae Xrs2, Schizosaccharomyces pombe and human Nbs1; Mms4/Eme1: S.cerevisiae and human Mms4, S.pombe and human Eme1). Three different pathways emanate from the postulated D-loop intermediate (4), the product of DNA strand invasion by the Rad51-ssDNA filament. DSBR (steps 5–8) engages both ends of the DSB to form a double Holliday junction intermediate (dHJ), which can be resolved into crossover and non-crossover products. SDSA (step 10) retracts the invading strand after DNA synthesis on the target duplex to anneal the newly synthesized strand with the tail of the second end, leading to localized conversion without crossover. BIR (step 11) was proposed to assemble a replication fork at the D-loop to copy the entire chromosome arm distal to the DSB site leading to long gene conversion events.
The proteins encoded by the RAD52 group of genes form the core of the HR machinery (Figure 1) (5–7). The Rad50-Mre11-Xrs2/Nbs1 complex, Exo1 and some unidentified nuclease(s) are involved in processing breaks to generate recombinogenic tailed substrates. The ssDNA binding protein, RPA, binds the ssDNA tails at the break site, to eliminate any possible secondary structure and likely to recruit other proteins to ssDNA. The mediator proteins, Rad52 and the Rad51 paralogs (Rad55-Rad57 in budding yeast and Rad51B, Rad51C, Rad51D, Xrcc2, Xrcc3 in humans), orchestrate the formation of the pre-synaptic RAD51 filament on RPA-coated ssDNA. The human breast cancer tumor suppressor protein Brca2 is thought to function at this step in pre-synapsis as well (15). During synapsis, the Rad51 filament performs homology search and DNA strand invasion. While the enzymatic steps and proteins involved in pre-synapsis and synapsis are comparably well understood from in vivo and in vitro studies (5–7,16), the enzymatic requirements for the later recombination steps in post-synapsis, including DNA synthesis, branch migration and junction resolution are less well defined. The potential roles of Rad54 protein are discussed later.
The RAD54 gene
The RAD54 gene was originally identified in three parallel S.cerevisiae mutant screens for ionizing radiation (IR) sensitive mutants that showed little sensitivity to ultraviolet (UV) radiation (17–19). Together with rad51 and rad52 mutants, rad54 mutants are the most IR-sensitive single mutants in budding yeast, also exhibiting extraordinary sensitivity to alkylating agents (e.g. methyl methanesulfonate), crosslinking agents (cis-platinum and mitomycin C), the topoisomerase I inhibitor camptothecin and a host of other agents inducing DSBs. Most Rad54-deficient cells cannot survive a single DSB introduced by the HO endonuclease at the MAT locus (20), but rad54 mutants grow almost as well as wild-type cells in the absence of induced DNA damage. These results indicate that DSBs occur only rarely during normal mitotic growth in budding yeast, suggesting that single-stranded gaps may act as initiating DNA lesion for spontaneous recombination.
RAD54 is required for spontaneous and induced mitotic recombination, and rad54 mutants show a reduction to the same extent as rad51 or rad52 mutants in recombination assays that require DNA strand invasion (21–23). While rad51 and rad54 mutants display highly similar phenotypes with respect to their damage sensitivities, recombination, chromosome loss and mutator phenotypes in mitotic cells, their meiotic phenotypes differ (1,6,7). Unlike rad51 cells that essentially do not generate viable meiotic products (spores), 25–65% of the spores of a rad54 meiosis are viable (21,24). Return-to-growth experiments and analysis of the meiotic products identified only relatively subtle meiotic recombination-defects in rad54 cells. This is likely the result of partial redundancy between Rad54 and the related Rdh54/Tid1 protein (see below) in meiosis (21). In the rad54 rdh54 double mutant, spore viability is reduced to the level of rad51 mutants (21). The redundancy between Rad54 and Rdh54/Tid1 is primarily noted during meiosis, where it may reflect a specific role of Rdh54/Tid1 in engaging in recombination between homologs, whereas Rad54 may play a more dominant role in sister chromatid interaction during meiosis (21,25,26). This functional specialization during meiosis of these two Snf2-related recombination factors may hold a key to understanding how cells direct meiotic recombination between homologs and suppress non-productive sister chromatid interactions.
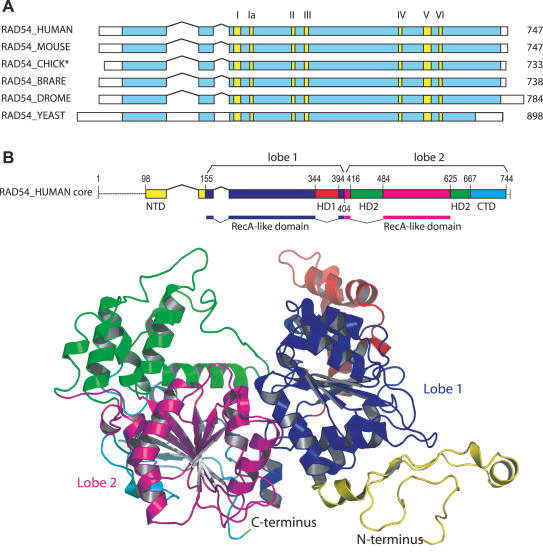
Bona fide Rad54 homologs appear to be present in all eukaryotes studied. The proteins not only share extensive sequence homology in the motor core domain (motifs I–VI in Figure 2A) but also significant similarity in the Rad54-specific N-terminal extension. Compared to other eukaryotic Rad54 proteins, budding yeast Rad54 shows two insertions in the N-terminal domain (Figure 2A). There are no Rad54 homologs in bacteria. A putative homolog has been identified in the archeaon Sulfolobus solfataricus but not other archaea (27). Genetic and biochemical evidence will be needed to support this notion, because due to the multitude of Snf2-related proteins it is difficult to assign homologous function based on sequence comparison.
Figure 2.
Rad54 protein structure and phylogenetic comparison. (A) Schematic alignment of Rad54 proteins from Homo sapiens (HUMAN), Mus musculus (MOUSE), Gallus gallus (CHICK), Zebrafish Brachydanio rerio/Danio rerio (BRARE), D.melanogaster (DROME) and S.cerevisiae (YEAST). The seven conserved motor motifs are highlighted in yellow. * The database sequence appears incomplete and some residues from the N-terminus of chicken Rad54 are missing. (B) Structural model of human Rad54 based on the X-ray crystal structure of zebrafish Rad54 (PDB code: 1Z3I) (47). Rad54 from human and zebrafish share 78.4% identity. The colors in the structural model reflect the colors in the primary structural scheme of the human Rad54 core. The N-terminal domain (NTD) and C-terminal domain (CTD) are shown in yellow and cyan, respectively. Snf2-specific helical domains are depicted in red (HD1) and green (HD2). The RecA-like α/β domains are shown in blue (lobe 1, containing motifs I, IA, II, III) and magenta (lobe 2, containing motifs IV, V, VI). The linear structural model (top) is aligned and at the same scale as the representations in A. The dotted lines at the N- and C-termini indicate residues not present in the crystal structure, The X-ray crystal structure of zebrafish Rad54 was used in structural alignment in ICMLite (http://www.molsoft.com). The molecular modeling and model evaluation was performed using the methods described in (109). The image of the structural model was generated using PyMol (http://www.pymol.org).
Genetic analyses of the RAD54 gene in mouse and chicken confirm the importance of Rad54 for HR and provide valuable insights into the cellular and organismic consequences of a recombination defect in vertebrates (28,29). Disruption of the mouse RAD51 gene causes embryonic lethality (30,31), whereas disruption of the mouse RAD52 gene does not cause DNA damage sensitivity (32). RAD54 knockout mice are viable and provide a critical tool for the analysis of HR in mammals (28,33). The discrepancy between the vertebrate and yeast system with regards to the viability of rad51 mutants and the phenotypes of the rad52 mutants is presently not well understood. At the cellular level, Rad54-deficiency causes sensitivity to IR and interstrand crosslinking agents (e.g. mitomycin C) in mice, whereas at the organismic level Rad54-deficient mice are not overtly IR-sensitive but display sensitivity to mitomycin C (28). This is likely a reflection of the more significant contribution of the NHEJ-pathway to DSB repair in mammals, because double mutants affecting both the HR and NHEJ pathways (rad54 scid, rad54 lig4) display synergistic sensitivities (34,35). HR, as assayed by gene targeting and DNA damage-induced sister chromatid exchange, is reduced but not eliminated in rad54 mouse ES cells (28,36). Rad54-deficient mice do not exhibit an overt meiotic recombination defect (28), similar to the situation in budding yeast. In summary, the genetic data in yeast and vertebrates identify a critical role of Rad54 protein in HR in eukaryotyes.
The Rad54 protein
Rad54 protein is a member of the Snf2-family of DNA-stimulated/dependent ATPases in the SF2 family of DNA helicases. In their core domain, all Rad54 proteins have the seven conserved Snf2-specific motifs that were proposed to be diagnostic of DNA helicases (3) (Figure 2A). However, Rad54 (and all other Snf2-related proteins) fail to catalyze strand displacement reactions typical for DNA helicases. Rad54 protein displays dsDNA-specific ATPase activity with a turnover of 600–1000 ATP molecules per Rad54 molecule per minute on protein-free duplex DNA (37–39). Typical DNA helicases display ssDNA-dependent/enhanced ATPase activity and use the energy of ATP hydrolysis to translocate on ssDNA (40). Instead, Rad54 protein uses the energy of ATP hydrolysis to translocate on dsDNA inducing topological changes. On circular duplex DNA, Rad54 introduces unconstrained positive and negative supercoils and displaces a triplex-forming oligonucleotide, typical for a translocating motor protein (41–44). Direct imaging of Rad54–dsDNA complexes by scanning force microscopy identified supercoiled domains anchored by Rad54 protein, providing further evidence for a translocation model (45). Single-molecule experiments directly visualized Rad54 translocation on dsDNA, demonstrating highly processive movement at 300 bp/s (46). The basic biochemical activities of Rad54 protein, ATP hydrolysis and induction of topological change, are significantly stimulated in the presence of the Rad51-ssDNA/dsDNA filaments (37,42,43), suggesting that Rad54 functions in concert with Rad51 in vivo.
The X-ray crystallographic structures of the core domain of the zebrafish Rad54 protein (47) and of the core domain of the putative Rad54 homolog of S.solfataricus, also as a co-crystal with duplex DNA (48), provide the first and exciting glimpses at Snf2-related proteins. Figure 2B shows a model of the human Rad54 protein based on the experimentally determined zebrafish Rad54 structure (47). The protein folds into a structure of two lobes, each consisting of a RecA-like α/β domain found in helicases (40), which is topologically and structurally similar to that of SF2 helicases [e.g. RecG (49)]. The Rad54 lobes contain Snf2-specific insertions (HD1 and HD2) in each lobe. The bi-lobal helicase domain structure is typical for SF2 DNA helicases and related SF1 helicases (40). This structural homology suggests that Rad54 (and the other Snf2-related proteins) use a mechanism to translocate on duplex DNA analogous to the inchworm mechanism proposed for helicases to translocate on ssDNA (47,48). In the Sulfolobus structure the relative orientation of the two lobes varies from that in zebrafish Rad54 and in SF2 helicases, such that the C-terminal lobe of the Sulfolobus protein is rotated along the x- and y-axis by about 90 and −110° respectively, compared to zebrafish Rad54. The domain (lobe) orientation of the DNA-free and DNA-bound forms of the Sulfolobus protein was very similar (48), suggesting that this difference is unlikely related to changes between the DNA-bound and unbound form of the proteins. The reasons for this structural difference between the Sulfolobus and zebrafish proteins are currently not understood. While the Sulfolobus protein is a structural homolog to Rad54 and displays dsDNA-specific ATPase activity as well as topological activities (48), its recombination function and in vivo role still remain to be tested. There is some uncertainty about the native sequence of the Sulfolobus protein; the difference is a 20 amino acid insertion/deletion between motifs IV and V (27,48), whose significance and impact is unclear. In summary, the biochemical, single-molecule, electron microscopic and X-ray structure data provide compelling evidence for a model, in which Rad54 translocates along duplex DNA.
MODELS FOR RAD54 FUNCTION
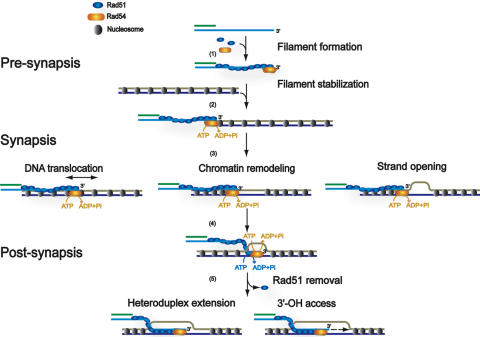
Starting with the seminal discovery that Rad54 stimulates Rad51 protein in DNA pairing reactions (38), much attention has been focused on Rad54 biochemistry, in particular in reconstituted recombination reactions with Rad51 protein and RPA using mostly protein-free (i.e. non-chromatin) templates. The physical interaction of Rad54 with Rad51 protein (41,50–52) suggested immediately a role of Rad54 either in the assembly or function of the Rad51-ssDNA filament, which performs the central homology search and strand invasion step in HR. The biochemical work identified functions of Rad54 at all three stages of recombination, pre-synapsis, synapsis and post-synapsis (12) (Figures 1 and 3), and is discussed below together with pertinent genetic, molecular and cytological data.
Figure 3.
Mechanistic models for Rad54 function in HR. The mechanistic models were derived from analysis of reconstituted in vitro recombination reactions and biochemical analysis of the Rad54 protein. For more details see text. Shown is one processed DSB end with a 3′-ending ssDNA tail that invades a nucleosomal duplex target DNA. Pre-synapsis: Rad54 was found to mediate formation or to stabilize Rad51 filaments on ssDNA. The pre-synaptic function does not require Rad54 ATPase activity and requires Rad51 binding to ATP but not hydrolysis. Synapsis: Rad54 augments the ability of Rad51-ssDNA filaments to form joint molecules, possibly involving translocating the Rad51-ssDNA filament along duplex DNA or inducing strand separation through induction of topological change. Rad54 also exhibits chromatin remodeling activity that may clear nucleosomes or other proteins from the pairing site. The synaptic function requires the Rad54 ATPase activity but not the Rad51 ATPase activity. Post-synapsis: Rad54 was identified to catalyze heteroduplex extension (branch migration) and can dissociate the Rad51–dsDNA product complex, possibly to allow DNA polymerase access to the invading 3′-OH end to prime DNA synthesis. Post-synapsis requires the ATPase activities of both the Rad54 and Rad51 proteins. The exact oligomeric structure of Rad54 in its interaction with the Rad51-ssDNA and Rad51–dsDNA filaments is not known, so it is unclear how many molecules of Rad54 are represented by the symbol drawn in the figure. Rad54 is likely to act as an oligomer on DNA (58), and an oligomeric Rad54 particle has been directly visualized at the terminus of Rad51–dsDNA filaments by electron microscopy (81). This figure is derived from Figure 2 of reference (12).
Pre-synaptic models
During pre-synapsis, mediator proteins function in the replacement of RPA with Rad51 on the single-stranded tails of the processed DSB (Figure 1). Rad54 was found to promote nucleation of Rad51 on RPA-coated ssDNA (53), as was previously reported for the Rad52 and Rad55-Rad57 proteins (7,54). This may be a reflection of Rad54 stabilizing the Rad51 filament by forming a co-complex with the Rad51-ssDNA filament (55,56). This pre-synaptic function of Rad54 is independent of its ATPase activity (57), as the Rad54-K341R mutant that is defective in ATP hydrolysis (58) functions as well as wild-type Rad54 protein in Rad51 filament stabilization (55). Genetic studies have demonstrated that the ATPase activity is crucial for in vivo Rad54 function and that the rad54-K341R mutant displays DNA damage sensitivities equivalent to the deletion mutant (58,59). Cytological studies in yeast, chicken DT40 cells, and mouse ES cells suggest that Rad54 is not necessary for the formation of Rad51 foci, which likely represent Rad51-ssDNA filaments and later recombination intermediates (16,60–63). Studies in mouse ES cells showed that the Rad51 foci formed in rad54−/− cells were not as stable as in wild-type ES cells, leading to loss of Rad51 foci using methanol/acetone fixation instead of para-formaldehyde (41,63). Together these data suggest that a pre-synaptic role of Rad54 is not sufficient to reflect the critical ATPase-dependent function of Rad54 in recombination. Yet the pre-synaptic function may be necessary and important to target Rad54 to the pairing site, where it can engage its ATPase activity on duplex DNA (42). A test of this model will require specific Rad54 mutants that are defective in their association with the Rad51-ssDNA filament.
Synaptic models
During synapsis, the Rad51 nucleoprotein filament searches for homology on duplex target DNA and promotes DNA strand invasion forming a D-loop intermediate (Figure 1). Stimulation of Rad51-mediated joint molecule formation by Rad54 has been observed in in vitro recombination reactions including the D-loop assay (ssDNA or tailed DNA and supercoiled circular duplex DNA) and DNA strand exchange reaction (circular ssDNA and linear duplex DNA) (38,42,43,55,58,64–67). The stimulation requires the ATPase activity of Rad54 protein and involves species-specific contacts between both proteins, because it is only observed when Rad51 and Rad54 from cognate species are used. This suggests that the observed stimulation bears biological significance. ATP-dependent translocation of Rad54 on duplex DNA is likely the critical biochemical activity, but this motor activity can be employed in different modes (12) (Figure 3). Rad54 targeted to the duplex by the Rad51 filament may clear the donor DNA of nucleosomes (see below) or other duplex-bound proteins, including non-productively bound Rad51 protein. Moreover, Rad54 translocation on duplex DNA may aid the homology search process to efficiently sample target DNA. Alternatively, the topological activity of Rad54 on duplex DNA induces negative supercoiling, which favors unpairing of duplex DNA, possibly helping joint molecule formation by Rad51 protein. Although, duplex DNA has no inherent polarity, Rad54 is positioned on duplex DNA through the incoming Rad51-ssDNA filament, which might determine its direction of translocation. The exact architectural disposition of the interaction of Rad54 with the Rad51-ssDNA filament during pre-synapsis, synapsis and post-synapsis remains to be determined and will provide valuable insights into the mechanism of Rad54 function.
Genetic analysis of the RAD52 epistasis group is consistent with a function of Rad54 at or after the Rad51 step, i.e. synapsis or post-synapsis (68). A particularly important synthetic lethal interaction of rad54 is the inviability of the rad54 srs2 double mutant (69). SRS2 encodes a 3′–5′ helicase that strips Rad51 from ssDNA (70,71), which provides a compelling mechanistic explanation for the anti-recombination function of Srs2 helicase (72,73). Importantly, only rad54 mutant cells, but not rad51, rad52, rad55 or rad57 mutants, are synthetically lethal with srs2 (69). The synthetic lethality of rad54 srs2 is suppressed by mutations in RAD51, RAD52, RAD55 or RAD57 (74,75). Such recombination-dependent lethality may be explained by a model, where the Rad51 filament itself or a joint molecule dependent on the Rad51 filament (D-loop) is a potentially lethal intermediate that can be reversed by Srs2 or alternatively requires Rad54 to move forward and complete recombination. Rad51, Rad52 and Rad55-Rad57 are critical for the assembly and structure of the Rad51 filament (1,5–7). Hence, these and other results from epistasis analysis of the RAD52 group (68) suggest that Rad54 may act after the assembly of the Rad51 filament during synapsis or post-synapsis. While these genetic data point to a critical role of Rad54 after pre-synapsis, the genetic analysis is unable to resolve synapsis and post-synapsis, and novel in vivo approaches are needed to resolve this question.
Post-synaptic models
Post-synapsis comprises the steps after D-loop formation and include priming of DNA synthesis from the invading 3′-OH end, branch migration, establishment and resolution of junction intermediates and the sealing of the strands by DNA ligase to restore two intact and contiguous duplex DNAs (Figure 1). Rad54 increases the rate of branch migration in an ATP-dependent fashion during the three-strand DNA strand exchange reaction (76). This activity requires species-specific protein interactions between the budding yeast Rad51 and Rad54 proteins, and is not observed when bacterial RecA protein or human Rad51 protein are used. It is unlikely that Rad54 acts like a junction motor analogous to the paradigmatic RuvB protein (8), as Rad54 does not display preference in binding DNA junctions (S. Kowalczykowski, personal communication).
Much attention has been focused on the assembly of protein complexes during recombination, in particular on the Rad51 filament, but these complexes also need to be disassembled to release their product DNA. In vitro this is often accomplished by treatment with proteinase K and detergent, which experimentally sidesteps this requirement. After DNA strand exchange, Rad51 is bound to the heteroduplex DNA product, and direct biochemical evidence shows that Rad51, unlike bacterial RecA protein, is extremely slow to turnover and release duplex DNA upon ATP hydrolysis (37,77,78). This is also reflected in the 200-fold lower dsDNA-dependent ATPase activity of Rad51 compared to RecA (79). Experiments with RecA have demonstrated the need for ATPase-dependent turnover of RecA to provide access of DNA polymerases to the invading 3′ end during post-synapsis (3′-OH access model in Figure 3) (80). A role of Rad54 in the turnover of Rad51 from product heteroduplex DNA has been suggested, because Rad54 was found to dissociate Rad51 from duplex DNA in an ATP-dependent fashion (56). This activity of Rad54 was accompanied by a significant 6-fold stimulation of the Rad54 ATPase by partial Rad51–dsDNA filaments (37,56) and involved species-specific protein interactions. These observations are consistent with a model that Rad54 translocates on duplex DNA (46) towards the Rad51 filament terminus to dissociate the Rad51–dsDNA complex. Indeed, Rad54 could be directly visualized by electron microscopy at the terminus of Rad51 filaments formed on dsDNA under conditions where protein-free dsDNA flanked the filament (81). The post-synaptic model of Rad54 being a turnover factor for the Rad51 product complex also provides a explanation for the biochemical difference in the ATPase activity between RecA and Rad51, and a rationale to explain, why bacteria that employ RecA protein have no need for a Rad54-like activity and, in fact, do not have a Rad54 homolog.
While the genetic data are unable to resolve the synaptic and post-synaptic phases of recombination, as discussed above, other in vivo observations may bear on this distinction. Meiotic or DNA damage-induced Rad51 foci in mouse, chicken DT40 cells and budding yeast exhibit a longer half-life in rad54 mutants (60–62,82). Unfortunately, the exact nature of cytologically observable Rad51 foci has not been determined. They may represent pre-synaptic Rad51 filaments and later recombination intermediates in synapsis (D-loops) or post-synapsis. Hence, these cytological data suggest a function of Rad54 after pre-synapsis, but cannot distinguish between a function in synapsis or post-synapsis.
Chromatin immunoprecipitation (ChIP) experiments have been used to monitor the recruitment of Rad51 protein to an HO endonuclease-induced DSB at the MAT locus and the HML donor locus in wild-type and rad54 mutant cells (53,57,83). Initially, two studies arrived at opposite conclusions as to, whether Rad54 had a role in Rad51 localization to the DSB (53,83). It appears now that Rad54 may have an ATP-independent role of localizing Rad51 close to the terminus of the DSB, whereas Rad51 readily binds more distant from the DSB in a Rad54-independent fashion (57,83). This ATP-independent function of Rad54 in pre-synapsis might be a reflection of the stabilization of the Rad51-ssDNA filament found in vitro, which was also independent of the Rad54 ATPase activity (55). It is inferred from these experiments that Rad51 forms functional filaments in rad54 cells, because Rad51 was found targeted to the duplex donor locus (HML) by ChIP (57,83). The noted difference in Rad51 localization to the HML donor site between wild-type and rad54 cells (53,57) may be a function of defects in filament assembly during pre-synapsis or defects in synapsis/post-synapsis, which may affect Rad51–DNA complexes during these phases of recombination. Although Rad51 is targeted to the donor locus in rad54 cells, it is unclear if D-loops are formed and several studies were unable to detect DNA synthesis from an invading 3′ end at the donor locus (53,57,83). This deficiency in directing DNA synthesis from the invading strand in the D-loop may be a consequence of an inability to form D-loops in the first place (synapsis defect) or an inability to recruit DNA polymerase to the invading 3′ end of the D-loop (post-synapsis defect) or both. While some ChIP data support a possible ATP-independent function of Rad54 in pre-synapsis (53,57), the ChIP experiments are unable to resolve the question whether the critical ATP-dependent function of Rad54 is in synapsis or post-synapsis.
Rad54 and chromatin remodeling
Chromatin represents the natural environment of nuclear DNA metabolism in eukaryotes and was found to negatively interfere with transcription. A similar inhibition may be expected for recombination, and the similarity of Rad54 to known chromatin remodeling factors immediately suggested that Rad54 might be a chromatin remodeling factor for recombinational repair. This activity is potentially relevant at the break site prior to end-processing (pre-synapsis) and at the pairing site on the template DNA, where Rad54 may not only clear nucleosomes but also other proteins bound to duplex DNA that would inhibit D-loop formation (synapsis) (Figures 1 and 3). Biochemical experiments using reconstituted nucleosomal templates have confirmed this expectation and shown that S.cerevisiae and D.melanogaster Rad54 remodel chromatin in vitro (44,84,85). Rad54 enhances the accessibility of nucleosomal DNA by restriction enzymes and can slide a single nucleosome in an ATP-dependent fashion. The efficiency of chromatin remodeling by Rad54 was below that of established chromatin remodeling factors, and Rad54 was unable to affect nucleosomal positioning in a nucleosomal array. This may reflect a requirement for protein-free DNA that is longer than the linker DNA in such arrays. Rad51 requires Rad54 protein to promote efficient D-loop formation on nucleosomal substrates in vitro (44,85). Nucleosomal remodeling was greatly stimulated by Rad51-ssDNA nucleoprotein filaments without the need for homology between the two DNAs, suggesting that nucleosomal remodeling precedes synapsis (84). These biochemical studies suggest a role of Rad54 in chromatin remodeling at the pairing site just prior to synapsis, where it is targeted by its interaction with the Rad51 nucleoprotein filament. This model was tested in vivo using HO endonuclease-initiated DSB repair in budding yeast (57). However, monitoring a positioned nucleosome on the HML donor site with micrococcal nuclease showed no difference between wild-type and rad54 cells (57), suggesting that Rad54 does not act by moving or removing this positioned nucleosome at the HML donor site. However, an effect of Rad54 on the accessibility of the HML donor site by the HO endonuclease was identified (57). Access of HO to the HML target site required the Rad54 ATPase activity, but it is unclear if this effect reflects chromatin remodeling or is an indirect consequence of forming recombination intermediates at HML. The chromatin model needs further testing in vivo and a specific interaction between Rad54 and the core histones, as shown for known chromatin remodeling factors (9–11), remains to be demonstrated.
Physical analysis of DSB-induced recombination involving the budding yeast MAT locus identified recombination events independent of the Rad54 (and Rad51, Rad55, Rad57) proteins. This led to the suggestion that Rad54, Rad51, Rad55, Rad57 were required for recombination involving chromatin substrates (86). However, later work showed that a number of recombination events involving repeat substrates occurs independent of the Rad54, Rad51, Rad55 and Rad57 proteins (1,6,7). It is likely now that these events are mediated by the single-strand annealing (SSA) and possibly BIR pathways of recombination (87). These pathways either do not require Rad51 and Rad54 outright or have Rad51-Rad54-independent sub-pathways (6,7), an explanation that is independent of an involvement of chromatin. The observation that Rad54 significantly stimulates Rad51-dependent recombination in vitro on protein-free (non-chromatin) templates also suggests that Rad54 function is not confined to chromatin.
OTHER SNF2-RELATED PROTEINS IN DNA REPAIR
The genome of the budding yeast S.cerevisiae predicts 17 Snf2-related proteins, belonging to various distinct subfamilies (A. Flaus and T. Owen-Hughes, manuscript in preparation). Seven of them, Rad54, Rdh54/Tid1, Rad5, Rad16 and Rad26/CS-B, as well as the Ino80 and Swr1 complexes, were identified to have specific functions during DNA repair (Table 1). Among the Snf2 paralogs, Tid1/Rdh54 (Rad54B in mammals?) shows the highest similarity to Rad54 in sequence, genetic function and biochemical properties (21,25,88,89). Tid1/Rdh54 appears to augment the function of Rad54 during DNA repair in mitotic cells and functions primarily during meiotic recombination, likely through its interaction with the meiosis-specific RecA homolog Dmc1 (21,25,90). However, Tid1/Rdh54 also has a specific but poorly understood function in adaptation from DNA damage not shared by Rad54 (91), which may be related to its specific localization at the kinetochore in undamaged cells (16).
Rad5 protein functions in the error-free sub-pathway of post-replication repair (RAD6 epistasis group) in budding yeast to bypass UV lesions, where it may be involved in remodeling protein complexes at stalled replication forks (92).
Rad16 protein participates in nucleotide excision repair, specifically of the non-transcribed strand of transcribed genes or transcriptionally silenced genes, but its specific function remains unclear (93).
CS-B (budding yeast Rad26), one of two human genes involved in Cockayne's syndrome, plays a role in transcription-coupled repair (94). In analogy with the function of the bacterial homolog, TRCF, CS-B was thought to remodel stalled RNA polymerase II complexes to allow access of repair proteins to the lesion (95), but CS-B was also found capable of remodeling nucleosomes in vitro (96).
Ino80 and Swr1, the catalytic subunits of two known chromatin remodeling complexes, were found to be recruited to the DSB site through a specific interaction with C-terminally phosphorylated histone H2A (γH2AX) (97–99). Ino80 is part of a 12 protein complex and a transcriptional regulator that displays efficient chromatin remodeling activity by shifting nucleosomes through specific interactions with histones (100,101). Swr1 functions in a histone exchange complex that replaces histone H2A with the H2A variant H2AZ (102). The genetic relationship between the Ino80 and Swr1 complexes and individual DNA repair pathways has not been fully elucidated and their specific protein–DNA substrates largely remain to be determined. The surprisingly high number of Snf2-family members that appear to function specifically in distinct DNA repair pathways suggests a significant degree of functional diversification that may reflect different protein–DNA complexes as target substrates for the individual enzymes or enzyme complexes.
CONCLUSION
Several biochemical models for the function of Rad54 protein during consecutive mechanistic stages in HR have been developed. While these models are not mutually exclusive, the biological significance of these putative individual roles of Rad54 needs to be tested in physical in vivo assays that can distinguish the various stages of recombination. While break formation and processing as well as the formation of the final repair product can be readily monitored, pairing intermediates during mitotic DSB repair (D-loops and Holliday junctions) have eluded detection so far. Such an assay has been successfully developed for the analysis of meiotic recombination (103,104) and will be critical to distinguish between the possible synaptic and post-synaptic roles of Rad54 in vivo. Likewise, the significance and function of the chromatin remodeling activity of Rad54 remain to be tested by in vivo analysis. What is the relationship of Rad54 with the confirmed chromatin remodeling factors Ino80 and Swr1 that are recruited to the break site; and are these factors also present at the target pairing site? Many of the Snf2-related proteins function in large multi-protein assemblies. It appears that Rad54 functions as a homo-multimeric assembly and no stable in vivo complex of Rad54 with other proteins has been reported. While the interaction of Rad54 with Rad51 has been analyzed in significant detail, its interaction with the structure-selective endonuclease Mus81-Mms4 (105) is poorly understood but may shed light on the function and in vivo substrates of Mus81-Mms4. Since the genetic identification of the RAD54 gene in the late 1960s, the cloning of the budding yeast gene in 1983 (106), the determination of its sequence in 1991 (107), the isolation of mammalian homologs in 1996 (108), the disruption of the mouse gene in 1997 (28) and the purification of the Rad54 protein in 1998 (38), much progress has been made to understand the function of the Rad54 gene and protein. Nevertheless, additional biochemical and in vivo analyses, including novel imaging techniques and physical recombination assays, will be needed to determine whether Rad54 really is the Swiss Army knife of HR.
Acknowledgments
This review was written for the 2005 conference on ‘Helicase and NTP-driven nucleic acid machines’ in Arolla (Switzerland), and the authors thank Patrick Linder and Stephen Kowalczykowski for organizing a stimulating meeting. The helpful comments of Tammy Doty, Kirk Ehmsen, Kristina Herzberg and Neil Hunter are highly appreciated. The authors thank Andrew Flaus, Tom Owen-Hughes, and Stephen Kowalczykowski for communicating unpublished data. Our work on Rad54 is supported by the National Institutes of Health (GM58015 to W.D.H.) and the Susan G. Komen Foundation (PDF0403213 to X.P.Z.). Funding to pay the Open Access publication charges for this article was provided by NIH grant GM58015.
Conflict of interest statement. None declared.
REFERENCES
- 1.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michel B., Grompone G., Flores M.J., Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorbalenya A.E., Koonin E.V. Helicases—amino acid sequence comparisons and structure function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 4.Pazin M.J., Kadonaga J.T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein–DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 5.Sung P., Trujillo K.M., Van Komen S. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- 6.Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 8.West S.C. Molecular views of recombination proteins and their control. Nature Rev. Mol. Cell. Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 9.Vignali M., Hassan A.H., Neely K.E., Workman J.L. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker P.B., Hörz W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 11.Martens J.A., Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 2003;13:136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 12.Tan T.L.R., Kanaar R., Wyman C. Rad54, a Jack of all trades in homologous recombination. DNA Repair. 2003;2:787–794. doi: 10.1016/s1568-7864(03)00070-3. [DOI] [PubMed] [Google Scholar]
- 13.Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 14.Malkova A., Ivanov E.L., Haber J.E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H.J., Li Q.B., Fan J., Holloman W.K., Pavletich N.P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 16.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Game J.C., Mortimer R.K. A genetic study of X-ray sensitive mutants in yeast. Mutat. Res. 1974;24:281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 18.Suslova N.G., Zakharov I.A. Genetic control of radiosensitivity in yeast, VII. Identification of the genes determining the sensitivity to X-rays. Genetika. 1970;6:158. [Google Scholar]
- 19.Snow R. Mutants of yeast sensitive to ultraviolet light. J. Bact. 1967;94:571–575. doi: 10.1128/jb.94.3.571-575.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmuckli-Maurer J., Heyer W.-D. The Saccharomyces cerevisiae RAD54 gene is important but not essential for natural homothallic mating-type switching. Mol. Gen. Genet. 1999;260:551–558. doi: 10.1007/s004380050928. [DOI] [PubMed] [Google Scholar]
- 21.Shinohara M., Shita-Yamaguchi E., Buerstedde J.M., Shinagawa H., Ogawa H., Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmuckli-Maurer J., Rolfsmeier M., Nguyen H., Heyer W.-D. Genome instability in RAD54 mutants of Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:1013–1023. doi: 10.1093/nar/gkg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeki T., Machida I., Nakai S. Genetic control of diploid recovery after g-irradiation in the yeast Saccharomyces cerevisiae. Mutat. Res. 1980;73:3314–3322. doi: 10.1016/0027-5107(80)90192-x. [DOI] [PubMed] [Google Scholar]
- 24.Schmuckli-Maurer J., Heyer W.D. Meiotic recombination in RAD54 mutants of Saccharomyces cerevisiae. Chromosoma. 2000;109:86–93. doi: 10.1007/s004120050415. [DOI] [PubMed] [Google Scholar]
- 25.Klein H.L. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbel A., Zenvirth D., Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz E.M., Haseltine C.A., Kowalczykowski S.C. DNA recombination and repair in Archaea. Adv. Appl. Microbiol. 2001;50:101–169. doi: 10.1016/s0065-2164(01)50005-2. [DOI] [PubMed] [Google Scholar]
- 28.Essers J., Hendriks R.W., Swagemakers S.M.A., Troelstra C., deWit J., Bootsma D., Hoeijmakers J.H.J., Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 29.Bezzubova O., Silbergleit A., YamaguchiIwai Y., Takeda S., Buerstedde J.M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54(−/−) mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 30.Lim D.S., Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rijkers T., VandenOuweland J., Morolli B., Rolink A.G., Baarends W.M., VanSloun P.P.H., Lohman P.H.M., Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essers J., Hendriks R.W., Wesoly J., Beerens C., Smit B., Hoeijmakers J.H.J., Wyman C., Dronkert M.L.G., Kanaar R. Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair. 2002;1:779–793. doi: 10.1016/s1568-7864(02)00110-6. [DOI] [PubMed] [Google Scholar]
- 34.Essers J., van Steeg H., de Wit J., Swagemakers S.M.A., Vermeij M., Hoeijmakers J.H.J., Kanaar R. Homologous and non-homologous recombination differentially affect DNA damage repair in mice. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills K.D., Ferguson D.O., Essers J., Eckersdorff M., Kanaar R., Alt F.W. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dronkert M.L.G., Beverloo H.B., Johnson R.D., Hoeijmakers J.H.J., Jasin M., Kanaar R. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiianitsa K., Solinger J.A., Heyer W.D. Rad54 protein exerts diverse modes of ATPase activity on duplex DNA partially and fully covered with Rad51 protein. J. Biol. Chem. 2002;277:46205–46215. doi: 10.1074/jbc.M207967200. [DOI] [PubMed] [Google Scholar]
- 38.Petukhova G., Stratton S., Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 39.Swagemakers S.M.A., Essers J., de Wit J., Hoeijmakers J.H.J., Kanaar R. The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 1998;273:28292–28297. doi: 10.1074/jbc.273.43.28292. [DOI] [PubMed] [Google Scholar]
- 40.Singleton M.R., Wigley D.B. Modularity and specialization in Superfamily 1 and 2 helicases. J. Bact. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan T.L.R., Essers J., Citterio E., Swagemakers S.M.A., de Wit J., Benson F.E., Hoeijmakers J.H.J., Kanaar R. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 42.Mazin A.V., Bornarth C.J., Solinger J.A., Heyer W.-D., Kowalczykowski S.C. Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 43.Van Komen S., Petukhova G., Sigurdsson S., Stratton S., Sung P. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 44.Jaskelioff M., Van Komen S., Krebs J.E., Sung P., Peterson C.L. Rad54p is a chromatin remodeling enzyme required for heteroduplex joint formation with chromatin. J. Biol. Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 45.Ristic D., Wyman C., Paulusma C., Kanaar R. The architecture of the human Rad54–DNA complex provides evidence for protein translocation along DNA. Proc. Natl Acad. Sci. USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amitani I., Baskin R.J., Kowalczykowski S. Direct visualization of a chromatin-remodeling protein, Rad54, translocating along single-molecules of double-stranded DNA. Mol. Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Thoma N.H., Czyzewski B.K., Alexeev A.A., Mazin A.V., Kowalczykowski S.C., Pavletich N.P. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nature Struct. Mol. Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 48.Dürr H., Körner C., Müller M., Hickmann V., Hopfner K.P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 49.Singleton M.R., Scaife S., Wigley D.B. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. doi: 10.1016/s0092-8674(01)00501-3. [DOI] [PubMed] [Google Scholar]
- 50.Clever B., Interthal H., Schmuckli-Maurer J., King J., Sigrist M., Heyer W.D. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang H., Xie Y.Q., Houston P., Stemke-Hale K., Mortensen U.H., Rothstein R., Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J. Biol. Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 52.Golub E.I., Kovalenko O.V., Gupta R.C., Ward D.C., Radding C.M. Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res. 1997;25:4106–4110. doi: 10.1093/nar/25.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolner B., van Komen S., Sung P., Peterson C.L. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 54.Sung P., Krejci L., Van Komen S., Sehorn M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 55.Mazin A.V., Alexeev A.A., Kowalczykowski S.C. A novel function of Rad54 protein—Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 2003;278:14029–14036. doi: 10.1074/jbc.M212779200. [DOI] [PubMed] [Google Scholar]
- 56.Solinger J.A., Kiianitsa K., Heyer W.-D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- 57.Wolner B., Peterson C.L. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- 58.Petukhova G., Van Komen S., Vergano S., Klein H., Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- 59.Clever B., Schmuckli-Maurer J., Sigrist M., Glassner B., Heyer W.-D. Specific negative effects resulting from elevated levels of the recombinational repair protein Rad54p in Saccharomyces cerevisiae. Yeast. 1999;15:721–740. doi: 10.1002/(SICI)1097-0061(19990630)15:9<721::AID-YEA414>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 60.Shinohara M., Gasior S.L., Bishop D.K., Shinohara A. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl Acad. Sci. USA. 2000;97:10814–10819. doi: 10.1073/pnas.97.20.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyazaki T., Bressan D.A., Shinohara M., Haber J.E., Shinohara A. In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair. EMBO J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takata M., Sasaki M.S., Sonoda E., Fukushima T., Morrison C., Albala J.S., Swagemakers S.M.A., Kanaar R., Thompson L.H., Takeda S. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Veelen L.R., Essers J., van de Rakt M., Odijk H., Pastink A., Zdzienicka M.Z., Paulusma C.C., Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. 2005. pp. , 34–49. [DOI] [PubMed]
- 64.Sigurdsson S., Van Komen S., Petukhova G., Sung P. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 2002;277:42790–42794. doi: 10.1074/jbc.M208004200. [DOI] [PubMed] [Google Scholar]
- 65.Solinger J.A., Lutz G., Sugiyama T., Kowalczykowski S.C., Heyer W.-D. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
- 66.Van Komen S., Petukhova G., Sigurdsson S., Sung P. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J. Biol. Chem. 2002;277:43578–43587. doi: 10.1074/jbc.M205864200. [DOI] [PubMed] [Google Scholar]
- 67.Mazin O.M., Mazin A.W. Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+ J. Biol. Chem. 2004;279:52041–52051. doi: 10.1074/jbc.M410244200. [DOI] [PubMed] [Google Scholar]
- 68.Rattray A.J., Symington L.S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palladino F., Klein H.L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 71.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 72.Aboussekhra A., Chanet R., Zgaga Z., Cassier Chauvat C., Heude M., Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aboussekhra A., Chanet R., Adjiri A., Fabre F. Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA protein. Mol. Cell. Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heude M., Chanet R., Fabre F. Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol. Gen. Genet. 1995;248:59–68. doi: 10.1007/BF02456614. [DOI] [PubMed] [Google Scholar]
- 75.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Solinger J.A., Heyer W.-D. Rad54 protein stimulates the postsynaptic phase of Rad51 protein-mediated DNA strand exchange. Proc. Natl Acad. Sci. USA. 2001;98:8447–8453. doi: 10.1073/pnas.121009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 78.Zaitseva E.M., Zaitsev E.N., Kowalczykowski S.C. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 79.Bianco P.R., Tracy R.B., Kowalczykowski S.C. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 80.Xu L., Marians K.J. A dynamic RecA filament permits DNA polymerase-catalyzed extension of the invading strand in recombination intermediates. J. Biol. Chem. 2002;277:14321–14328. doi: 10.1074/jbc.M112418200. [DOI] [PubMed] [Google Scholar]
- 81.Kiianitsa K., Solinger J.A., Heyer W.D. Terminal association of the Rad54 protein with the Rad51-dsDNA filament. Proc. Natl Acad. Sci. USA. 2006;103:9767–9772. doi: 10.1073/pnas.0604240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wesoly J., Agarwal S., Sigurdsson S., Bussen W., Van Komen S., Qin J.A., van Steeg H., van Benthem J., Wassenaar E., Baarends W.M., et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sugawara N., Wang X., Haber J.E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 84.Alexeev A., Mazin A., Kowalczykowski S.C. Rad54 protein possesses chromatin-remodeling activity stimulated by a Rad51-ssDNA nucleoprotein filament. Nature Struct. Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 85.Alexiadis V., Kadonaga J.T. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 2003;16:2767–2771. doi: 10.1101/gad.1032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sugawara N., Ivanov E.L., Fishman Lobell J., Ray B.L., Wu X., Haber J.E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 87.Bärtsch S., Kang L.E., Symington L.S. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hiramoto T., Nakanishi T., Sumiyoshi T., Fukuda T., Matsuura S., Tauchi H., Komatsu K., Shibasaki Y., Inui H., Watatani M., et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 89.Petukhova G., Sung P., Klein H. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 2000;14:2206–2215. doi: 10.1101/gad.826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dresser M.E., Ewing D.J., Conrad M.N., Dominguez A.M., Barstead R., Jiang H., Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee S.E., Pellicioli A., Malkova A., Foiani M., Haber J.E. The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol. 2001;11:1053–1057. doi: 10.1016/s0960-9822(01)00296-2. [DOI] [PubMed] [Google Scholar]
- 92.Broomfield S., Hryciw T., Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. DNA Repair. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 93.Prakash S., Prakash L. Nucleotide excision repair in yeast. Mutat. Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 94.Troelstra C., Van Gool A., Dewit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.J. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 95.Selby C.P., Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 96.Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H.J., Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morrison A.J., Highland J., Krogan N.J., Arbel-Eden A., Greenblatt J.F., Haber J.E., Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 98.van Attikum H., Fritsch O., Hohn B., Gasser S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 99.Downs J.A., Allard S., Jobin-Bobitaille O., Javaheri A., Auger A., Bouchard N., Kron S.J., Jackson S.P., Cote J. Binding of chromatin-modifying activities to phopshorylated histone H2A at DNA damage sites. Mol. Cell. Biol. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 100.Shen X., Mizuguchi G., Harmiche A., Wu C. A chromatin remodelling complex involved in transciption and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 101.Shen X., Ranallo R., Choi E., Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- 102.Mizuguchi G., Shen X.T., Landry J., Wu W.H., Sen S., Wu C. ATP-Driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 103.Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 104.Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double strand break to double Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 105.Interthal H., Heyer W.D. MUS81 encodes a novel Helix-hairpin-Helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 106.Calderon I.L., Conotopoulou C.R., Mortimer R.K. Isolation and characterization of yeast DNA repair genes. II. Isolation of plasmids that complement the mutations rad50-1, rad51-1, rad54-3 and rad55-3. Curr. Genet. 1983;7:93–100. doi: 10.1007/BF00365632. [DOI] [PubMed] [Google Scholar]
- 107.Emery H.S., Schild D., Kellogg D.E., Mortimer R.K. Sequence of RAD54, a Saccharomyces cerevisiae gene involved in recombination and repair. Gene. 1991;104:103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- 108.Kanaar R., Troelstra C., Swagemakers S.M.A., Essers J., Smit B., Franssen J.H., Pastink A., Bezzubova O.Y., Buerstedde J.M., Clever B., et al. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol. 1996;6:828–838. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 109.Zhang X.P., Lee K.I., Solinger J.A., Kiianitsa K., Heyer W.D. Gly-103 in the N-terminal domain of Saccharomyces cerevisiae Rad51 protein is critical for DNA binding. J. Biol. Chem. 2005;280:26303–26311. doi: 10.1074/jbc.M503244200. [DOI] [PubMed] [Google Scholar]