Abstract
Phototropins (phot1 and phot2, formerly designated nph1 and npl1) are blue-light receptors that mediate phototropism, blue light-induced chloroplast relocation, and blue light-induced stomatal opening in Arabidopsis. Phototropins contain two light, oxygen, or voltage (LOV) domains at their N termini (LOV1 and LOV2), each a binding site for the chromophore flavin mononucleotide (FMN). Their C termini contain a serine/threonine protein kinase domain. Here, we examine the kinetic properties of the LOV domains of Arabidopsis phot1 and phot2, rice (Oryza sativa) phot1 and phot2, and Chlamydomonas reinhardtii phot. When expressed in Escherichia coli, purified LOV domains from all phototropins examined bind FMN tightly and undergo a self-contained photocycle, characterized by fluorescence and absorption changes induced by blue light (T. Sakai, T. Kagawa, M. Kasahara, T.E. Swartz, J.M. Christie, W.R. Briggs, M. Wada, K. Okada [2001] Proc Natl Acad Sci USA 98: 6969–6974; M. Salomon, J.M. Christie, E. Knieb, U. Lempert, W.R. Briggs [2000] Biochemistry 39: 9401–9410). The photocycle involves the light-induced formation of a cysteinyl adduct to the C(4a) carbon of the FMN chromophore, which subsequently breaks down in darkness. In each case, the relative quantum efficiencies for the photoreaction and the rate constants for dark recovery of LOV1, LOV2, and peptides containing both LOV domains are presented. Moreover, the data obtained from full-length Arabidopsis phot1 and phot2 expressed in insect cells closely resemble those obtained for the tandem LOV-domain fusion proteins expressed in E. coli. For both Arabidopsis and rice phototropins, the LOV domains of phot1 differ from those of phot2 in their reaction kinetic properties and relative quantum efficiencies. Thus, in addition to differing in amino acid sequence, the phototropins can be distinguished on the basis of the photochemical cycles of their LOV domains. The LOV domains of C. reinhardtii phot also undergo light-activated spectral changes consistent with cysteinyl adduct formation. Thus, the phototropin family extends over a wide evolutionary range from unicellular algae to higher plants.
Plants use light not only as an energy source for photosynthesis but also as a signal to indicate the properties of their surrounding environment. UV-A (320–390 nm) and blue (390–500 nm) light regulate a wide variety of responses in higher plants. These responses include phototropism, chloroplast relocation, inhibition of hypocotyl elongation, circadian timing, regulation of gene expression, and stomatal opening (Briggs and Huala, 1999; Lin, 2000; Christie and Briggs, 2001; Briggs et al., 2001b). Four blue-light receptors have been identified from the model higher plant Arabidopsis: cryptochrome 1 (Ahmad et al., 1993), cryptochrome 2 (Hoffman et al., 1996; Lin et al., 1996, 1998), phototropin 1 (phot1; Huala et al., 1997; Christie et al., 1998, 1999), and phototropin 2 (phot2; Jarillo et al., 1998).
In Arabidopsis, phot1 and phot2 (formerly known as nph1 and npl1, respectively; see Briggs et al., 2001a) have been shown to serve as photoreceptors mediating phototropism (Huala et al., 1997; Christie et al., 1998), blue light-mediated chloroplast movement (Kagawa et al., 2001; Sakai et al., 2001; Jarillo et al., 2001), and blue light-induced stomatal opening (Kinoshita et al., 2002). Phot1 and phot2 function in a light fluence-dependent manner to regulate these two responses (Sakai et al., 2001). Phot1 mediates phototropism in response to low- and high-fluence rates of unilateral blue light; phot2 does so only in response to high-fluence rate unilateral blue light.
The situation is slightly different with respect to light-activated chloroplast movement. Chloroplasts exhibit different responses depending on the light intensity. Under low-light intensities, chloroplasts move to the periclinal surfaces of mesophyll cells (accumulation response), presumably maximizing light capture for photosynthesis. Under high-light intensities, chloroplasts move away from the periclinal surfaces of these cells to the anticlinal surface (avoidance response), presumably minimizing photodamage to the photosynthetic machinery. Genetic evidence indicates that phot1 mediates the accumulation response at both low- and high-fluence rates of blue light (Kagawa et al., 2001; Sakai et al., 2001), whereas phot2 mediates the accumulation response at low-fluence rates and the avoidance response at high-fluence rates of blue light (Kagawa et al., 2001; Jarillo et al., 2001; Sakai et al., 2001).
The phototropins contain a Ser/Thr protein kinase domain at their C termini and two light, oxygen, or voltage (LOV) domains (LOV1 and LOV2) at their N termini (Huala et al., 1997). The LOV domains function as binding sites for the chromophore flavin mononucleotide (FMN; Christie et al., 1999) and belong to the PAS domain (originally characterized in PER, ARNT, and SIM proteins) superfamily (Taylor and Zhulin, 1999). LOV1 and LOV2 are approximately 100 amino acids in length and are separated by a variable intervening sequence. Both LOV domains undergo a self-contained photocycle (Salomon et al., 2000). The spectral properties of the longest-lived intermediate in the photocycle of LOV1 and LOV2 are consistent with those reflecting the formation of a flavin-cysteinyl adduct (Miller et al., 1990). A highly conserved Cys residue found in all presently characterized phototropin LOV domains is essential for the formation of this photoproduct. The light reaction likely causes binding of the sulfur covalently to the C(4a) carbon of the isoalloxazine ring of the FMN chromophore after photoexcitation of the FMN. The recently solved crystal structure of the LOV2 domain of phy3 from the fern Adiantum capillus-veneris is consistent with this model (Crosson and Moffat, 2001). Phy3 is a chimeric photoreceptor with a phytochrome chromophore-binding domain its N terminus and a full-length phototropin sequence (with LOV1, LOV2, and kinase domains) at its C terminus.
After photoproduct formation, the cysteinyl adduct decays spontaneously back to the ground state in the dark. The kinetic parameters, relative quantum efficiencies, and rate constants for dark recovery differ between LOV1 and LOV2 of oat phot1, although the same photochemistry likely occurs for both domains (Salomon et al., 2000; see Briggs et al., 2001a). A conformational change in the LOV domain induced by formation of the cysteinyl adduct is hypothesized to lead to the signaling state and activation of the C-terminal kinase domain (Salomon et al., 2000; Swartz et al., 2001). The LOV domains of phot1 and phot2 undergo this photocycle (Salomon et al., 2000; Sakai et al., 2001), and both full-length photoreceptors, when expressed in insect cells, undergo light-activated autophosphorylation (Christie et al., 1998; Sakai et al., 2001).
On the basis of sequence homology, a number of phototropin genes have been identified from plants, including Arabidopsis, rice (Oryza sativa), pea (Pisum sativum), spinach (Spinacia oleracea), oat (Avena sativa), the fern A. capillus-veneris, and the green alga Chlamydomonas reinhardtii (see Briggs et al., 2001a). The phototropin genes from angiosperms have been divided by phylogenetic analysis into two groups, the PHOT1 group and the PHOT2 group (Kanegae at al., 2000; Briggs et al., 2001a). PHOT2 genes have been identified from Arabidopsis, rice, and spinach; the remaining angiosperm PHOT genes identified so far belong to the PHOT1 group. The fact that two types of phototropins appear to be evolutionarily conserved suggests that there may be some common physical and/or chemical properties within each group and differences in these properties between the two groups. In this study, we investigate this possibility, comparing the kinetic properties of the chromophore-binding LOV domains of the phot1 and phot2 from Arabidopsis and rice. We also present results from experiments investigating the kinetic properties of the LOV domains from C. reinhardtii phototropin.
RESULTS
Absorption Characteristics of the LOV Domains
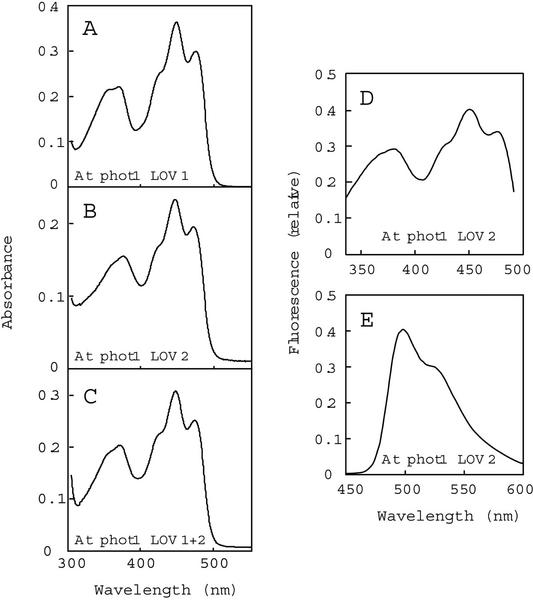
To compare the photochemical properties of the LOV domains from Arabidopsis, rice, and C. reinhardtii, we constructed LOV1, LOV2, and LOV1+2 domains as fusion proteins to a calmodulin-binding protein (CBP) and purified them as described under “Materials and Methods.” We estimate purity between 80% and 90% based on Coomassie-stained SDS-PAGE. Figure 1, A through C, shows the absorption spectra of the LOV-domain fusion proteins from Arabidopsis phot1. The spectra exhibit features of LOV domains described previously for oat and Arabidopsis phot1 (Christie et al., 1999; Salomon et al., 2000) and A. capillus-veneris phy3 (Christie et al., 1999), with absorption peaks in the UV-A and blue regions of the spectrum and prominent vibrational bands in the blue (Swartz et al., 2001). The fusion proteins from Arabidopsis phot2, rice phot1, rice phot2, and C. reinhardtii phot all exhibit essentially the same spectra as those shown in Figure 1 for the Arabidopsis phot1 chromopeptides (data not shown). Molar extinction coefficients, ranging from 11,100 to 15,200 L mol−1 cm−1, calculated as described under “Materials and Methods” are shown in Table I.
Figure 1.
Absorption and fluorescence spectra of the Arabidopsis phot1 LOV domains purified from E. coli extracts. A through C, Absorption spectra of LOV1 (A), LOV2 (B), and LOV1+2 (C). The concentrations of FMN bound for each fusion protein were 28, 16, and 25 μm, respectively. D, Fluorescence excitation spectrum for Arabidopsis phot1 LOV2. Fluorescence emission was monitored at 520 nm. E, Fluorescence emission spectrum for Arabidopsis phot1 LOV2. Excitation was at 390 nm. At, Arabidopsis.
Table I.
Spectral properties of the various LOV-domain fusion proteins
| Construct | Wavelength of Peak | Extinction at Peak Wavelength | Kppr (× 102) | Photoproduct at Photoequilibrium | Relative Quantum Efficiencya |
|---|---|---|---|---|---|
| nm | 1 mol−1 cm−1 | s−1 | % | ||
| Atb phot1 LOV1 | 448.5 | 13,900 | 0.38 | 37 | 0.035 |
| At phot1 LOV2 | 446.5 | 14,300 | 3.9 | 91 | 0.34 |
| At phot1 LOV1+2 | 447.5 | 13,700 | 1.9 | 55 | |
| At phot2 LOV1 | 447.5 | 14,700 | 1.5 | 62 | 0.13 |
| At phot2 LOV2 | 445.5 | 13,500 | 2.9 | 52 | 0.27 |
| At phot2 LOV1+2 | 446.5 | 13,400 | 2.4 | 55 | |
| Os phot1 LOV1 | 449.5 | 11,200 | 0.23 | 33 | 0.026 |
| Os phot1 LOV2 | 446.5 | 15,200 | 3.7 | 87 | 0.30 |
| Os phot1 LOV1+2 | 448.0 | 11,800 | 1.6 | 41 | |
| Os phot2 LOV1 | 448.0 | 11,100 | 2.9 | 83 | 0.33 |
| Os phot2 LOV2 | 446.5 | 13,300 | 3.1 | 81 | 0.29 |
| Os phot2 LOV1+2 | 447.0 | 13,300 | 2.9 | 77 | |
| Cr LOV1 | 447.0 | 13,800 | 3.3 | 73 | 0.30 |
| Cr LOV2 | 445.5 | 13,300 | 3.7 | 69 | 0.35 |
| Cr LOV1+2 | 446.5 | 13,200 | 3.6 | 75 |
Approximate efficiencies (Q) were calculated from the equation Q = Kppf ε−1 I−1, where Kppf is the initial rate of photoproduct formation in s−1, ε is the molar extinction coefficient in 1 mol−1 cm−1, and I is the fluence rate of the actinic light in μmol m−2 s−1.
At, Arabidopsis; Os, rice; Cr, C. reinhardtii.
Figure 1, D and E, shows the fluorescence excitation and emission spectra, respectively, of the LOV2-domain fusion protein from Arabidopsis phot1. The highest peaks are at 450 nm in the excitation spectrum and 490 nm for the emission spectrum, with a shoulder near 535 nm in the latter. The other LOV-domain proteins show fluorescence excitation and emission spectra similar to those shown in Figure 1, D and E (data not shown). These spectral properties are consistent with those previously reported for the LOV domains from oat and Arabidopsis phot1 (Christie et al., 1999; Salomon et al., 2000), Arabidopsis phot2 (Sakai et al., 2001), and full-length Arabidopsis phot1 expressed in insect cells (Christie et al., 1998). Note that the ratio of the 350 to 450 nm peaks differs between the absorption and fluorescence excitation spectrum. The difference arises because the high-fluence-rate light required to elicit the fluorescence emission actually is sufficient to drive some of the chromopeptide to the cysteinyl-adduct form, differentially bleaching the two spectral regions.
Photoproducts of the LOV Domains Show Reduced Fluorescence
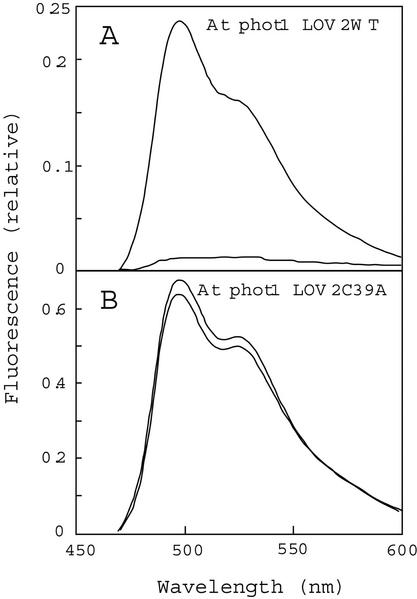
The LOV1 and LOV2 domains from oat phot1 have been shown to undergo a self-contained photocycle (Salomon et al., 2000). Blue-light irradiation induces photoproduct formation, followed by dark regeneration to the initial blue-light-absorbing form (Salomon et al., 2000). Replacement of a highly conserved Cys residue with Ala (C39A) or Ser (C39S) in these LOV domains abolishes the formation of the cysteinyl adduct without reducing FMN binding (Salomon et al., 2000). This Cys corresponds to amino acid residue 39 within each LOV domain and residue 512 in full-length Arabidopsis phot1 LOV2. Irradiation of the Arabidopsis phot1 LOV2-domain fusion protein with high-fluence-rate white light (to obtain maximal photoproduct formation) greatly reduces its fluorescence emission (Fig. 2A, compare with upper and lower traces). This fluorescence decrease is consistent with the decrease shown elsewhere for the formation of a flavin-cysteinyl adduct (Miller et al., 1990; Salomon et al., 2000). During subsequent dark incubation, the fluorescence recovers completely (see below). Saturating photoexcitation virtually eliminates any fluorescence change in the same Arabidopsis phot1 LOV2 fusion protein in which the conserved Cys residue has been mutated to an Ala (Fig. 2B, compare with upper and lower traces), consistent with the results obtained with the oat phototropin LOV domains cited above. The present results support the hypothesis that this Cys is essential for the photocycle (Salomon et al., 2000) and that formation of the cysteinyl adduct strongly quenches flavin fluorescence.
Figure 2.
The changes in the fluorescence emission spectrum induced by saturating white-light irradiation. A, Emission spectrum for the Arabidopsis LOV2 domain before (upper line) and after (lower line) a 10-s irradiation with white light (5,000 μmol m−2 s−1). B, Emission spectra for the mutant Arabidopsis LOV2 C39A domain from phot1 before (upper line) and after (lower line) the white light irradiation. In both A and B, the excitation wavelength was 450 nm. At, Arabidopsis.
The wild-type LOV-domain fusion proteins undergo their photocycle many times without any detectable change in their absorption properties, fluorescence yields on excitation at 450 nm, or the kinetics of photoproduct formation and decay (Salomon et al., 2000; data not shown). Thus, the light-induced decrease in fluorescence at 490 nm and the subsequent recovery of fluorescence in darkness provide a convenient assay to analyze both photoproduct formation and its subsequent dark decay. This assay was used in the experiments reported below.
Photoproduct-Formation Kinetics for the LOV Domains of Arabidopsis phot1 and phot2
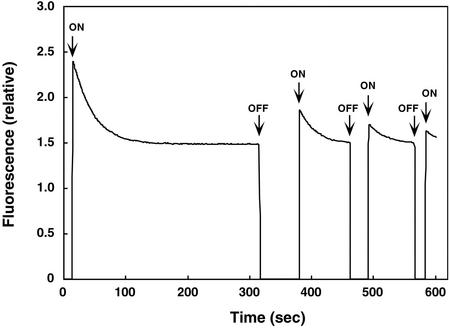
First, we determined the rate constants of photoproduct formation for the various LOV-domain fusion proteins from Arabidopsis phot1 and phot2. Formation of the photoproduct was monitored by recording the decrease in fluorescence induced by irradiating with blue light (Fig. 3). After a dark pre-incubation period of 10 min, we irradiated the LOV-domain fusion proteins with excitation light at 450 nm (starting at 13 s in Fig. 3) and monitored fluorescence emission at 490 nm where fluorescence emission is maximal (Fig. 2). Fluorescence emission was initially at a maximum intensity and then gradually decreased, approaching a steady-state level after approximately 300 s (Fig. 3), presumably reflecting adduct formation. The rate of decrease in fluorescence is proportional to the quantum yield, because the decrease is associated with adduct formation, a process that is orders of magnitude faster than the return of the pigment to the ground state in darkness (Swartz et al., 2001).
Figure 3.
Time course for fluorescence changes for the Arabidopsis phot2 LOV1 domain. Excitation was at 450 nm and emission was monitored at 490 nm. Before the start of the experiment, the sample was incubated in darkness for 10 min. The time scan was started at 0 s in the dark, and the shutter for the excitation beam was opened at 13 s (first ON). The decay curve for fluorescence emission was used to calculate photoproduct formation. When the fluorescence reached steady state (316 s), the shutter was closed (first OFF). After an additional predetermined time in darkness (74 s), the shutter was reopened. The magnitude of the immediate fluorescence increase reflects the amount of recovery of the initial fluorescent state. As soon as the signal reached steady state again (462 s), the shutter was closed. It was then re-opened to make a lights-on measurement after another predetermined period of time (in this case 29 s), etc. In this way, a time course for the regeneration of a fluorescent species could be followed.
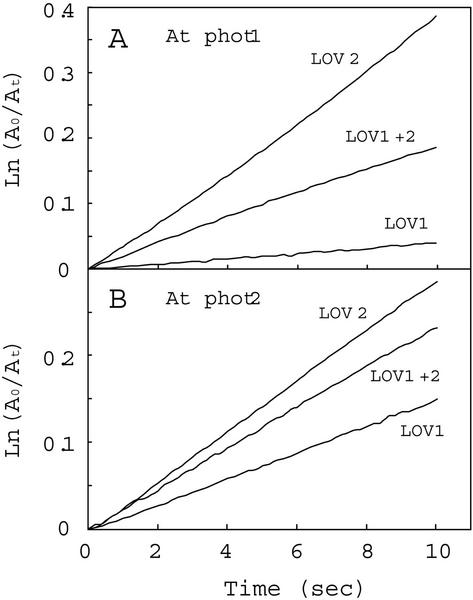
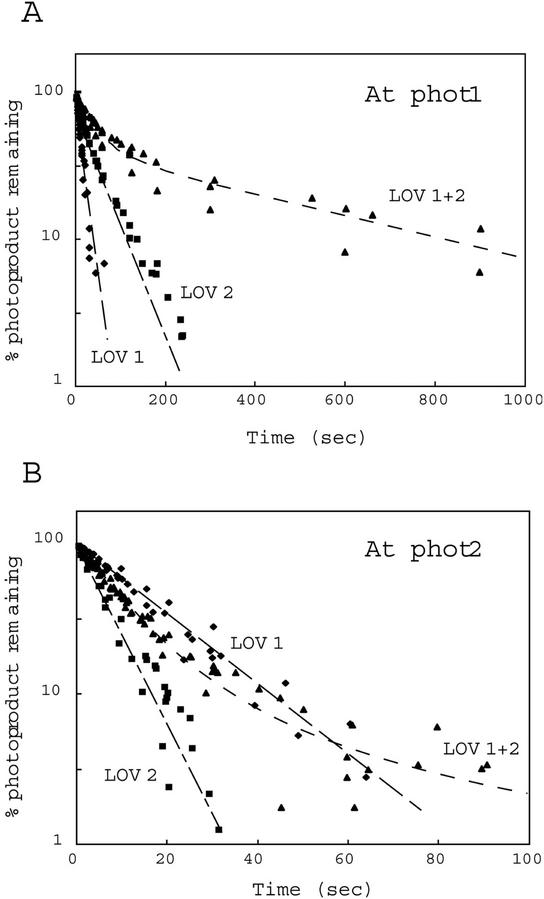
Figure 4 shows semilogarithmic plots of photoproduct formation versus time. For both Arabidopsis phot1 and phot2, the rates of photoproduct formation obtained for the LOV2 domains were faster than those observed for the LOV1 domains. Moreover, the LOV1+2 domains showed rates of photoproduct formation intermediate between those of the individual LOV1 and LOV2 domains (Fig. 4; Table I). The data for the tandem fusion protein within the first 10 s largely reflect an early rapid phase only. Longer exposures show a significant decrease in the rate of photoproduct formation (data not shown).
Figure 4.
Reaction kinetics for photoproduct formation. The kinetics for photoproduct formation for the Arabidopsis phot1 LOV-domain fusion proteins were monitored at 4°C. A0 represents the amount of unreacted LOV-domain protein as indicated by the fluorescence detected immediately on opening the excitation lamp shutter. At represents the corresponding amount of unreacted protein at time t, calculated from the amount of fluorescence at time t. A, LOV-domain proteins from phot1; B, LOV-domain proteins from phot2. At, Arabidopsis.
The relative quantum efficiencies for each photoreaction were calculated from the rate of fluorescence decrease (Kppf), the molar extinction coefficients, and the fluence rate of the blue light (80 μmol m−2 s−1; Table I). The calculated values were 0.035 for LOV1 and 0.34 for LOV2 from Arabidopsis phot1, a 10-fold difference similar to that reported previously for oat phot1 LOV1 and LOV2 (0.045 and 0.44, respectively; Salomon et al., 2000). In contrast, the relative quantum efficiencies for Arabidopsis phot2 LOV1 and LOV2 (0.13 and 0.27, respectively) differed only by a factor of 2.
Dark-Regeneration Kinetics for the Arabidopsis LOV-Domain Photoproducts
The light-inducible fluorescence changes were in all cases reversible in the dark (Fig. 3). After a given fluorescence decrease reached a steady state, the shutter for the excitation lamp was closed, and the sample was allowed to recover in the dark for a specific length of time (e.g. 316–380 s and 462–491 s in Fig. 3). With longer dark incubations, the intensity of the fluorescence detected immediately upon re-opening the excitation shutter became progressively greater, indicating a decrease in the amount of photoproduct remaining and a concomitant increase in the photosensitive LOV-domain dark state. Figure 5, A and B, shows semilogarithmic plots for photoproduct remaining versus time in the dark after a saturating light exposure. These plots are nearly log-linear for the single LOV domains from both Arabidopsis phototropins, indicating that regeneration of the dark form is first order in all four cases. However, there are major quantitative differences between the phot1 and phot2 LOV domains: The regeneration rate for phot2 LOV2 at room temperature was 6-fold faster than phot1 LOV2 (t1/2, for regeneration 5 and 29 s, respectively), whereas the regeneration rates for the LOV1 domains were almost the same (t1/2, 13 and 10 s for phot2 LOV1 and phot1 LOV1, respectively; Table II). With phot1, the dark decay of LOV1 was more rapid than that for LOV2; whereas for phot2, the situation was reversed (Fig. 5; Table II).
Figure 5.
Reaction kinetics for the dark recovery of the Arabidopsis LOV-domain fusion proteins from both phot1 (A) and phot2 (B). The recovery kinetics were monitored at room temperature. The amount of photoproduct remaining after dark incubation is plotted against the duration of dark incubation. The fluorescence intensity difference between that obtained immediately on opening the excitation lamp shutter after a 10-min dark incubation and that obtained when the fluorescence intensity reached steady state was used to determine the initial amount of photoproduct. The amount of photoproduct remaining after a certain duration of dark incubation was calculated as the initial amount of photoproduct minus the amount of recovery during the dark incubation. Half-lives calculated from the fitted curves are shown in Table III. At, Arabidopsis.
Table II.
Half-lives for dark decay of the cysteinyl adduct following photoexcitation of the single LOV domain fusion proteins
| Photoreceptor | Construct | t1/2 |
|---|---|---|
| s | ||
| At phot1a | LOV1 | 10 |
| At phot1 | LOV2 | 29 |
| At phot2 | LOV1 | 13 |
| At phot2 | LOV2 | 5 |
| Os phot1 | LOV1 | 27 |
| Os phot1 | LOV2 | 43 |
| Os phot2 | LOV1 | 28 |
| Os phot2 | LOV2 | 14 |
| Cr Phot | LOV1 | 168 |
| Cr Phot | LOV2 | 20 |
At, Arabidopsis; Os, rice; Cr, C. reinhardtii.
Photoproduct-Formation Kinetics for the LOV Domains of Rice phot1 and phot2
The genes encoding rice phot1 and phot2 were recently cloned, and phylogenetic analysis indicated that the rice PHOT1 and PHOT2 genes showed high sequence homology to the Arabidopsis PHOT1 and PHOT2, respectively (Kanegae et al., 2000; see Briggs et al., 2001). Therefore, it was of interest to determine whether the kinetic differences between the LOV domains from Arabidopsis phot1 and phot2 were also observed for the rice photoreceptors. The kinetic properties of the LOV domains from the two rice phototropins were determined by the same methods that were used for the Arabidopsis phototropins.
For rice phot1, the relative quantum efficiency values for photoproduct formation were 0.026 for LOV1 and 0.30 for LOV2 (Table I). This order-of-magnitude difference (12-fold) in relative quantum efficiency between LOV1 and LOV2 is very close to that reported above for Arabidopsis phot1 (10-fold) and elsewhere for oat phot1 (10-fold, Salomon et al., 2000). In contrast, there was only a small difference in relative quantum efficiencies of photoproduct formation between LOV1 and LOV2 for rice phot2, similar to the small difference reported above for Arabidopsis phot1.
Dark-Regeneration Kinetics of the Rice LOV-Domain Photoproducts
As for the Arabidopsis phot1 and phot2 LOV-domain fusion proteins, semilogarithmic plots for dark regeneration for all of the LOV domains from rice phot1 and phot2 were linear (data not shown; see Fig. 5). Furthermore, the relationships observed for the dark decay of the rice phototropin LOV domains were also strikingly similar to those of the Arabidopsis chromopeptides (Table II). The rice phot2 LOV2 domain decayed more rapidly than the phot1 LOV2 domain (just over 3-fold), whereas the LOV1 domains from rice phot1 and phot2 showed almost identical dark-regeneration kinetics.
Kinetic Properties of Fusion Proteins Containing Both LOV Domains
For both Arabidopsis and rice phot1 and phot2, the LOV1+2 fusion proteins showed apparent quantum efficiencies, based on the initial rate of photoproduct formation, that are relatively high (0.17–0.27). Especially for the phot1 LOV1+2 proteins from both species, the relatively high apparent quantum efficiencies suggest that the initial slope might be determined largely by LOV2 rather than by LOV1. However, interactions between the domains (and/or the intervening sequences) could affect the quantum efficiencies of either or both LOV domains. Hence, further studies are required to clarify this aspect of the photochemistry of these two-domain fusion proteins.
As the LOV1+2 fusion proteins contain both LOV domains, it is not surprising that their dark regeneration is not log-linear but rather shows fast and slow components (Fig. 5). However, the overall decay kinetics were far slower than for either LOV domain alone (Fig. 5, A and B; compare with Tables II and III). For both species, dark decay for the phot1 LOV1+2 fusion protein was far slower than for the phot2 LOV1+2 fusion protein (Table III).
Table III.
Half-lives, s, for the two components in the dark decay of the cysteinyl adduct following photoexcitation for LOV1+2 fusion proteins and full-length phototropins
| Peptide or Holoprotein | t1/2 |
|---|---|
| s | |
| At phot1 LOV1+2a | 30 and 393 |
| At phot2 LOV1+2 | 6.5 and 28 |
| At full-length phot1 | 70 and 760 |
| At full-length phot2 | 15 and extremely longb |
| Os phot1 LOV1+2 | 28 and 137 |
| Os phot2 LOV1+2 | 13 and 60 |
| Cr phot LOV1+2 | 17 and 200 |
At, Arabidopsis; Os, rice; Cr, C. reinhardtii.
Data of insufficient resolution to calculate half-lives.
Kinetic Properties of Full-Length Arabidopsis phot1 and phot2 Expressed in Insect Cells
The LOV domains of phot1 and phot2 exhibit different photochemical properties and reaction kinetics. It was, therefore, important to investigate these parameters for the full-length phot1 and phot2 proteins. Because we have been unable to express the full-length phototropin proteins in E. coli to date, we used a recombinant baculovirus/insect cell system to express phot1 and phot2 (Christie et al., 1998). When expressed in insect cells, Arabidopsis phot1 and phot2 bind FMN and undergo autophosphorylation in response to blue-light irradiation (Christie et al., 1998; Sakai et al., 2001). Although the concentration of phototropin in soluble fractions from insect cells is too low for accurate absorption measurements, it is sufficient for the sensitive fluorescence technique used in obtaining the data in Figures 3 through 5. Thus, soluble extracts from insect cells expressing either phot1 or phot2 provide a convenient system to analyze the reaction kinetics of the full-length phototropin proteins.
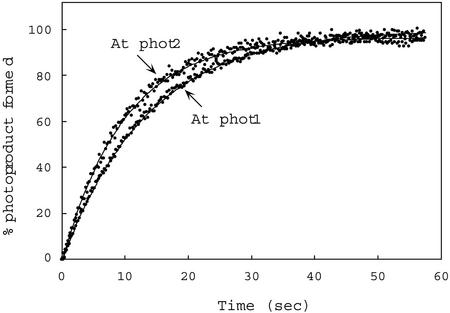
As before, both photoproduct formation and dark regeneration for full-length phot1 and phot2 were determined by monitoring light-induced fluorescence changes. The rates of photoproduct formation determined for these holoproteins were very similar (t1/2 = 14 and 11 s, respectively, Fig. 6) and are somewhat shorter than those obtained for the Arabidopsis phot1 and phot2 LOV1 + 2 fusion peptides expressed in E. coli (t1/2, 36.5 and 29.2 s, respectively, calculated from kreg values; Table II). Because crude soluble extracts were used instead of purified protein preparations, relative quantum efficiencies were not calculated for the full-length photoreceptors.
Figure 6.
Time course for photoproduct formation for full-length Arabidopsis phot1 and phot2 produced in insect cells from recombinant baculovirus. Measurements were made at room temperature. Data points calculated as for Figure 4. Protein concentrations for the crude soluble extracts were 18.4 mg/mL (phot1) and 23.4 mg/mL (phot2). At, Arabidopsis.
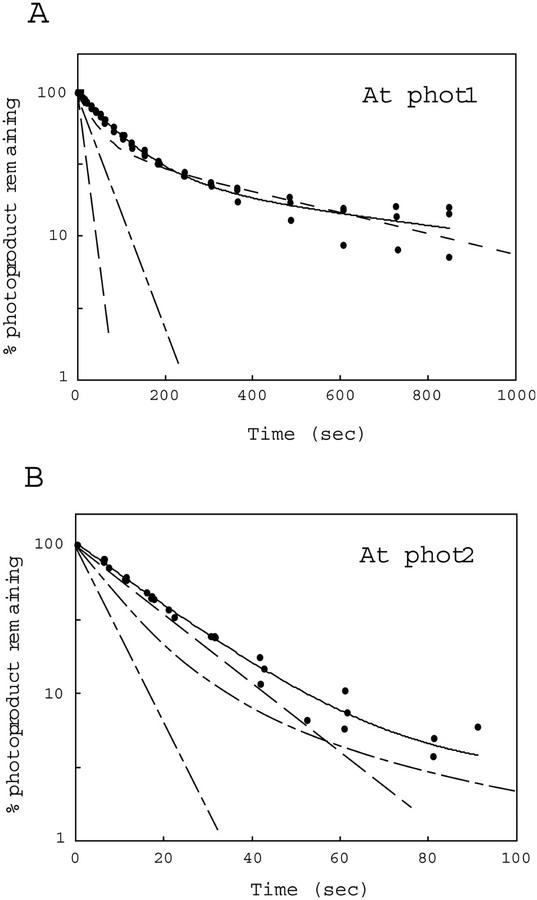
Dark-regeneration kinetics for the phot1 and phot2 holoproteins were somewhat slower than those for the phot1 and phot2 LOV1+2 peptides expressed in E. coli (Table III). Nevertheless, the dark regeneration rate for full-length phot1 (t1/2 for the main component 70 s) is significantly slower than that for full-length phot2 (t1/2 for the main component 15 s). These values are consistent with the dark-decay properties exhibited by the Arabidopsis phot1 and phot2 LOV1+2 domains expressed in E. coli (compare with Figs. 5 and 7). These findings suggest that the properties of the photocycle observed for the phot1 and phot2 LOV1+2 fusion peptides are similar to those observed for the full-length proteins. However, for full-length phot1 and phot2, photoproduct formation appears to be somewhat more rapid and the dark decay somewhat slower than that for the LOV1+2 peptides.
Figure 7.
Reaction kinetics for the dark regeneration of full-length Arabidopsis phot1 and phot2 expressed in insect cells transfected with recombinant baculovirus. Remaining photoproduct was calculated as for Figure 5. Data points and solid line represent the dark recovery for the full-length photoreceptors. Dashed lines are from Figure 5 and represent the dark recovery of the various shorter chromopeptides. Protein concentrations of samples are the same as for Figure 6. At, Arabidopsis.
Kinetic Properties of the LOV-Domain Fusion Proteins from C. reinhardtii Phototropin
A gene encoding a phototropin homolog has been isolated and characterized from the motile alga C. reinhardtii (A. Onodera, N. Mochizuki, and A. Nagatani, unpublished data). It is similar to the higher plant phototropins in having two LOV domains in its N-terminal half and a Ser/Thr kinase in its C-terminal half. It differs in that there are only a few amino acids upstream from the start of the LOV1 domain instead of 110 to 178 (phot1 proteins) or 86 to 116 (phot2 proteins). We expressed the LOV domains of the C. reinhardtii phot both singly and in combination in E. coli as described above for the rice and Arabidopsis phototropins. The photochemical and reaction-kinetic properties of C. reinhardtii LOV-domain fusion proteins were studied in a similar fashion. With relatively high apparent quantum efficiencies for the LOV domains expressed separately (0.30 and 0.35 for LOV1 and LOV2, respectively) and an equally high apparent quantum efficiency (0.34) for the LOV1+2 peptides (Table I), the C. reinhardtii phot LOV domains most closely resemble those of the rice and Arabidopsis phot2 LOV1, LOV2, and LOV1+2 proteins. Likewise, the more rapid regeneration rate for LOV2 compared with LOV1 observed for C. reinhardtii phot (Table II) mostly resembles the properties of the higher plant phot2 LOV domains rather than the phot1 LOV domains. The LOV1 decay rate for the C. reinhardtii protein was the slowest measured in this study. The dark regeneration rate for the C. reinhardtii LOV1+2 peptide (Table III) was also very slow. In this case, the rate is similar to the rates determined for the higher plant phot1 LOV1+2 fusion protein than those of the phot2 LOV1+2 fusion protein (Table III).
DISCUSSION
Examination of the relative quantum efficiencies for the LOV1 and LOV2 domains between phot1 and phot2 from higher plants shows that there are major differences between the two photoreceptor groups. For the three phot1 LOV domains thus far investigated (Arabidopsis, rice, and oat), the relative quantum efficiency for light-activated formation of the cysteinyl adduct for LOV2 is approximately 10-fold higher than that for LOV1 (Table IV). In contrast, the ratio between LOV2 and LOV1 is much smaller for the phot2 examined. All five phototropin LOV2 domains examined here share a high relative quantum efficiency (0.27–0.44). The difference in the quantum-efficiency ratio between the LOV1 and LOV2 domains of phot1 and phot2 is because the relative quantum efficiencies for the phot1 LOV1 domains are much lower (0.026–0.044; Table I) than those of phot2 LOV1 domains. The relative quantum efficiency values of the phot2 LOV1 domains (0.13 and 0.33 for Arabidopsis and rice phot2, respectively; Table I) approach those of the LOV2 domains. The somewhat higher quantum efficiency reported for the oat phot1 LOV2 domain (0.44) may reflect the approximate methods used in both studies (Salomon et al., 2000).
Table IV.
Ratio of relative quantum efficiencies and dark-regeneration rate constants between LOV1 and LOV2 domains of phot1 and phot2
| Photoreceptor | Quantum Efficiency Ratio LOV2:LOV1 | Rate Constant Ratio LOV2:LOV1 |
|---|---|---|
| At phot1a | 9.7 | 0.34 |
| Os phot1 | 11.5 | 0.62 |
| Oat phot1b | 9.8 | 0.43 |
| At phot2 | 2.1 | 2.5 |
| Os phot2 | 0.9 | 2.0 |
At, Arabidopsis; Os, rice.
Values are from Salomon et al. (2000).
Examination of the rate constants for dark regeneration also shows clear differences between the two photoreceptor groups. For phot1 from all three species examined, dark regeneration for LOV2 is relatively slow (t1/2 between 27 and 46 s; Table II; Salomon et al., 2000). For phot2, the regeneration rate for LOV2 is relatively fast (t1/2 5 and 14 s for Arabidopsis and rice phot2, respectively). Thus, dark regeneration of LOV2 is faster than that for LOV1 in phot1, but significantly slower than for LOV1 in phot2 (last column, Table IV).
It is too early to explain the differing behavior of the various LOV domains on the basis of their amino acid sequences. The amino acid sequence identity between LOV1 and LOV2, whether within a given phototropin or between phototropins from different plant species, is always approximately 40% (Christie et al., 1999). Hence, there are numerous differences between LOV1 and LOV2, and between the LOV domains of phot1 and phot2. Further site-directed mutagenesis experiments will be required to resolve the molecular basis for the photocycle differences reported above. Recent resolution of the LOV2 domain structure from the chimeric photoreceptor phy3 from A. capillus-veneris (Crosson and Moffat, 2001) indicates that many conserved amino acid residues play an important role in forming a tight FMN-binding pocket through hydrogen bonding and van der Waals forces. These amino acids are in close proximity to the isoalloxazine ring of the FMN chromophore and may contribute to the photochemical differences between LOV1 and LOV2. Only one of these conserved amino acids differs between LOV1 and LOV2: a Leu (Leu-83) in LOV1 and a Phe (Phe-83) in LOV2. However, mutating the Phe in LOV2 to a Leu does not change the relative quantum efficiency or dark-regeneration rate dramatically (J.M. Christie, T.E. Swartz, and W.R. Briggs, unpublished data). Whether the relatively slow photocycle of the LOV2 domain has some functional significance remains to be determined. The C. reinhardtii phot is unique in having few amino acid residues upstream from LOV1 and the slowest dark decay measured for any LOV1 fusion peptide. The long lifetime of this particular cysteinyl adduct may also have some functional significance. At present, there is no known biological function for this putative C. reinhardtii photoreceptor.
Phylogenetic analysis based on amino acid sequence divides the phototropins into two distinct groups (phot1 and phot2; Kanegae et al., 2000; Briggs et al., 2001a). The members of each group share kinetic properties related to formation of the cysteinyl adduct, but the two groups are distinct from each other. These distinctions hold both for the light reactions and for dark regeneration of the phototropin LOV domains. However, the kinetic behavior of the individual LOV1 and LOV2 domains, while distinguishing the phot1 group from the phot2 group, is markedly different from that of the LOV1+2 peptides and the full-length photoreceptors. The results from experiments measuring light-induced fluorescence changes in full-length Arabidopsis phot1 and phot2 produced in insect cells (Figs. 6 and 7, compare with Fig. 5 and Table I) indicate photoactivation and dark-regeneration kinetics similar to those reported here for the Arabidopsis phot1 and phot2 LOV1+2 peptides containing both the LOV domains and their intervening sequences. Full-length phot1 and phot2 from the insect cells are likely correctly folded as the phosphorylation kinetics and photosensitivity of the phot1 protein have been shown to be identical to those of the native photoreceptor (Christie et al., 1998). The behavior of the LOV1+2 fusion proteins is, hence, likely to reflect the behavior of the chromophore domains in the full-length photoreceptor more closely than the isolated single LOV-domain fusion proteins. Thus, the LOV1+2 fusion proteins provide a reasonable model system for further experimentation.
In fusion proteins containing both LOV domains, there must be interactions occurring that affect the regeneration kinetics of the two individual domains. The kinetics for phot1 and phot2 (either LOV1+2 or full-length proteins) show both rapid and slow phases of dark decay. However, the rapid phase is in all cases slower than the rate of the more rapid of the two LOV domains measured separately, and the slower phase is slower than the rate of the slower LOV domain measured separately. Thus, in the LOV1+2 fusion proteins or the full-length photoreceptors, there are evidently interactions between the LOV1 and LOV2 domains and between the LOV domains and the intervening sequences, or both, that retard the dark regeneration to the ground state. This interaction is not observed when the two LOV domains are expressed separately and then mixed before assay for photoproduct formation or dark decay. The rapid phase of dark decay for both tandem constructs is similar (though not identical) to that of LOV2 alone for both phototropins and, hence, may reflect the behavior of LOV2 within the larger peptide containing both LOV1 and LOV2. If so, then it is the dark decay of LOV1 that is greatly retarded in the tandem fusion proteins. Further experiments are needed to address this suggestion.
The long dark-regeneration times reported here for full-length phot1 match the dark-regeneration times for three other processes: the loss of the influence of a light flash with increasing time in darkness after the irradiation of membrane preparations in phosphorylation assays of native phot; the reappearance of photosensitivity for induction of phosphorylation in vivo; and the recovery of phototropic sensitivity after a saturating light treatment (Palmer et al., 1993; see Briggs et al., 2001a). In all three cases, return to the dark state requires many minutes. It is likely that formation of the cysteinyl adducts activates the kinase domain and other downstream events and that decay of the adducts restores the membrane system to its dark state. Once adduct formation is saturated, further irradiation has no effect, and it is only upon adduct decay that photosensitivity reappears. However, recovery of photosensitivity in vivo must involve different steps, because in that case, the photoreceptors are phosphorylated and not just converted to the cysteinyl adducts.
It is not clear how these kinetic differences between the two phototropin groups may be related to the different physiological functions identified to date for phot1 and phot2. Both photoreceptors play a role in light-induced chloroplast movement and phototropism in Arabidopsis; phot2 functions largely under high-light conditions, whereas phot1 functions under a broad range of light intensities (Kagawa et al., 2001; Sakai et al., 2001). A direct consequence of these differences is that under equal steady-state illumination, there will be differences in photostationary equilibrium values between the two photoreceptors. For both phot2 LOV1+2 constructs, the relatively rapid dark recovery rate will yield steady-state levels of the cysteinyl adduct under continuous illumination lower than for the phot1 LOV1+2 constructs, with their slower regeneration rates. As a consequence, higher fluences are needed to drive phot2 LOV1+2 to the same photostationary equilibrium value as phot1 LOV1+2. This kinetic difference could explain why phot2 mediates responses mainly to higher fluence rates than does phot1 (Sakai et al., 2001).
Kanegae et al. (2000) have reported other differences between phot1 and phot2 in rice. For example, phot1 is strongly expressed in coleoptiles but only weakly expressed in leaves. phot2 is conversely more strongly expressed in leaves than in coleoptiles. In both leaves and coleoptiles, phot2 appears to be up-regulated by light, whereas phot1 is down-regulated. Likewise, phot2 is up-regulated by light in Arabidopsis (Jarillo et al., 2001). It will be of interest to learn whether these or other differences between the two photoreceptors appear in other plant species. It will also be of interest to determine whether the differences in photosensitivity between phot1 and phot2 are related to differences in expression, to differences in photochemical properties, or to both. Although phot1 has been localized to the plasma membrane (see Briggs et al., 2001b), the intracellular localization of phot2 is currently unknown.
The present study supports the conclusion that phot1 and phot2 belong to distinct phylogenetic groups. Regardless of whether one organizes the characterized phototropins on the basis of amino acid sequence (Kanegae et al., 2000; Briggs et al., 2001a), differential light sensitivity (Kagawa et al., 2001; Sakai et al., 2001), or the kinetic properties of their photocycles (the present study), the same groupings prevail. Thus, in addition to reflecting differences in gene expression for the two phototropins (see Jarillo et al., 2000; Kagawa et al., 2001; Kanegae et al., 2001), it seems likely that the physiological differences in light sensitivity and dark regeneration rates are related to differences in the photocycles between phot1 and phot2. Further experimentation will be needed to test this hypothesis.
MATERIALS AND METHODS
Construction and Purification of the LOV Fusion Proteins
Construction, expression, and purification procedures were essentially the same as those described previously (Christie et al., 1999). DNA fragments encoding LOV1 and LOV2 domains, and the combined LOV1+2 domains (including the intervening sequence) of the various phototropins were cloned into the bacterial expression vector pCAL-n-EK (Stratagene, La Jolla, CA) and expressed as a fusion protein to the CBP. The following CBP N-terminal fusion proteins were prepared: for Arabidopsis, phot1 LOV1 (amino acids 180–325), phot1 LOV2 (amino acids 448–599), phot1 LOV1+2 (amino acids 180–599), phot2 LOV1 (amino acids 125–261), phot2 LOV2 (amino acids 371–512), and phot2 LOV1+2 (amino acids 125–512); for rice (Oryza sativa), phot1 LOV1 (amino acids 119–264), phot1 LOV2 (amino acids 386–530), phot1 LOV1+2 (amino acids 119–530), phot2 LOV1 (amino acids 85–230), phot2 LOV2 (amino acids 361–512), and phot LOV1+2 (amino acids 85–512); and for Chlamydomonas reinhardtii, phot LOV1(amino acids 12–145), phot LOV2 (amino acids 205–333), and phot LOV1+2 (amino acids 12–333). The transformed BL21(DE3) pLysS Escherichia coli cells were grown at 30°C in Luria-Bertani medium supplemented with ampicillin (100 μg ml−1) and chloramphenicol (30 μg ml−1) to an A600 of approximately 0.5. Protein expression was carried out in darkness for 3 h in the presence of 1 mm isopropyl β-d-galactopyranoside. The cells were harvested by centrifugation, frozen at −20°C, thawed at 4°C, and then resuspended in 30 mL of 50 mm Tris-HCl (pH 8.0) buffer containing 150 mm NaCl, 10 mm 2-mercaptoethanol, 1 mm magnesium acetate, and 2 mm CaCl2. After sonication for disruption, the samples were centrifuged at 200,000g for 30 min at 4°C. The CPB fusion proteins were purified from the supernatants on calmodulin resin according to the instructions from Stratagene.
Expression of the Full-Length phot1 and phot2 Genes in Insect Cells
Full-length phot1 and phot2 genes were expressed in insect cells as described previously (Christie et al., 1998; Sakai et al., 2001). Three-day-old cells (10 mL) infected with recombinant virus were harvested and suspended in 250 μL of 1× phosphorylation buffer as described by Sakai et al. (2001). Cells were lysed by sonication and samples were centrifuged at 14,000g for 5 min at 4°C. Supernatant fractions were used for analysis.
Spectral Analysis
Absorption and fluorescence spectra for the LOV-domain fusion proteins were obtained in a DU-70 spectrophotometer (Beckman Instruments, Fullerton, CA) and an Alphascan spectrofluorimeter (Photon Technology International, Monmouth Junction, NJ), respectively. The FMN to protein ratio was previously calculated to be 1:1 (w/v) for purified LOV1 and LOV2 and 2:1 (w/v) for LOV1+2 for oat phototropin fusion proteins (Christie et al., 1999). Therefore, to calculate extinction coefficients of the LOV-domain fusion proteins, we used the absorption at peak wavelength (Table I) and the concentration of FMN released from each fusion protein upon denaturation as the protein concentration. The proteins were denatured with 10% (w/v) trichloroacetic acid, vortexed, and centrifuged to remove the precipitated protein. The optical density of the clear supernatant was measured at 450 nm. Unless otherwise stated, protein samples for spectral analysis were used at a concentration of 10 μm FMN bound in each preparation. Photoproduct formation was measured with all samples at 4°C with the exception of the full-length proteins produced in insect cells where the irradiation was done at room temperature. For measurement of relative quantum efficiencies, the fluorescence excitation beam (450 nm, one-half bandwidth approximately 10 nm) set at 80 μmol m−2 was used to drive the reaction and the initial rate of fluorescence decrease after the onset of light used to determine the rate of photoproduct formation. The entire sample was irradiated for these measurements. Exponential fitting was performed with the plotting program Cricket Graph. Half-lives for dark decay of the photoproduct were obtained directly from the exponential fits. The time resolution of the spectrofluorometer was sufficiently short (<200 ms) that it did not interfere with the rather slow changes that we observed.
Other Analytical Procedures
Protein concentrations were determined by the method of Bradford as described in the instructions accompanying the Bio-Rad protein assay kit. Identification of flavin molecules was carried out by TLC analysis as described previously (Christie et al., 1998, 1999). The concentration of FMN associated with each of the CBP-LOV-domain fusion proteins was determined as described previously (Christie et al., 1999).
ACKNOWLEDGMENT
We thank Dr. Roberto Bogomolni for many useful discussions and for his careful review of the manuscript.
Footnotes
This research was supported by the National Science Foundation (grant nos. 9601164 and 9940546 to W.R.B.). This is Carnegie Institution of Washington, Department of Plant Biology publication no. 1,500.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.002410.
LITERATURE CITED
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Beck C, Cashmore AR, Christie JM, Hughes J, Jarillo JA, Kagawa T, Kanegae H, Liscum E, Nagatani A et al. The phototropin family of photoreceptors. Plant Cell. 2001a;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM, Salomon M. Phototropins: a new family of flavin-binding blue light receptors in plants. Antioxidants Redox Signaling. 2001b;3:775–788. doi: 10.1089/15230860152664975. [DOI] [PubMed] [Google Scholar]
- Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- Christie JM, Briggs WR. Blue light sensing in higher plants. J Biol Chem. 2001;276:11457–11460. doi: 10.1074/jbc.R100004200. [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman PD, Batschauer A, Hays JB. PHH1, a novel gene from Arabidopsis thaliana, that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol Gen Genet. 1996;253:259–265. doi: 10.1007/s004380050321. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han I-S, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Ahmad M, Cashmore AR. NPL1 (accession no. AF053941): a second member of the NPH serine/threonine kinase family of Arabidopsis. Plant Physiol. 1998;117:719. [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimizaki K-I. phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2002;414:656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, Tabata S, Okada K, Wada M. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- Kanegae H, Tahin M, Savazzini F, Yamamoto K, Yano M, Sasaki T, Kanegae T, Wada M, Takano M. Rice NPH1 homologues, OsMPH1a and OsNPH1b, are differently photoregulated. Plant Cell Physiol. 2000;41:415–423. doi: 10.1093/pcp/41.4.415. [DOI] [PubMed] [Google Scholar]
- Lin C. Plant blue-light receptors. Trends Plant Sci. 2000;5:337–342. doi: 10.1016/s1360-1385(00)01687-3. [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Chan J, Cashmore AR. CRY2: a second member of the Arabidopsis cryptochrome gene family (accession no. U43397) Plant Physiol. 1996;110:1047. [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen C, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SM, Massey V, Ballou D, Williams CH, Jr, Distefano MD, Moore MJ, Walsh CT. Use of a site-directed triple mutant to trap intermediates: demonstration that the flavin C(4a) adduct and reduced flavin are kinetically competent intermediates in mercuric ion reductase. Biochemistry. 1990;29:2831–2841. doi: 10.1021/bi00463a028. [DOI] [PubMed] [Google Scholar]
- Palmer JM, Short TW, Briggs WR. Correlation of blue light-induced phosphorylation to phototropism in Zea mays L. Plant Physiol. 1993;102:1219–1225. doi: 10.1104/pp.102.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domain of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni R. The photocycle of a flavin-binding domain of the blue-light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]