Abstract
Spironolactone (SPIR) binds to cytoplasmic mineralocorticoid receptors (MR) and functions as an aldosterone antagonist. Recently, the drug was shown to have an early suppressive effect on several immunoactive and proinflammatory cytokines.
To elucidate the mechanism behind this, the four MR-binding steroids SPIR, canrenone, 7α-thiomethyl-spironolactone and aldosterone (ALDO) were investigated for effects on lipopolysaccharide- and phytohemagglutinin-A-activated human blood mononuclear cells. Gene expression was examined after 4 h using microarrays, and SPIR affected 1018 transcripts of the (=) 22,000 probed. In contrast, the SPIR-related steroids affected 17 or fewer transcripts. Combining SPIR and ALDO resulted in 940 affected transcripts, indicating that SPIR has an early gene-regulatory effect independent of MR.
The affected genes encode a large number of signalling proteins and receptors, including immunoinflammatory response genes and apoptosis and antiapoptosis genes. Apoptosis was evident in CD3-, CD14- and CD19-positive cells, but only after 18 h of exposure to SPIR.
The transcriptional network involving the differentially regulated genes was examined and the results indicate that SPIR affects genes controlled by the transcription factors NF-κB, CEBPβ and MYC.
These observations provide new insight into the non-MR-mediated effects of SPIR.
Keywords: Mineralocorticoid receptors, immunotherapy, inflammation, pharmacology, gene expression, autoimmunity, cytokines, programmed cell death
Introduction
The steroids, aldosterone (ALDO) and spironolactone (SPIR), as well as the two SPIR metabolites, 7α-thiomethyl-spironolactone (7TS) and canrenone (CAN), bind to the mineralocorticoid receptor (MR) located in the cytoplasm of many cell types including blood mononuclear cells (MNC) (Armanini et al., 1985; Zennaro et al., 1995; 1997). While the metabolic effect of ALDO is mediated through translocation of the ALDO–MR complex to the nucleus and subsequent gene activation, SPIR, 7TS and CAN function as ALDO antagonists in that the SPIR–, 7TS– and CAN–MR complexes remain largely in the cytoplasm (Lombes et al., 1994; Los & Colby, 1994). About 90% of SPIR is bound to proteins in the circulation; maximum free plasma levels of about 0.5 μM are reached 2–4 h after oral administration of 100–200 mg SPIR and the drug is cleared from the circulation after about 8 h (Overdiek et al., 1985; Gardiner et al., 1989; Jankowski et al., 1996; Takamura et al., 1997). In contrast, the major metabolites of SPIR, 7TS, 6β-hydroxy-7α-thiomethyl-spironolactone (6HTS) and CAN, can be detected at higher maximum levels and for up to 24 h after a single oral administration of the drug. A daily dose of 100 mg SPIR causes accumulation of 7TS, 6HTS and CAN, and peak serum levels of 7TS are 5–6 times higher than those of SPIR (Gardiner et al., 1989; Jankowski et al., 1996). It has been argued that some of the in vivo effects of SPIR are partially attributable to 7TS, 6HTS and/or CAN (Los & Colby, 1994; Klauber et al., 1996).
SPIR's ability to bind MR and thus function as an ALDO antagonist has been used for more than 40 years to treat patients with hyperaldosteronism. In recent years, however, clinical investigations indicate that SPIR may have effects beyond inhibition of signal transduction through MR. For example, SPIR increases the survival of patients with congestive heart failure, improves endothelial dysfunction and reduces turnover of cardiac and vascular collagen, all due to effects thought to be at least partly unassociated with the drugs anti-ALDO function (Pitt et al., 1999; Zannad et al., 2000; Doggrell & Brown, 2001). It has also been demonstrated that SPIR has an early suppressive effect on the production by human blood MNC of inflammatory cytokines, including tumour necrosis factor (TNF)-α and -β, interferon (IFN)-γ, interleukin (IL)-6 and granulocyte–macrophage colony-stimulating factor (GM-CSF), and the drug alleviated inflammatory symptoms in patients with rheumatoid arthritis (RA) and juvenile idiopathic arthritis (Bendtzen et al., 2003; Hansen et al., 2004).
In an effort to clarify the cellular signalling of the MR-binding steroids, we investigated the gene-regulatory effects of SPIR, 7TS, CAN and ALDO using human blood MNC. Surprisingly, SPIR, but not the SPIR-related steroids, affected a large number of genes, including apoptosis-related genes, which resulted in late apoptosis affecting both monocytes, and T- and B lymphocytes.
Methods
Cell cultures
MNC from three healthy and unmedicated individuals, one female and two male subjects, aged 23–61 years, were isolated by density gradient centrifugation on Ficoll-Hypaque (Lymphoprep, Nycomed, Oslo, Norway). The cells, 2.0 × 106 ml−1, were cultured at 37°C in a humidified 5% CO2 atmosphere in 1640 RPMI media (Biological Industries, Kibbutz Beit Haemek, Israel), containing 10% autologous serum. Unless otherwise stated, the monocytes and lymphocytes were stimulated with 100 ng ml−1 of lipopolysaccharide (LPS; Escherichia coli 055 : B5; Difco Laboratories, Detroit, MI, U.S.A.) plus 20 μg ml−1 of phytohaemagglutinin-A (PHA; Difco).
Gene transcription
MNC, 10 ml, 2 × 106 ml−1, were incubated at 37°C with PBS, 100 μM SPIR, 100 μM 7TS, 100 μM CAN, 100 μM ALDO or 100 μM SPIR+100 μM ALDO. After 15 min, LPS and PHA were added and the cells were further incubated at 37°C. After 4 h, the cells were isolated by centrifugation and the cells were lysed directly in the tubes by adding Trizol (Invitrogen, Tåstrup, Denmark). Total RNA was then prepared for analysis using human genome U133A GeneChips according to the Affymetrix GeneChip Expression Analysis Manual 2001 (Affymetrix, Santa Clara, CA, U.S.A.). The chips were scanned on a Hewlett Packard 2500 Gene Array Scanner from Affymetrix. Data analyses were performed using Data Mining Tools 3.1 (DMT) from Affymetrix and DNA-Chip Analyser 1.3 (dCHIP) (Schadt et al., 2001). The signal intensity of the perfect match probes was corrected using the intensity of the mismatch probes (PM/MM analysis).
Comparative analyses were initially performed for the five samples SPIR, 7TS, CAN, ALDO and SPIR+ALDO using the PBS sample from the respective individual as baseline. Using DMT, a differentially regulated gene was scored as either increased (I) or decreased (D) with a change of at least 50%, that is, with signal log ratios >0.59 or <−0.59, respectively, P<0.0025. Furthermore, all I and D transcripts had to be scored as present in the treated and the nontreated baseline samples, respectively. Using dCHIP, a differentially regulated gene had to fulfil the following criteria: (a) an over 1.5 normalised ratio between baseline (B) and experimental samples (E), that is, E/B>1.5 or B/E>1.5, using lower 90% confidence bound of change, and (b) a more than 50 difference in signals between B and E, that is, E−B>50 or B−E>50. Genes scored as differentially regulated in all three replicates and in both DMT and dCHIP were analysed further. Gene ontology (GO) classifications were carried out using the GO browser in the Affymetrix NetAffx analysis centre (Liu et al., 2003). Pathway analysis and visualisation were carried out using the Ingenuity pathways analysis (Ingenuity, Mountain View, CA, U.S.A.; see http://www.ingenuity.com/), as described (Kasamatsu et al., 2005).
Apoptosis
MNC, 2.0 × 106 ml−1, were incubated with either PBS, 10 μM SPIR, 100 μM SPIR, 100 μM 7TS, 100 μM CAN, 100 μM ALDO or 100 μM SPIR+100 μM ALDO and assessed for apoptosis after timed intervals from 4 to 22 h. Analyses were carried out on both unstimulated and LPS+PHA-activated MNC, and 5 mM EDTA was added to loosen adhesive cells 10 min prior to harvest. Cell surface receptor antibodies were CD14 PerCP (monocytes), CD3 PerCP (T cells) and CD19 PE (B cells), all from BD Biosciences (Erembodegem, Belgium). Apoptosis was detected as phosphatidylserine externalisation and DNA fragmentation using annexin-V and propidium iodide (PI) staining and terminal deoxynucleotidyl transferase dUTP nick-end labelling flow cytometry kits (TUNEL APO-BRDU kit; BD Biosciences) according to the manufacturer's instructions. FACScalibur with CellQuest software v. 4.0.2 (BD Biosciences) was used for data acquisition and WinList 32 (Verity Software House, Topsham, ME, U.S.A.) was used for data analysis. Annexin-V and PI 10,000 events were gated in each experiment when staining with CD3, CD14, CD19. In the TUNEL assay, 10,000 events were gated in each experiment according to the manufacturer's instructions.
Cytokines
MNC, 2.0 × 106 ml−1, were incubated with either PBS, 100 μM SPIR or 100 μM 7TS 15 min prior to LPS+PHA challenge. After 22 h, the supernatants were collected and stored at −20°C until cytokine quantifications using Luminex 100 IS (Luminexcorp, Austin, TX, U.S.A.). StarStation v.1.1B (Applied Cytometry Systems, Dinnington, U.K.) was used for data acquisition and analysis. The concentrations of IL-1β, IL-2, IL-6, IL-10, TNF-α, IFN-γ and GM-CSF in cell cultures were measured simultaneously using the Human Cytokine Panel I Kit (Biosource, Nivelle, Belgium), and the levels of TNF-β, IL-1α and IL-15 were measured simultaneously using Beadlyte® Human Multi-Cytokine Standard 6 (Upstate, Lake Placid, NY, U.S.A.). All the samples were diluted 1+1 before assay, and the detection limit was 20 pg ml−1.
Drugs and chemicals
SPIR and CAN were purchased from Sigma (St Louis, MO, U.S.A.), ALDO was purchased from Research Plus (Bayonne, NJ, U.S.A.) and 7TS was purchased from SynFine Research (Richmond Hill, Ontario, Canada) and kindly donated by Novo Nordisk (Måløv, Denmark). A 10 mM PBS suspension was prepared at 37°C before each assay and added to the cells at 1 : 100 dilution yielding a final total (free and protein-bound) concentration of 100 μM.
Results
Effects of SPIR- and ALDO-related steroids on mRNA expression
As shown in Table 1, SPIR affected 1018 transcripts in activated MNC at 4 h: 831 were downregulated and 187 were upregulated. CAN did not differentially regulate any gene, and ALDO affected only 17 transcripts. SPIR and ALDO together affected 940 transcripts, 792 of which were also influenced by SPIR alone. 7TS was tested only once and affected only 17 transcripts (results not shown).
Table 1.
Numbers of affected transcripts in in vitro activated human MNC treated for 4 h with mineralocorticosteroids
| Donors | Data analysis | SPIR | CAN | ALDO | SPIR+ALDO |
|---|---|---|---|---|---|
| 1 |
DMT |
2987 |
61 |
200 |
2793 |
| |
dCHIP |
2673 |
270 |
346 |
2761 |
| |
Both |
1673 |
6 |
73 |
1575 |
| |
|
|
|
|
|
| 2 |
DMT |
3519 |
26 |
138 |
3483 |
| |
dCHIP |
3458 |
148 |
268 |
3206 |
| |
Both |
2104 |
4 |
38 |
2202 |
| |
|
|
|
|
|
| 3 |
DMT |
2942 |
58 |
131 |
3004 |
| |
dCHIP |
3017 |
191 |
439 |
2734 |
| |
Both |
1606 |
17 |
55 |
1679 |
| |
|
|
|
|
|
| 1+2+3 |
DMT |
1871 |
0 |
27 |
1718 |
| |
dCHIP |
1525 |
0 |
21 |
1309 |
| Both | 1018 | 0 | 17 | 940 |
MNC from three healthy individuals were incubated with 100 μM of SPIR, CAN, ALDO, or SPIR+ALDO, and the monocytes and lymphocytes were activated with 100 ng ml−1 LPS plus 20 μg ml−1 PHA. Cellular mRNA was analysed using U133A GeneChips, and LPS+PHA-activated cells added PBS alone were used as baseline for each analysis.
SPIR effects on apoptosis- and immune-related genes
All genes scored as differentially regulated by SPIR (see Table 1) were divided into groups based on their GO annotation, see http://www.iir.suite.dk/IIR/PubS/+PubS.htm. Many genes had more then one GO annotation and several genes were represented in more than one GO group. Of the 1018 transcripts affected, those involved in apoptosis and the I-κB/NF-κB cascade, as well as in immune and inflammatory responses and in cytokine/chemokine activities, were significantly over-represented (Table 2). The SPIR-regulated genes involved in apoptosis are both apoptosis-inducing and -inhibiting. To investigate which transcription factor(s) were responsible for the gene-regulatory effects of SPIR, the differentially regulated genes were analysed with the Ingenuity database. The analysis showed that NF-κB, CEBPβ and MYC control the expression of many of the genes differentially regulated by SPIR (Supplementary Figures S1 and S2; see http://www.iir.suite.dk/IIR/PubS/+PubS.htm). The affected genes controlled by NF-κB are highlighted in Table 3. The table also shows that SPIR influenced the expression of several cytokines and chemokines as well as their receptors. Thus, genes encoding many clinically important proinflammatory cytokines were downregulated by SPIR, including TNFα, and the drug also downregulated the TNF receptor encoding genes TNFRSF1A and TNFRSF1B. Within the IL-1 system of cytokines, SPIR downregulated IL1α and IL1R1, whereas the genes encoding two IL-1 decoy receptors, the single Ig IL-1R-related molecule (SIGIIR) and IL1R2, were upregulated. In contrast, IL1RN encoding the IL-1 receptor antagonist (IL-1Ra) was downregulated by SPIR.
Table 2.
Numbers of apoptosis- and immune-related transcripts affected by SPIR
| Upregulated | Downregulated | P | |
|---|---|---|---|
| All transcripts |
187 |
831 |
|
| Apoptosis |
7 |
73 |
<0.001 |
| Antiapoptosis |
1 |
25 |
<0.001 |
| Induction of apoptosis |
2 |
19 |
0.007 |
| I-κB/NF-κB cascade |
1 |
25 |
<0.001 |
| Immune response |
24 |
120 |
<0.001 |
| Inflammatory response |
7 |
30 |
<0.001 |
| Cytokine binding |
1 |
20 |
<0.001 |
| Cytokine activity |
1 |
32 |
<0.001 |
| Chemokine activity | 0 | 11 | <0.001 |
Genes scored as differentially regulated by SPIR were divided into groups based on their GO annotation, and the χ2 test was performed for each group to calculate if there was an over-representation of affected transcripts in a specific group. The numbers of up- and downregulated genes in the different GO groups are shown together with the P-values.
Table 3.
SPIR-induced differential gene regulation in in vitro activated human MNC
| (a) Signalling proteins and receptors | Fold change DMT | Fold change dCHIP | |
|---|---|---|---|
|
Cytokine activity | |||
|
IL1R2 |
Interleukin 1 receptor type II |
3.7 |
4.3 |
|
CSF3R |
Colony-stimulating factor 3 receptor granulocyte |
2.9 |
2.7 |
|
SIGIRR |
Single Ig IL-1R-related molecule |
2.3 |
2.1 |
|
IL2Rβ |
Interleukin 2 receptor beta |
−1.7 |
−2.6 |
|
IL1R1 |
Interleukin 1 receptor type I |
−2.3 |
−2.6 |
|
IL15 |
Interleukin 15 |
−2.6 |
−2.7 |
|
IL10Rα |
Interleukin 10 receptor alpha |
−2.6 |
−3.6 |
|
IL15Rα |
Interleukin 15 receptor alpha |
−2.6 |
−4.7 |
|
IL18R1 |
Interleukin 18 receptor 1 |
−2.7 |
−3.2 |
|
IL2Rα |
Interleukin 2 receptor alpha |
−2.8 |
−3.5 |
|
TNFRSF1A |
Tumour necrosis factor receptor 1A |
−2.9 |
−4.6 |
|
TNFRSF1B |
Tumour necrosis factor receptor 1B |
−3.2 |
−4.3 |
|
IL10 |
Interleukin 10 |
−3.4 |
−4.6 |
|
IL6 |
Interleukin 6 |
−3.4 |
−4.5 |
|
TNFα |
Tumour necrosis factor alpha |
−3.6 |
−5.1 |
|
Il1RN |
Interleukin 1 receptor antagonist |
−3.9 |
−6.3 |
|
IL1α |
Interleukin 1 alpha |
−5.4 |
−8.5 |
|
IFNγ |
Interferon gamma |
−9.4 |
−15.5 |
|
TNFSF14 |
Tumour necrosis factor 14 |
−14.4 |
−297.4 |
|
G-CSF3 |
Granulocyte colony-stimulating factor 3 |
−16.1 |
−589.2 |
|
IL2 |
Interleukin 2 |
−29.2 |
−53.4 |
|
IL17 |
Interleukin 17 |
−56.0 |
−390.1 |
| |
|
|
|
|
Chemokine activity | |||
|
CXCL5 |
Chemokine ligand 5 |
−2.5 |
−3.5 |
|
CCR1 |
Chemokine receptor 1 |
−4.2 |
−3.8 |
|
CXCL9 |
Chemokine ligand 9 |
−8.8 |
−12.9 |
|
CCL7 |
Chemokine ligand 7 |
−10.4 |
−11.1 |
|
CXCL10 |
Chemokine ligand 10 |
−20.2 |
−38.3 |
|
CXCL11 |
Chemokine ligand 11 |
−22.5 |
−29.5 |
|
CCL2 |
Chemokine ligand 2 |
−51.7 |
−45.2 |
| |
|
|
|
|
Cell adhesion | |||
|
VEGF |
Vascular endothelial growth factor |
2.1 |
1.9 |
|
ICAM1 |
Intercellular adhesion molecule 1 |
−4.0 |
−5.3 |
| |
|
|
|
|
(b) Intercellular signalling and transcription | |||
|
CUTL1 |
Cut-like 1 CCAAT displacement protein |
9.8 |
2.8 |
|
STAT1 |
Signal transducer and activator of transcription 1 |
−1.9 |
−2.5 |
|
NR3C1 |
Nuclear receptor subfamily 3 group C member 1 |
−1.9 |
−2.3 |
|
NFκBIE |
NF-κB inhibitor E |
−2.2 |
−3.9 |
|
MAP3K8 |
Mitogen-activated protein kinase kinase kinase 8 |
−2.2 |
−2.5 |
|
CEBPγ |
CCAAT/enhancer-binding protein (C/EBP), gamma |
−2.6 |
−3.9 |
|
NF-κBIα |
NF-κB inhibitor alpha |
−2.7 |
−4.4 |
|
SOCS1 |
Suppressor of cytokine signalling 1 |
−2.8 |
−3.6 |
|
PC4 |
Activated RNA polymerase II transcription cofactor 4 |
−3.2 |
−5.5 |
|
CEBPδ |
CCAAT enhancer-binding protein delta |
−3.5 |
−4.9 |
|
CEBPβ |
CCAAT enhancer binding protein beta |
−8.6 |
−13.6 |
|
MYC |
MYC transcription factor |
−10.3 |
−12.6 |
| |
|
|
|
|
(c) Immune response | |||
|
ALOX5 |
Arachidonate 5-lipoxygenase |
2.8 |
2.5 |
|
S100A12 |
S100 calcium-binding protein A12 |
2.7 |
2.4 |
|
IRF1 |
Interferon regulatory factor 1 |
−1.9 |
−3.1 |
|
APOL3 |
Apolipoprotein L 3 |
−2.0 |
−3.1 |
|
RIPK2 |
Receptor-interacting serine-threonine kinase 2 |
−2.2 |
−1.9 |
|
IRF2 |
Interferon regulatory factor 2. |
−2.5 |
−3.4 |
|
IRF4 |
Interferon regulatory factor 4 |
−2.9 |
−4.4 |
|
PTX3 |
Pentaxin-related gene rapidly induced by IL-1 beta |
−3.4 |
−3.9 |
|
SOD2 |
Superoxide dismutase 2 mitochondrial |
−4.4 |
−5.5 |
| |
|
|
|
|
(d) Apoptosis | |||
|
NALP1 |
NACHT, leucine-rich repeat and PYD containing 1 |
2.9 |
2.4 |
|
BAG1 |
BCL2-associated athanogene |
−1.7 |
−1.9 |
|
TNFAIP3 |
TNF alpha-induced protein 3 (A20) |
−1.7 |
−3.0 |
|
CASP1 |
Caspase 1 apoptosis-related cysteine protease |
−1.9 |
−2.3 |
|
TNFSF6 |
Tumour necrosis factor 6 |
−2.4 |
−4.4 |
|
TNFSF10 |
Tumour necrosis factor 10 |
−2.7 |
−3.4 |
|
CASP4 |
Caspase 4 apoptosis-related cysteine protease |
−2.8 |
−3.0 |
|
CFLAR |
CASP8 and FADD-like apoptosis regulator |
−2.9 |
−4.5 |
|
TNFRSF6 |
Tumour necrosis factor receptor 6 |
−3.4 |
−3.3 |
|
TRAF1 |
TNF receptor-associated factor 1 |
−3.9 |
−6.7 |
|
TRAF4 |
TNF receptor-associated factor 4 |
−4.5 |
−4.4 |
| IER3 | Immediate early response 3 | −11.1 | −17.7 |
SPIR affected 1018 transcripts and a few of the encoding genes are shown here; for a complete list, see http://www.iir.suite.dk/IIR/PubS/+PubS.htm. The table shows the gene prefix and full name together with the average fold changes of the three independent GeneChip experiments. The fold change was calculated using DMT and dCHIP. For D genes, the scale ranges from [−∞ → −1], where −1 represents no change and −2 represents a reduction to 1/2 the baseline expression. For I genes, the scale ranges from [1 → ∞], where 1 represents no change and 2 represents a doubling in expression. Genes transcribed by NF-κB are in bold.
Interestingly, SPIR downregulated both IL2 and IL15, the products of which control T- and B-cell activation and proliferation. IFNγ, another important T cell-activating cytokine, was also downregulated.
Some of the most profoundly reduced transcripts were those of IL-17 and granulocyte-colony-stimulating factor (G-CSF), both involved in neutrophil recruitment and host defences against certain bacteria. Several chemokine transcripts were also strongly suppressed, again in line with the clinically observed anti-inflammatory effect of the drug.
SPIR-induced apoptosis
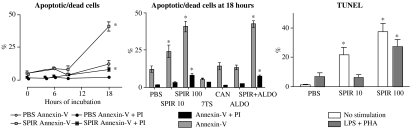
As SPIR affected several genes involved in cell survival, annexin-V, PI and TUNEL assays were carried out to investigate if SPIR induces apoptosis in MNC in vitro. The results show that <15% of the cells were apoptotic or dead after 4 and 6 h, and SPIR did not affect the proportions of annexin-V-positive MNC at these time points (Figure 1). After 18 h, however, there was a significant increase in both annexin-V-positive (apoptotic) and annexin-V+PI double-positive (dead) cells in the SPIR-treated cultures. A lower concentration of SPIR, 10 μM, also induced apoptosis at 18 h, although the percentage of apoptotic cells was lower compared to that obtained with 100 μM SPIR. CAN, 7TS and ALDO had no effect after 18 h of incubation, and ALDO did not inhibit SPIR's ability to induce apoptosis.
Figure 1.
SPIR-induced apoptosis in MNC in vitro. MNC from three healthy individuals were incubated with SPIR, at 10 and 100 μM, 7TS, CAN, ALDO, or SPIR+ALDO. Apoptosis and cell death were measured at various times using annexin-V/PI; the TUNEL assays were carried out after 22 h of incubation. Apoptotic cells are annexin-V positive and dead cells are annexin-V+PI double-positive. The cells were not challenged with LPS+PHA unless noted. Data represent mean values±s.d., n=3. A paired Student's t-test was used for comparisons. *P<0.05 versus cells added with PBS alone.
Results obtained by the TUNEL assay further support the notion that SPIR induces late apoptosis in MNC. The data also show that while stimulation with LPS+PHA by itself failed to significantly affect cell viability, the apoptotic effect of 10 μM SPIR was lower in activated MNC compared to nonactivated MNC.
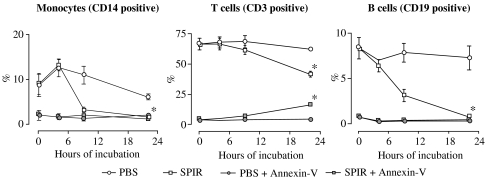
Figure 2 shows that there was a reduction in CD14-, CD3- and CD19-positive cells and a slight increase in CD3-positive, annexin-V double-positive cells after 9 h with SPIR. Almost all CD14- and CD19-positive cells disappeared after 22 h of incubation with the drug.
Figure 2.
SPIR-induced apoptosis in MNC in vitro. MNC from the same individuals as in Figure 1 were incubated with SPIR, and apoptosis was measured at various times using the annexin-V assay together with phenotype markers specific for monocytes (CD14 positive), T cells (CD3 positive) and B cells (CD19 positive). Data are shown as in Figure 1.
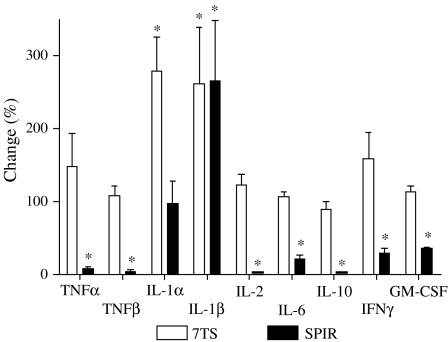
SPIR effects on production of specific cytokines
Some of the detected GeneChip expression patterns were verified at the protein level. Figure 3 shows the significant decrease in the extracellular levels of TNF-α, TNF-β, IL-2, IL-6, IL-10, IFN-γ and GM-CSF resulting from SPIR treatment. This is in agreement with the results of the gene expression experiments (Table 3). SPIR did not affect the concentration of IL-1α, whereas 7TS markedly increased the level of this cytokine. Even though IL1β was unaffected by SPIR (Table 3), both SPIR and 7TS increased the level of IL-1β. 7TS did not influence the concentrations of any of the other tested cytokines. The levels of IL-15 were below the detection limit in all samples.
Figure 3.
SPIR effects on production of selected cytokines. MNC from six healthy individuals were incubated with PBS, SPIR or 7TS and challenged with LPS+PHA. After 22 h at 37°C, supernatant cytokine concentrations were measured. The extracellular levels of cytokines in cultures of LPS+PHA-activated MNC without SPIR (controls) were set to 100%, and the experimental data were in each setup normalised to these controls. The control levels in pg ml−1±s.d. were: 19,173±1342 (TNF-α), 173±65 (TNF-β), 1500±1255 (IL-1α), 3619±3301 (IL-1β), 4218±2899 (IL-2), 18,143±2567 (IL-6), 3644±2549 (IL-10), 1531±1153 (IFN-γ) and 115±25 (GM-CSF). Data represent mean values±s.d., n=6. The Wilcoxon signed rank test was used for comparisons. *P<0.05 versus control.
We also investigated the effect of two androgen receptor antagonists, flutamide and cyproterone acetate, and they did not affect the extracellular levels of TNF-α, IL-2, IL-6, IL-10, IFN-γ and GM-CSF (results not shown).
To see whether SPIR selectively affected the cytokine release process, MNC were lysed and the intra- and extracellular levels of the above-mentioned cytokines were assessed. SPIR showed the same pattern of responses as seen in the supernatants (results not shown).
Discussion
The gene-regulatory effects of the four MR-binding steroids ALDO, SPIR, 7TS and CAN were investigated on in vitro activated (LPS+PHA) human blood MNC. While ALDO affected 17 transcripts, SPIR influenced 1018 transcripts in the same cells and under the same culture conditions. 7TS, the immediate in vivo metabolite of SPIR, regulated only 17 transcripts, and the other metabolite, CAN, failed to affect any of the more than 22,000 transcripts investigated. ALDO did not inhibit SPIR's gene-regulatory effects. All four steroids bind MR, but only ALDO causes translocation of the ALDO–MR complex to the nucleus (Lombes et al., 1994). These data suggest, therefore, that SPIR affects transcription of many genes through cellular pathways other than MR.
SPIR is also known to bind to androgen-, glucocorticoid- and progesterone receptors with antagonist and agonist functions, respectively (Cumming, 1990; Couette et al., 1992). However, binding to these receptors is very weak (Garthwaite & McMahon, 2004), and it is unlikely that this binding can explain the effects observed in the present study. In addition, two known androgen receptor antagonists, flutamide and cyproterone acetate, did not affect TNF-α, IL-2, IL-6, IL-10, IFN-γ or GM-CSF, the production of which was profoundly suppressed by SPIR.
GO analyses of the SPIR-induced effects showed that there was a significant over-representation of differentially regulated genes involved in immune responses, including inflammatory and cytokine/chemokine responses. While it has previously been shown that SPIR affects the ex vivo expression of several proinflammatory cytokines and that SPIR seems to have an anti-inflammatory potential in vivo (Bendtzen et al., 2003; Hansen et al., 2004), we now show that SPIR also affects early transcription of genes encoding chemokines and certain cytokine receptors, and cell adhesion molecules. Most of these genes were downregulated.
TNFSF14, also known as LIGHT, and the genes encoding the TNF receptors, TNFRSF1A and TNFRSF1B, were also downregulated by SPIR after 4 h. This is of potential clinical interest, because they are all known to be upregulated in patients with RA (Ware, 2005), and the data thus substantiate previous observations that SPIR alleviates inflammatory symptoms in RA patients (Bendtzen et al., 2003).
SPIR downregulated IL2, IL2R, IL6, IL15, IL15R and IFNγ, all of central importance in T-cell responses, and suppression of production of these cytokines was verified at the protein level. In the case of IL-15, however, protein expression was below the detection limit, most likely due to strict regulation of IL-15 translation (McInnes & Gracie, 2004).
It is interesting that there are two decoys for IL-1α/β signalling among the few genes upregulated by SPIR, namely IL1R2 and SIGIRR (Bourke et al., 2003; Wald et al., 2003). Thus, SPIR downregulates functional IL-1 receptors and upregulates cellular protection against IL-1 agonists. IL-1β expression was upregulated at the protein level but not at the mRNA level, in agreement with previous observations (Bendtzen et al., 2003; Hansen et al., 2004). Whether this is a late consequence of apoptosis induction is unknown (Hogquist et al., 1991).
Two of the genes most affected by SPIR were IL17 and G-CSF. This is of potential clinical interest as IL-17 is thought to play a major role in immunoinflammatory diseases and cancer (Kolls & Linden, 2004). IL-17 signalling is also critical for pulmonary neutrophil recruitment and host defence against Gram-negative bacteria through the coordinated release of G-CSF and CXC chemokines (McAllister et al., 2005), many of which were also downregulated by the drug.
Genes involved in both pro- and antiapoptotic processes were also significantly affected by SPIR: 73 of the 80 affected transcripts were downregulated after only 4 h of exposure. Despite this, unequivocal apoptosis appeared to be a late event in MNC challenged with SPIR. Even though staining with annexin-V is capable of detecting apoptosis as early as 1.5 h after induction (Martin et al., 1995), apoptosis revealed by annexin-V staining was not observed with certainty until after 18 h of culture. Since most of the cytokines measured here are produced and secreted within 2–6 h of cell activation, it appears unlikely that SPIR-induced suppression of cytokine production is solely a consequence of cell death.
Separate experiments using annexin-V staining together with flow cytometry of phenotypically marked cells revealed that SPIR induced apoptosis in all major cell types present in the MNC cultures. Monocytes and B cells, however, were affected faster than T cells.
The mechanism by which SPIR induces apoptosis in MNC is unknown, but it is unlikely that the effect is mediated through the death domain of the TNF superfamily of receptors because the drug suppressed the expression of TNFSF1A/TNFα, TNFRSF1A/TNF receptor type 1, TNFSF6/FasL, TNFRSF6/Fas and TNFSF10/TRAIL already after 4 h. 7TS, CAN and ALDO, all tested at 100 μM, did not induce apoptosis in MNC, and ALDO was unable to inhibit SPIR-induced apoptosis, suggesting that induction of apoptosis was independent of MR signalling.
Our data indicate that SPIR affects the I-κB/NF-κB cascade and suppresses the activities of the transcription factors NF-κB, MYC, CEBPβ and the homologues CEBPδ and CEBPγ. NF-κB is an important transcription factor during inflammatory and apoptotic responses, and it transcribes several of the genes differentially regulated by SPIR, including TNFα, TNFAIP3, TNFSF6, TNFRSF6, IL2, IL2Rα, IL6, IL15, IL17, GCSF3, CXCL5, CXCL10, CXCL11, CCL2, ICAM1, NF-κBIA, IER3, TRAF4, and IRF1 and -2. The expression of NF-κB was not differentially regulated, but NF-κBIA and TNFAIP3, both of which inhibit NF-κB, were downregulated. NF-κB transcribes both of them, and both act as negative feedback regulators of NF-κB (Auphan et al., 1995; Lee et al., 2000; Wertz et al., 2004). MYC controls processes governing both cell proliferation and apoptosis, and hence immunoinflammation (Levens, 2002). Interestingly, SPIR downregulated MYC and, accordingly, suppressed several of the genes under positive control of MYC and upregulated four of the genes under negative control of MYC. SPIR also suppressed the transcripts of CEBPβ, -δ and -γ, which regulates some of the same genes as NF-κB, and the drug upregulated the CCAAT displacement protein, CUTL1, that blocks transcription by CEBPβ (Snyder et al., 2001).
We have not yet determined whether SPIR inhibits these transcription factors directly or indirectly, but comparisons of the gene-regulatory effects of SPIR and those of NF-κB-, MYC- and CEBPβ inhibitors are topics of current studies.
In conclusion, the data extend recent findings that SPIR inhibits in vitro production of proinflammatory cytokines from human blood MNC. Suppression of several cytokines/chemokines of known clinical importance was substantiated at the gene and protein levels. Both monocytes and T- and B cells were affected. In addition, SPIR affected a host of genes encoding factors involved in apoptotic processes resulting in late MNC apoptosis. Examination of the affected transcripts indicates that SPIR downregulates many genes transcribed by NF-κB, MYC and CEBPβ. All of these effects were independent of the classical MR-mediated anti-aldosterone effect of SPIR. The findings may have pharmacological interest in that systemic and/or local administration of SPIR may be used as an inexpensive treatment of autoimmune and inflammatory diseases.
Acknowledgments
The assistance from Morten Svenson, Lone F. Bovin and Jacob Nersting is gratefully acknowledged. The Danish Research Council and Direktør Jens Aage Sørensen og hustru Edith Sørensens Mindefond provided financial support. Novo-Nordisk supplied 7α-thiomethyl-spironolactone.
Abbreviations
- 6HTS
6β-hydroxy-7α-thiomethyl-spironolactone
- 7TS
7α-thiomethyl-spironolactone
- ALDO
aldosterone
- CAN
canrenone
- dCHIP
DNA-chip analyser 1.3
- DMT
data mining tools 3.1
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- GO
gene ontology
- IFN
interferon
- IL
interleukin
- LPS
lipopolysaccharide
- MNC
mononuclear cells
- MR
mineralocorticoid receptor
- PBS
phosphate-buffered saline
- PHA
phytohaemagglutinin-A
- PI
propidium iodide
- RA
rheumatoid arthritis
- SPIR
spironolactone
- TNF
tumour necrosis factor
References
- ARMANINI D., WITZGALL H., STRASSER T., WEBER P.C. Mineralocorticoid and glucocorticoid receptors in circulating mononuclear leukocytes of patients with primary hyperaldosteronism. Cardiology. 1985;72 (Suppl. 1):99–101. doi: 10.1159/000173953. [DOI] [PubMed] [Google Scholar]
- AUPHAN N., DIDONATO J.A., ROSETTE C., HELMBERG A., KARIN M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- BENDTZEN K., HANSEN P.R., RIENECK K., THE SPIRONOLACTONE/ARTHRITIS STUDY GROUP Spironolactone inhibits production of proinflammatory cytokines, including tumor necrosis factor a and interferon-g, and has potential in the treatment of arthritis. Clin. Exp. Immunol. 2003;134:151–158. doi: 10.1046/j.1365-2249.2003.02249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOURKE E., CASSETTI A., VILLA A., FADLON E., COLOTTA F., MANTOVANI A. IL-1beta scavenging by the type II IL-1 decoy receptor in human neutrophils. J. Immunol. 2003;170:5999–6005. doi: 10.4049/jimmunol.170.12.5999. [DOI] [PubMed] [Google Scholar]
- COUETTE B., MARSAUD V., BAULIEU E.E., RICHARD-FOY H., RAFESTIN-OBLIN M.E. Spironolactone, an aldosterone antagonist, acts as an antiglucocorticosteroid on the mouse mammary tumor virus promoter. Endocrinology. 1992;130:430–436. doi: 10.1210/endo.130.1.1309341. [DOI] [PubMed] [Google Scholar]
- CUMMING D.C. Use of spironolactone in treatment of hirsutism. Cleve. Clin. J. Med. 1990;57:285–287. doi: 10.3949/ccjm.57.3.285. [DOI] [PubMed] [Google Scholar]
- DOGGRELL S.A., BROWN L. The spironolactone renaissance. Expert Opin. Investig. Drugs. 2001;10:943–954. doi: 10.1517/13543784.10.5.943. [DOI] [PubMed] [Google Scholar]
- GARDINER P., SCHRODE K., QUINLAN D., MARTIN B.K., BOREHAM D.R., ROGERS M.S., STUBBS K., SMITH M., KARIM A. Spironolactone metabolism: steady-state serum levels of the sulfur-containing metabolites. J. Clin. Pharmacol. 1989;29:342–347. doi: 10.1002/j.1552-4604.1989.tb03339.x. [DOI] [PubMed] [Google Scholar]
- GARTHWAITE S.M., MCMAHON E.G. The evolution of aldosterone antagonists. Mol. Cell. Endocrinol. 2004;217:27–31. doi: 10.1016/j.mce.2003.10.005. [DOI] [PubMed] [Google Scholar]
- HANSEN P.R., RIENECK K., BENDTZEN K. Spironolactone inhibits production of proinflammatory cytokines by human mononuclear cells. Immunol. Lett. 2004;91:87–91. doi: 10.1016/j.imlet.2003.11.008. [DOI] [PubMed] [Google Scholar]
- HOGQUIST K.A., NETT M.A., UNANUE E.R., CHAPLIN D.D. Interleukin 1 is processed and released during apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8485–8489. doi: 10.1073/pnas.88.19.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANKOWSKI A., SKOREK-JANKOWSKA A., LAMPARCZYK H. Simultaneous determination of spironolactone and its metabolites in human plasma. J. Pharm. Biomed. Anal. 1996;14:1359–1365. doi: 10.1016/s0731-7085(96)01767-0. [DOI] [PubMed] [Google Scholar]
- KASAMATSU A., ENDO Y., UZAWA K., NAKASHIMA D., KOIKE H., HASHITANI S., NUMATA T., URADE M., TANZAWA H. Identification of candidate genes associated with salivary adenoid cystic carcinomas using combined comparative genomic hybridization and oligonucleotide microarray analyses. Int. J. Biochem. Cell Biol. 2005;37:1869–1880. doi: 10.1016/j.biocel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- KLAUBER N., BROWNE F., ANAND-APTE B., D'AMATO R.J. New activity of spironolactone. Inhibition of angiogenesis in vitro and in vivo. Circulation. 1996;94:2566–2571. doi: 10.1161/01.cir.94.10.2566. [DOI] [PubMed] [Google Scholar]
- KOLLS J.K., LINDEN A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- LEE E.G., BOONE D.L., CHAI S., LIBBY S.L., CHIEN M., LODOLCE J.P., MA A. Failure to regulate TNF-induced NF-kB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVENS D. Disentangling the MYC web. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5757–5759. doi: 10.1073/pnas.102173199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU G., LORAINE A.E., SHIGETA R., CLINE M., CHENG J., VALMEEKAM V., SUN S., KULP D., SIANI-ROSE M.A. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOMBES M., BINART N., DELAHAYE F., BAULIEU E.E., RAFESTIN-OBLIN M.E. Differential intracellular localization of human mineralocorticosteroid receptor on binding of agonists and antagonists. Biochem. J. 1994;302:191–197. doi: 10.1042/bj3020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOS L.E., COLBY H.D. Binding of spironolactone metabolites in vivo to renal mineralocorticoid receptors in guinea pigs. Pharmacology. 1994;48:86–92. doi: 10.1159/000139166. [DOI] [PubMed] [Google Scholar]
- MARTIN S.J., REUTELINGSPERGER C.P., MCGAHON A.J., RADER J.A., VAN SCHIE R.C., LAFACE D.M., GREEN D.R. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCALLISTER F., HENRY A., KREINDLER J.L., DUBIN P.J., ULRICH L., STEELE C., FINDER J.D., PILEWSKI J.M., CARRENO B.M., GOLDMAN S.J., PIRHONEN J., KOLLS J.K. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J. Immunol. 2005;175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCINNES I.B., GRACIE J.A. Interleukin-15: a new cytokine target for the treatment of inflammatory diseases. Curr. Opin. Pharmacol. 2004;4:392–397. doi: 10.1016/j.coph.2004.04.003. [DOI] [PubMed] [Google Scholar]
- OVERDIEK H.W., HERMENS W.A., MERKUS F.W. New insights into the pharmacokinetics of spironolactone. Clin. Pharmacol. Ther. 1985;38:469–474. doi: 10.1038/clpt.1985.206. [DOI] [PubMed] [Google Scholar]
- PITT B., ZANNAD F., REMME W.J., CODY R., CASTAIGNE A., PEREZ A., PALENSKY J., WITTES J. The effect of spironolactone on morbidity and mortality in patients with severe heart failure. Randomized Aldactone Evaluation Study Investigators. N. Engl. J. Med. 1999;341:709–717. doi: 10.1056/NEJM199909023411001. [DOI] [PubMed] [Google Scholar]
- SCHADT E.E., LI C., ELLIS B., WONG W.H. Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell Biochem. 2001;84 (Suppl. 37):120–125. doi: 10.1002/jcb.10073. [DOI] [PubMed] [Google Scholar]
- SNYDER S.R., WANG J., WARING J.F., GINDER G.D. Identification of CCAAT displacement protein (CDP/cut) as a locus-specific repressor of major histocompatibility complex gene expression in human tumor cells. J. Biol. Chem. 2001;276:5323–5330. doi: 10.1074/jbc.M009454200. [DOI] [PubMed] [Google Scholar]
- TAKAMURA N., MARUYAMA T., AHMED S., SUENAGA A., OTAGIRI M. Interactions of aldosterone antagonist diuretics with human serum proteins. Pharm. Res. 1997;14:522–526. doi: 10.1023/a:1012168020545. [DOI] [PubMed] [Google Scholar]
- WALD D., QIN J., ZHAO Z., QIAN Y., NARAMURA M., TIAN L., TOWNE J., SIMS J.E., STARK G.R., LI X. SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 2003;4:920–927. doi: 10.1038/ni968. [DOI] [PubMed] [Google Scholar]
- WARE C.F. Network communications: lymphotoxins, LIGHT, and TNF. Annu. Rev. Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- WERTZ I.E., O'ROURKE K.M., ZHOU H., EBY M., ARAVIND L., SESHAGIRI S., WU P., WIESMANN C., BAKER R., BOONE D.L., MA A., KOONIN E.V., DIXIT V.M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- ZANNAD F., ALLA F., DOUSSET B., PEREZ A., PITT B. Limitation of excessive extracellular matrix turnover may contribute to survival benefit of spironolactone therapy in patients with congestive heart failure: insights from the randomized aldactone evaluation study (RALES). Rales Investigators. Circulation. 2000;102:2700–2706. doi: 10.1161/01.cir.102.22.2700. [DOI] [PubMed] [Google Scholar]
- ZENNARO M.C., FARMAN N., BONVALET J.P., LOMBES M. Tissue-specific expression of alpha and beta messenger ribonucleic acid isoforms of the human mineralocorticoid receptor in normal and pathological states. J. Clin. Endocrinol. Metab. 1997;82:1345–1352. doi: 10.1210/jcem.82.5.3933. [DOI] [PubMed] [Google Scholar]
- ZENNARO M.C., KEIGHTLEY M.C., KOTELEVTSEV Y., CONWAY G.S., SOUBRIER F., FULLER P.J. Human mineralocorticoid receptor genomic structure and identification of expressed isoforms. J. Biol. Chem. 1995;270:21016–21020. doi: 10.1074/jbc.270.36.21016. [DOI] [PubMed] [Google Scholar]