Abstract
Anthrax toxin receptors 1 and 2 (ANTXR1 and ANTXR2) have a related integrin-like inserted (I) domain which interacts with a metal cation that is coordinated by residue D683 of the protective antigen (PA) subunit of anthrax toxin. The receptor-bound metal ion and PA residue D683 are critical for ANTXR1-PA binding. Since PA can bind to ANTXR2 with reduced affinity in the absence of metal ions, we reasoned that D683 mutant forms of PA might specifically interact with ANTXR2. We show here that this is the case. The differential ability of ANTXR1 and ANTXR2 to bind D683 mutant PA proteins was mapped to nonconserved receptor residues at the binding interface with PA domain 2. Moreover, a D683K mutant form of PA that bound specifically to human and rat ANTXR2 mediated killing of rats by anthrax lethal toxin, providing strong evidence for the physiological importance of ANTXR2 in anthrax disease pathogenesis.
Synopsis
The bacterium that causes anthrax produces a toxin which is largely responsible for the symptoms and death associated with this disease. The toxin acts by first docking onto specific proteins, called receptors, located on the host cell surface, and it is then taken up into cells where it can act on its cellular substrates. There are two known receptors for the toxin, anthrax toxin receptors 1 and 2 (ANTXR1 and ANTXR2). However, the physiological importance of each receptor in host organisms is not yet understood. To address this issue directly, the authors designed a form of the toxin which binds specifically to ANTXR2 but not to ANTXR1. They show that this ANTXR2-specific form of the toxin is capable of killing rats following intravenous injection. These studies provide direct evidence for the physiological importance of ANTXR2 in anthrax toxin action in a model host organism.
Introduction
The spore-forming bacterium Bacillus anthracis causes anthrax and is classified as one of seven Centers for Disease Control and Prevention category A agents that are considered major threats as bioweapons [1]. B. anthracis secretes a toxin which contributes to bacterial virulence and causes many of the disease symptoms. Anthrax toxin is an AB-type toxin, with a single receptor-binding B-moiety, protective antigen (PA), and two catalytic A-moieties, lethal factor (LF) and edema factor (EF). LF is a zinc-dependent metalloprotease that cleaves members of the mitogen-activated protein kinase kinase family (all MKKs except MEK5) [2–4], whereas EF is a calmodulin and calcium–dependent adenylate cyclase [5,6]. LF and PA combine to form lethal toxin, and EF and PA combine to form edema toxin. These toxins are responsible for disabling host innate and adaptive immune responses, causing vascular leakage, and leading to the death of animals and cultured cells [7–16].
Following binding of PA to cell surface receptors and internalization of toxin complexes, EF and LF are translocated into the cytoplasm through a heptamerized PA pore that forms at endosomal low pH [17–20]. There are two known cell surface receptors for PA, ANTXR1 (anthrax toxin receptor/tumor endothelial marker 8; ATR/TEM8) and ANTXR2 (capillary morphogenesis gene 2; CMG2) [21,22]. These receptors are expressed in various human tissues [22–24], but there is evidence that ANTXR1 may be preferentially expressed in cancer cells and tumor endothelium [25–29]. The relative importance of either receptor for anthrax disease pathogenesis has not been established.
PA interacts with both receptors through a common von Willebrand factor A/integrin-like inserted (I) domain that contains a metal ion adhesion site (MIDAS) with five metal ion coordinating residues [30]. Similar to binding of ligands to α-integrins, binding of PA to its receptors involves direct coordination of a divalent cation in the MIDAS by a carboxylate-containing side chain from PA (residue D683) [31–34] (Figure 1). Previous studies have shown that PA residue D683 is critical for intoxication of cells via ANTXR1 [34].
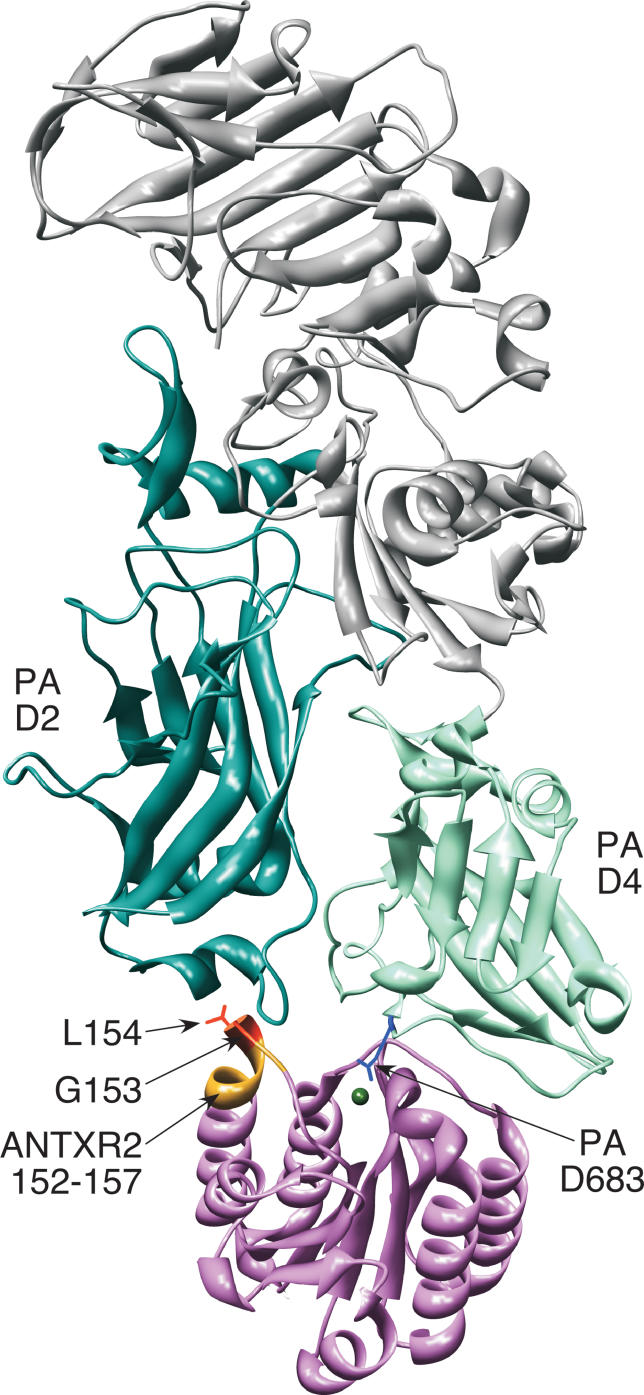
Figure 1. ANTXR2 Binding to PA Domains 2 and 4.
Ribbon model of the PA-ANTXR2 complex generated with UCSF Chimera [32]. PA domains 2 (D2) and 4 (D4) are shown in dark cyan and aquamarine, respectively, and the remainder of PA is depicted in gray. The ANTXR2 I domain is depicted in pink with its chelated metal ion in green. The ANTXR2 G153 and L154 residues (orange), which contact PA domain 2, and PA D683 residue (blue), which binds the receptor cation, are shown in stick representation. The ANTXR2 152–157 region is shown in yellow.
The ANTXR2 I domain binds to PA domains 2 and 4, and the surface area of the protein interface is much larger (approximately 2,000 Å2) than that of α-integrin–ligand interactions (approximately 1,300 Å2) [31,32]. The large contact surface correlates with a very tight ANTXR2 I domain–PA binding affinity (KD = 170 or 780 pM in Mg2+ or Ca2+, respectively) [35], compared with the affinity of α-integrin–ligand interactions that are usually in the micromolar to millimolar range [36]. ANTXR2 I domain contact with PA domain 2, the domain that forms pores and translocates EF/LF, has been proposed to act as a molecular clamp that prevents pore formation until the complex is trafficked to a low pH endosomal compartment [20,31,32,37] (Figure 1).
Despite strong amino acid sequence homology between ANTXR1 and ANTXR2, there are striking differences in the PA binding activities of the two receptors. When the PA heptamer is bound to ANTXR2, a pH value of approximately 5.0 is required to allow efficient pore formation, whereas approximately pH 6 is required for PA pore formation when the toxin is bound to ANTXR1 [20,37]. Also, the metal-dependent PA binding affinity of the ANTXR1 I domain is weaker than that of ANTXR2 by approximately 1,000-fold (KD = 130 nM or 1.1 μM in Mg2+ or Ca2+, respectively) [38]. Indeed, the ANTXR1 I domain–PA binding affinity in the presence of metal ions is similar to the ANTXR2 I domain–PA binding affinity in the absence of metal ions (KD = 960 nM in EDTA/EGTA) [35].
Because PA-ANTXR2 binding appeared to be less dependent on interactions mediated by the metal ion, we reasoned that the PA–ANTXR2 interaction should be less sensitive to mutations of PA residue D683. Here we show that this is the case and that PA proteins with D683 mutations bind specifically to human and rodent ANTXR2 and mediate intoxication via these receptors. We also show that a single region of nonconserved receptor residues that bind PA domain 2 is responsible for the differential abilities of ANTXR1 and ANTXR2 to bind D683 mutant forms of PA. Moreover, we demonstrate that a D683K mutant form of PA mediates lethal toxin killing of rats, implicating ANTXR2 as a physiologically important anthrax toxin receptor.
Results
PA Residue D683 Is Not Essential for Binding or Intoxication via ANTXR2
Previously, PA residue D683, which contacts the receptor-bound metal ion (Figure 1), was shown to be critically important for the interaction of PA with ANTXR1: mutation of this residue to Asn completely abrogated receptor binding [34]. To test the dependence of ANTXR2-PA binding on PA D683, this residue was replaced with either an asparagine or a lysine. The resultant PAD683N and PAD683K proteins were tested for their abilities to bind to a soluble human ANTXR2 protein (sANTXR2) and a full-length human ANTXR2–enhanced green fluorescent protein (EGFP) fusion protein expressed on PA receptor-deficient Chinese hamster ovary (CHO)-R1.1 cells. Additionally, cells expressing ANTXR2-EGFP were tested for their ability to be intoxicated with PAD683N or PAD683K and LFN-DTA, a recombinant protein composed of the N-terminal PA-binding portion of LF fused to the catalytic A chain of diphtheria toxin, which kills CHO cells. For control purposes, these experiments were also performed with CHO-R1.1 cells engineered to express a human ANTXR1-EGFP fusion protein.
Surface plasmon resonance (SPR) analysis, performed as described previously [35], revealed that PAD683N binds tightly to the sANTXR2 I domain, KD = 28 (±1.7) and 17 (±0.68) nM in the presence of Ca2+ and Mg2+, respectively. This binding was still metal ion dependent because the KD value was 240 (±27) nM in the presence of EDTA, a metal chelator. PAD683K bound the soluble receptor with similar properties, KD = 26 (±0.81) and 33 (±3.1) nM, in Ca2+ and Mg2+, respectively, and was 94 (±8.8) nM when EDTA was added.
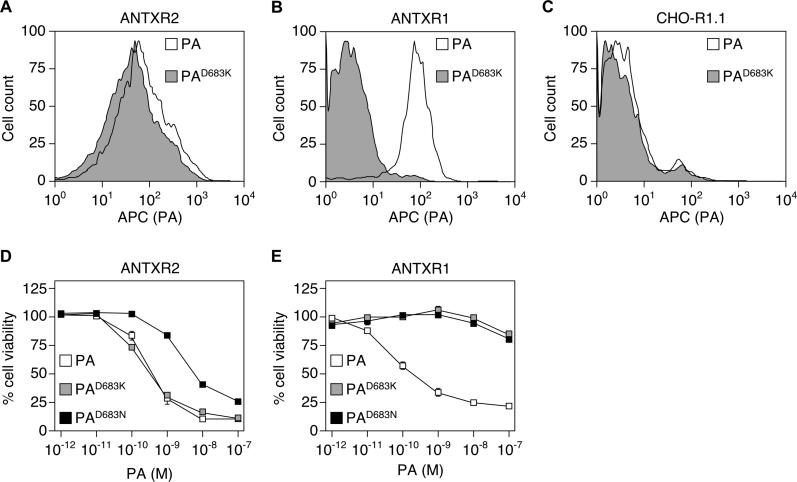
Flow cytometric analysis performed with one of the altered proteins, PAD683K, confirmed that it bound to cells expressing ANTXR2-EGFP but not ANTXR1-EGFP (Figure 2A, 2B, and 2C). Consistently, both PAD683N and PAD683K supported the intoxication of cells expressing human ANTXR2-EGFP (Figure 2D) but not ANTXR1-EGFP (Figure 2E). Indeed, cells expressing ANTXR2-EGFP were just as susceptible to PAD683K-dependent killing as they were to wild-type PA-dependent killing (Figure 2D). We conclude that PA residue D683 is not critical for toxin action mediated by ANTXR2 in the range of toxin concentrations tested.
Figure 2. PAD683N and PAD683K Bind and Support Intoxication via ANTXR2.
CHO-R1.1 cells stably expressing human ANTXR2-EGFP (A), human ANTXR1-EGFP (B), and control CHO-R1.1 cells expressing no receptors (C) were analyzed by flow cytometry after incubation with 100 nM purified PA or PAD683K proteins, followed by an anti-PA serum, and an APC-conjugated secondary antibody. Triplicate samples of CHO-R1.1 cells stably expressing human ANTXR2-EGFP (D) or human ANTXR1-EGFP (E) were incubated with 10−10 M LFN-DTA, and with increasing amounts of either purified PA, PAD683N, or PAD683K proteins. Cell viability was measured by CellTiter-Glo reagent and is represented as the percentage of signal seen with cells incubated with LFN-DTA alone (100% viable).
ANTXR2 Residues That Interact with PA Domain 2 Specify the D683 Mutant PA Interaction
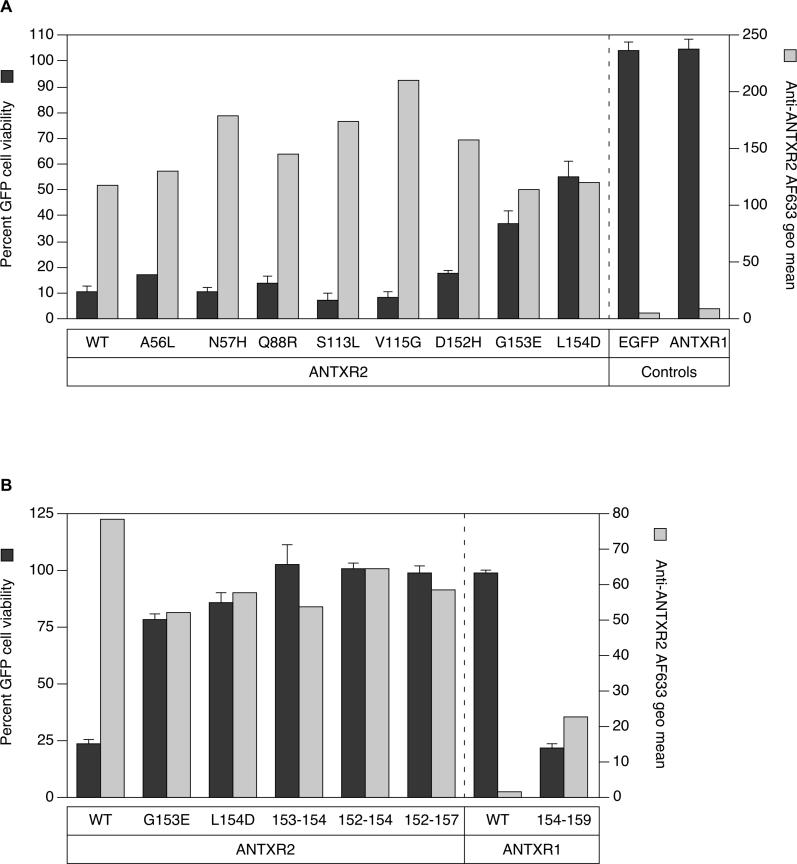
Based on the co-crystal structures of ANTXR2 bound to PA (Figure 1), there are eight contact residues that would be different at the toxin-binding interface in ANTXR1: A56L, N57H, Q88R, S113L, V115G, D152H, G153E, and L154D [31,32] (Figure S1). We reasoned that the differential abilities of the receptors to bind mutant PA proteins would be due to one or more of these variable amino acids. To test this idea, each of these amino acid substitutions was introduced independently into ANTXR2-EGFP and the altered receptors were expressed in transiently transfected CHO-R1.1 cells. Cell surface expression of the mutant receptors was confirmed by flow cytometry with a polyclonal chicken anti-ANTXR2 serum and AlexaFluor-633–conjugated secondary antibody (Figure 3A). These cells were also subjected to intoxication with PAD683N and LFN-DTA. Cell viability was measured in a flow cytometry–based assay where the percentage of live, EGFP-positive cells remaining after toxin challenge was compared with cells incubated with LFN-DTA alone (no toxin killing). Cells expressing cytoplasmic EGFP or ANTXR1-EGFP were included as negative controls.
Figure 3. ANTXR2 Contact with PA Domain 2 Confers Sensitivity to PAD683N-Mediated Intoxication.
(A) Triplicate samples of CHO-R1.1 cells transiently expressing ANTXR2, or ANTXR2 proteins with alterations in PA contacts to nonconserved ANTXR1 residues were incubated with 10−10 M LFN-DTA and 10−7 M PAD683N, or LFN-DTA only, and analyzed by flow cytometry. The average percentage of live EGFP-positive cells in the samples with both parts toxin is divided by that in the LFN-DTA only samples to determine percent cell viability (black bars). Individual samples of the same cells were also incubated with an anti-ANTXR2 antibody, followed by an AlexaFluor-633–conjugated secondary antibody and analyzed by FACS to determine the receptor cell surface expression level. The geometric mean of AlexaFluor-633 fluorescence after staining cells with antibodies (gray bars) is depicted next to the cell viability data for comparison purposes.
(B) Receptor sequences responsible for contacting PA domain 2 were exchanged between ANTXR1 and ANTXR2 proteins in an analysis similar to (A). Cells expressing ANTXR1154–159 bound anti-ANTXR2 antibodies, suggesting that an epitope for the polyclonal antibody mapped to this site. Saturable binding studies performed with transfected cells and with increasing amounts of an AlexaFluor-633–labeled monomeric PA protein confirmed that the mutant 154–159 ANTXR1 protein is expressed on the cell surface at levels similar to wild-type ANTXR1 and ANTXR2 (unpublished data).
The largest defects in PAD683N-mediated intoxication were seen with cells expressing G153E or L154D mutant ANTXR2 receptors (Figure 3A). Residues G153 and L154 are located in a surface region of ANTXR2 that contacts PA domain 2 (Figure 1). To further investigate the role of this region in the D683 mutant PA interaction, we made reciprocal exchanges of the residues located between positions 152 and 157 (DGLVPS) in the β4-α4 loop region of the ANTXR2 I domain and the corresponding residues of ANTXR1 (154 to 159; HEDLFF) in the context of ANTXR1-EGFP and ANTXR2-EGFP proteins, respectively. These studies revealed that replacement of both G153 and L154 residues in ANTXR2 with the residues from ANTXR1 was sufficient to make cells completely resistant to PAD683N intoxication (Figure 3B). Additionally, replacement of ANTXR1 residues 154 to 159 with the corresponding ANTXR2 residues gave rise to a recombinant receptor that could support PAD683N-mediated intoxication (Figure 3B).
PAD683K Supports LF-Mediated Intoxication in Rats
To address the possible role of ANTXR2 in lethal toxin killing of Fischer 344 rats, we first confirmed that D683 mutant forms of PA can interact with rat ANTXR2 (rANTXR2) but not rat ANTXR1. To this end, we attempted to isolate a full-length rat ANTXR1 cDNA clone but were not successful. However, the rat ANTXR1 I domain differs from that of its human counterpart by only two amino acids (residues K72 and R136 of the human protein are, in the rat protein, arginine and serine, respectively). Therefore, we constructed a rat version of the ANTXR1-EGFP protein (designated as ANTXR1R72,S136-EGFP by replacing these two human-specific residues with those of rat ANTXR1. Also, cDNAs encoding the rANTXR2 open reading frame were isolated from rat liver and brain tissue RNAs; both cDNAs had an identical DNA sequence. Comparison with the human ANTXR2 (hANTXR2) revealed that the I domains of these proteins differ by ten amino acid acids, two of which are PA-contact residues: Arg111 and Ser113 in hANTXR2 are alanine and glutamine in rANTXR2, respectively (Figure S1). CHO-R1.1 cells engineered to express rANTXR2-EGFP were susceptible to PAD683K-dependent killing, albeit slightly less efficiently than the killing observed with wild-type PA (Figure 4A and 4B). By contrast, cells engineered to express either hANTXR1-EGFP or ANTXR1R72,S136-EGFP were resistant to PAD683K-dependent killing. Taken together, these observations confirm that PAD683K interacts specifically with ANTXR2, both in humans and in rats.
Figure 4. PAD683K Supports Lethal Toxin-Mediated Killing of Rats via rANTXR2.
(A) Triplicate samples of CHO-R1.1 cells transiently expressing the different receptor-EGFP proteins were incubated with 10−10 M LFN-DTA and with either 10−8 M wild-type PA or 10−7 M PAD683K or without PA. These amounts of wild-type and mutant PA proteins were used because they gave rise to maximal levels of killing when incubated with cells expressing human ANTXR2 (Figure 1D). Cell viability was measured by counting the percentage of live EGFP-positive cells in the sample by flow cytometry and is expressed as described in the Figure 3A legend.
(B) Triplicate samples of CHO-R1.1 cells transiently expressing ANTXR1R72,S136-EGFP were incubated with toxin and analyzed as in (A) except that increasing amounts of either the wild-type or mutant PA protein were used.
(C) Groups of five rats each were injected by jugular vein cannula with 8 μg of LF and either 100 μg of PAD683K or 10 to 40 μg of PA and time until death postinjection was recorded. One rat in the 40 μg PA group died immediately after injection from pulmonary embolism and was excluded from analysis. Each data point represents a single animal, and the vertical line represents the mean for each group. P-values were determined by Student's unpaired t-test. Control rats injected with PBS all survived (unpublished data).
In an initial experiment, it was found that PAD683K could support lethal toxin-killing of male Fischer 344 rats, albeit several-fold less efficiently than that seen with wild-type PA: the mean time to death (TTD) (±SD) seen with three animals injected intravenously with 8 μg of LF and 10 μg of wild-type PA was 93.3 (±10.2) minutes, similar to that seen with two animals injected with four times as much PAD683K and the same amount of LF, i.e., TTD = 98.5 (±10.6) minutes. This several-fold decrease in the efficacy of PAD683K was also apparent from comparing the mean TTD of two animals injected with 200 μg of this mutant protein and LF (TTD = 68.5 [±5.0] minutes compared to those of three animals in each group that received LF with either 20, 30, or 40 μg of wild-type PA [mean TTD = 60.3 ± 2.9; 57.0 ± 2.0; and 54.7 ± 1.5 minutes, respectively]).
To investigate the relative efficacy of PAD683K in more detail, a subsequent experiment was performed with rats injected with the same amount of LF and with either a fixed amount of PAD683K (100 μg) or varying amounts of wild-type PA (ranging from 10 to 40 μg). Rats injected with LF and 100 μg of PAD683K died on average approximately 67 min following toxin administration (Figure 4C). By comparison, rats injected with LF and either 20 μg of PA or 10 μg of PA died an average of approximately 64 and 89 min after toxin injection, respectively (Figure 4C). The difference in TTD between the rats treated with 100 μg of PAD683K was statistically significant when compared with those injected with 10 μg of PA (P = 0.0001) but not from those treated with 20 μg of PA (P = 0.104) (Figure 4C). Based on these dosage effects, we conclude that PAD683K can mediate anthrax lethal toxin killing of rats, albeit with an approximately 5-fold reduced efficiency compared with wild-type PA.
Discussion
This report demonstrates that D683 mutant forms of PA bind selectively to ANTXR2 and that this selectivity is dependent on amino acid residues of ANTXR2 that contact domain 2 of PA. Moreover, we have shown that a D683K mutant form of PA is capable of mediating lethal toxin killing of rats, implicating an important physiological role for ANTXR2 in this process.
We have demonstrated that the ability of ANTXR2 to bind PA proteins with mutations at the D683 site maps to the receptor surface region containing residues G153 and L154 that engage PA domain 2. Replacing the ANTXR2 G153, which allows a polypeptide backbone turn, and L154, which participates in hydrophobic interactions with PA, with the negatively charged Glu and Asp residues from ANTXR1 resulted in resistance to cellular intoxication with PAD683N. By contrast, exchange of the HEDLFF154–159 sequence from ANTXR1 for the corresponding DGLVPS152–157 sequence from ANTXR2 conferred upon ANTXR1 the ability to support PAD683N-mediated intoxication.
Prior to this report it was unclear if ANTXR2 was important for lethal toxin killing in vivo. Our results showing that PAD683K supports lethal toxin-mediated killing of rats provides direct evidence for the physiological importance of this receptor in this rodent model system. There are several potential explanations for the reduced efficacy of PAD683K in vivo, compared to wild-type PA. First, this result could reflect the slightly reduced levels of PAD683K intoxication observed in cultured cells expressing rANTXR2 relative to those expressing hANTXR2. This difference may be due to the combined effects of residue differences at positions 111 and 113 of these proteins. The hANTXR2 R111 residue interacts with a negatively charged environment in PA domain 4, and the S113 residue makes H-bond contacts with PA domain 4 in the crystal structure (unpublished data) [31,32]. The rANTXR2 Ala and Gln residues at these respective locations may not be able to participate in these interactions. A second possibility is that cell type–specific differences downstream of initial PA binding might dictate different activities of PA proteins with altered residues at the D683 site. Indeed, we observed a 10-fold difference in intoxication activities of the PAD683N and PAD683K proteins despite similar ANTXR2 binding affinities (Figure 2D and 2E). Third, the reduced activity of the PAD683K protein in vivo might be indicative of a role for ANTXR1 in intoxication of rats by wild-type lethal toxin.
The PAD683K protein is the first engineered toxin that can discriminate between the two anthrax toxin receptors, binding specifically to ANTXR2. This ANTXR2-specific form of PA can now be used to study the role of ANTXR2 in mediating the effects of lethal toxin in other cell types, and it can also be engineered into the bacterium to probe the importance of this receptor for various aspects of anthrax disease pathogenesis following infection with B. anthracis spores.
Materials and Methods
DNA constructs and cell lines.
The ANTXR2-EGFP, ANTXR1-EGFP, and sANTXR2-mycHis fusion constructs have already been described (CMG2489-EGFP, ATR/TEM8 sv2-EGFP, and sCMG2-mycHis, respectively) [22,38]. QuikChange mutagenesis (Stratagene, La Jolla, California, United States) was performed with the oligonucleotide primers in Table S1 to generate various mutant versions of the ANTXR2-EGFP and ANTXR1-EGFP fusion constructs in the retroviral vector pLEGFP.N1 [22]. The QuikChange mutagenesis method was also used with the primers listed in Table S1 to generate the PAD683K allele in the PA-Pet22b construct [39] and the ANTXR1R72,S136-EGFP construct. To isolate rat ANTXR2 (rANTXR2) cDNA, rat liver and brain RNA were prepared by homogenizing tissue from adult male Fischer 344 rats with TriZOL reagent (Invitrogen, Carlsbad, California, United States) according to the manufacturer's instructions, followed by purification with RNeasy mini spin columns (Qiagen, Valencia, California, United States). The rANTXR2 open reading frame was isolated using a nested, touch-down PCR protocol on 3' RACE cDNA products (Clontech, Palo Alto, California, United States) with a Universal Primer Mix (Clontech) and primer 5' CCC GAG CCC AAG GGA CTG TGA GC 3' for the first round, and primers 5' ata gtc gac AC AGG ATG GTG GCC GGT CGG TCC C and 5' aa tag atc tgg TTG ATG TGG AAC TCG GGA GAA GTT TAT GC in the second round PCR. Primers included engineered SalI and BamHI restriction sites (underlined), respectively, for cloning rANTXR2 as an EGFP fusion protein in the pLEGFP.N1 plasmid. The open reading frames of all constructs were confirmed by sequencing. The PAD683N-Pet22b plasmid [34] was a gift from Jeremy Mogridge.
PA receptor–deficient CHO-R1.1 cells and CHO-R1.1 cells that were engineered to stably express recombinant receptor-EGFP fusion proteins, CMG2489-EGFP or ATR/TEM8 sv2-EGFP, have been described previously [22,34]. For transient expression of receptors, approximately 5 × 106 CHO-R1.1 cells was transfected with 1 μg of receptor-EGFP fusion construct and 5 μg of pBSII KS(–) or pTRE2hyg plasmids as carrier DNA and LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions. Cells were split and intoxicated at 24 h post-transfection, and analyzed for cell viability or receptor cell surface expression at 48 h post-transfection.
PA and sANTXR2 protein production.
The sANTXR2-mycHis protein was purified from the extracellular supernatants of human 293 Freestyle cells (Invitrogen) stably expressing this protein as described elsewhere [38]. The wild-type and altered forms of PA were prepared from the periplasm of E. coli BL21 cells that had been transformed with the PA-Pet22b, PAD683N-Pet22b, or PAD683K-Pet22b plasmids, as described previously [40]. The PA proteins were purified by FPLC with HiTrap QFF and Superose 12 (Amersham, Little Chalfont, United Kingdom) or HiLoad Superdex 200 (Amersham) columns, and the relative protein purity was determined by ImageQuant analysis (Amersham) of Coomassie-stained protein samples following SDS-PAGE.
Cell surface receptor expression analysis.
Anti-ANTXR2 antibodies were raised in chickens against the sANTXR2-mycHis protein [22,38] that should include ANTXR2 residues 34 to 232 as a mature protein (Aves Labs, Tigard, Oregon, United States). Antibodies were affinity purified against the same protein antigen by FPLC with a HiTrap NHS-activated HP column (Amersham). For flow cytometric analysis of ANTXR2 expression on the surfaces of transfected cells, cells were incubated with a 1:100 dilution of the anti-ANTXR2 antibody, followed by a 1:1,000 dilution of an AlexaFluor-633–conjugated goat anti-chicken antibody (Molecular Probes, Eugene, Oregon, United States). All FACS data were collected on an LSR flow cytometer (BD Biosciences, San Diego, California, United States) and analyzed with a FloJo software package (Tree Star, Ashland, Oregon, United States). The geometric mean of AlexaFluor-633 fluorescence after incubation with primary and secondary antibody was graphed after subtracting background binding of cells incubated with secondary antibody. Cell surface expression of human ANTXR1-EGFP, ANTXR2-EGFP, ANTXR1R72,S136-EGFP, and rat ANTXR2 proteins shown in Figure 4 was confirmed by flow cytometric analysis using an AlexaFluor-633–conjugated monomeric form of PA83 (unpublished data).
PA binding and in vitro intoxication assays.
PA binding to cells was monitored by flow cytometric analysis following incubations of cells for 2 h on ice with 100 nM PA or PAD683K, then with a 1:2,000 dilution of an anti-PA rabbit polyclonal serum, and a 1:500 dilution of an APC-conjugated anti-rabbit antibody (Molecular Probes) [22]. Intoxication was monitored by incubating samples of cells with LFN-DTA [41] and either PA, PAD683K, or PAD683N proteins. Cell viability was measured 45 to 50 h later using the CellTiter-Glo reagent (Promega, Madison, Wisconsin, United States). In the case of transiently expressed receptors, samples of cells were incubated with LFN-DTA and either PA, PAD683K, or PAD683N proteins and analyzed by FACS 20 to 24 h later. Cell viability was determined by dividing the percentage of live, EGFP-positive cells in the samples with complete toxin by that in the LFN-DTA only control (100% cell viability). In all intoxications, percent cell viability is the average from three samples, ± SD.
Binding affinity analysis.
All experiments were performed at 25 °C using the Biacore 2000 system and sensor chips with immobilized PA proteins as described previously [35]. Concentrations of sANTXR2 proteins used ranged from 24 nM to 4.8 μM in HBS (10 mM HEPES [pH 7.6], 150 mM NaCl) with either 1 mM CaCl2, 1 mM MgCl2, or 2 mM EDTA at pH 7.6. Serial injections were made at 10 μl/min, followed by a 40-μl buffer injection to allow for off-rate measurements. All kinetic data were analyzed using Origin Software. The equilibrium dissociation constants were calculated from kinetic measurements of the association and dissociation rate constants according to KD = koff/kon, and the errors were propagated. All results described are the average values of two independent trials.
Rat intoxication challenge.
Animal lethal toxin challenges were performed according to protocols approved by the Scripps Institutional Animal Care and Use Committee. Male Fischer 344 rats (180 to 200 g; Harlan) were anesthetized with isofluoranes and inoculated with 500 μl of a toxin mixture through a jugular vein cannula. The toxin mixture was prepared for each group by mixing 8 μg of LF (List Biological Laboratories, Campbell, California, United States) with 10 to 40 μg of purified PA or with 100 μg of PAD683K in a 500-μl volume per rat. Rats recovered from anesthesia within 5 min and were monitored for symptoms of intoxication and death (defined by cessation of respiration). The Student's unpaired t-test (Prism) was used for statistical analysis.
Supporting Information
Sequences were aligned by the ClustalW method (MacVector). The actual hANTXR1 and hANTXR2 protein isoform sequences aligned are indicated above. The dark line above the sequence indicates the I domain region that binds PA. The residues of the MIDAS are indicated with green boxes. The residues implicated in PA contact in the hANTXR2-PA co-crystal structure are indicated with red boxes, and the residues important for binding PAD683N, as discussed in this report, are indicated with purple boxes. Asterisks (*) and periods (.) below the sequence designate conserved and similar residues, respectively, at that site.
(2.5 MB TIF)
(43 KB DOC)
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accessions numbers used in this paper are rat ANTXR1 I domain (DQ789143) and cDNAs encoding the rANTXR2 open reading frame isolated from rat liver and brain tissue RNAs (DQ486884 for each). The Protein Data Bank (http://www.rcsb.org/pdb) accession number used in this paper is UCSF Chimera (IT6B).
Acknowledgments
We thank Jeremy Mogridge for providing reagents; Kenneth A. Bradley, Jared D. Evans, and Patricia L. Ryan for their helpful discussions; and John A. Naughton for help with manuscript preparation.
Abbreviations
- ANTXR1 and ANTXR2
anthrax toxin receptors 1 and 2, respectively
- CHO
Chinese hamster ovary
- EF
edema factor
- EGFP
enhanced green fluorescent protein
- hANTXR2
human ANTXR2
- LF
lethal factor
- MIDAS
metal ion adhesion site
- PA
protective antigen
- rANTXR2
rat ANTXR2
- sANTXR2
soluble human ANTXR2 protein
- SPR
surface plasmon resonance
- TTD
time to death
Footnotes
¤ Current address: Department of Microbiology and Immunology, Vanderbilt University Medical Center, Nashville, Tennessee, United States of America
Competing interests. RJC and JATY hold equity on PharmAthene, Inc. (Annapolis, Maryland, United States).
Author contributions. HMS, DJW, DBL, MM, RJC, and JATY conceived and designed the experiments. HMS, DJW, JMM, DT, and MM performed the experiments. HMS, DJW, JMM, MM, DT, and JATY analyzed the data. JMM and GJAR contributed reagents/materials/analysis tools. HMS and JATY wrote the paper.
Funding. The authors acknowledge support for this work from National Institutes of Health grant AI56013.
References
- Darling RG, Catlett CL, Huebner KD, Jarrett DG. Threats in bioterrorism. I: CDC category A agents. Emerg Med Clin North Am. 2002;20:273–309. doi: 10.1016/s0733-8627(02)00005-6. [DOI] [PubMed] [Google Scholar]
- Vitale G, Bernardi L, Napolitani G, Mock M, Montecucco C. Susceptibility of mitogen-activated protein kinase kinase family members to proteolysis by anthrax lethal factor. Biochem J. 2000;352((Pt 3)):739–745. [PMC free article] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Chopra AP, Boone SA, Liang X, Duesbery NS. Anthrax lethal factor proteolysis and inactivation of MAP-kinase-kinase. J Biol Chem. 2003;278:9402–9406. doi: 10.1074/jbc.M211262200. [DOI] [PubMed] [Google Scholar]
- Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, et al. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415:396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- Leppla SH. Anthrax toxin edema factor: A bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lingappa J, Leppla SH, Agrawal S, Jabbar A, et al. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature. 2003;424:329–334. doi: 10.1038/nature01794. [DOI] [PubMed] [Google Scholar]
- Kirby JE. Anthrax lethal toxin induces human endothelial cell apoptosis. Infect Immun. 2004;72:430–439. doi: 10.1128/IAI.72.1.430-439.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Haines D, Young HA, Leppla SH. Bacillus anthracis lethal toxin induces TNF-alpha-independent hypoxia-mediated toxicity in mice. J Clin Invest. 2003;112:670–682. doi: 10.1172/JCI17991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. Effects of anthrax toxin components on human neutrophils. Infect Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Greten FR, Li ZW, Karin M. Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science. 2002;297:2048–2051. doi: 10.1126/science.1073163. [DOI] [PubMed] [Google Scholar]
- Voth DE, Hamm EE, Nguyen LG, Tucker AE, Salles II, et al. Bacillus anthracis oedema toxin as a cause of tissue necrosis and cell type-specific cytotoxicity. Cell Microbiol. 2005;7:1139–1149. doi: 10.1111/j.1462-5822.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- Webster JI, Tonelli LH, Moayeri M, Simons SS, Jr, Leppla SH, et al. Anthrax lethal factor represses glucocorticoid and progesterone receptor activity. Proc Natl Acad Sci U S A. 2003;100:5706–5711. doi: 10.1073/pnas.1036973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoved AM, Miller GF, Moayeri M, Kakkar R, Shen Y, et al. Bacillus anthracis edema toxin causes extensive tissue lesions and rapid lethality in mice. Am J Pathol. 2005;167:1309–1320. doi: 10.1016/S0002-9440(10)61218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozes Y, Moayeri M, Wiggins JF, Leppla SH. Anthrax lethal toxin induces ketotifen-sensitive intradermal vascular leakage in certain inbred mice. Infect Immun. 2006;74:1266–1272. doi: 10.1128/IAI.74.2.1266-1272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassam A, Der SD, Mogridge J. Differentiation of human monocytic cell lines confers susceptibility to Bacillus anthracis lethal toxin. Cell Microbiol. 2005;7:281–292. doi: 10.1111/j.1462-5822.2004.00458.x. [DOI] [PubMed] [Google Scholar]
- Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrami L, Lindsay M, Parton RG, Leppla SH, Van Der Goot FG. Membrane insertion of anthrax protective antigen and cytoplasmic delivery of lethal factor occur at different stages of the endocytic pathway. J Cell Biol. 2004;166:645–651. doi: 10.1083/jcb.200312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Collier RJ. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol Microbiol. 1993;10:647–653. doi: 10.1111/j.1365-2958.1993.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Rainey GJ, Wigelsworth DJ, Ryan PL, Scobie HM, Collier RJ, et al. Receptor-specific requirements for anthrax toxin delivery into cells. Proc Natl Acad Sci U S A. 2005;102:13278–13283. doi: 10.1073/pnas.0505865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonuccelli G, Sotgia F, Frank PG, Williams TM, de Almeida CJ, et al. Anthrax toxin receptor (ATR/TEM8) is highly expressed in epithelial cells lining the toxin's three sites of entry (lung, skin, and intestine) Am J Physiol Cell Physiol. 2005;288:C1402–C1410. doi: 10.1152/ajpcell.00582.2004. [DOI] [PubMed] [Google Scholar]
- Hanks S, Adams S, Douglas J, Arbour L, Atherton DJ, et al. Mutations in the gene encoding capillary morphogenesis protein 2 cause juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am J Hum Genet. 2003;73:791–800. doi: 10.1086/378418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, et al. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649–6655. [PubMed] [Google Scholar]
- Davies G, Cunnick GH, Mansel RE, Mason MD, Jiang WG. Levels of expression of endothelial markers specific to tumour-associated endothelial cells and their correlation with prognosis in patients with breast cancer. Clin Exp Metastasis. 2004;21:31–37. doi: 10.1023/b:clin.0000017168.83616.d0. [DOI] [PubMed] [Google Scholar]
- Davies G, Mason MD, Martin TA, Parr C, Watkins G, et al. The HGF/SF antagonist NK4 reverses fibroblast- and HGF-induced prostate tumor growth and angiogenesis in vivo. Int J Cancer. 2003;106:348–354. doi: 10.1002/ijc.11220. [DOI] [PubMed] [Google Scholar]
- De Preter K, Pattyn F, Berx G, Strumane K, Menten B, et al. Combined subtractive cDNA cloning and array CGH: an efficient approach for identification of overexpressed genes in DNA amplicons. BMC Genomics. 2004;5:11. doi: 10.1186/1471-2164-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda A, Carson-Walter EB, Seaman S, Barber TD, Stampfl J, et al. TEM8 interacts with the cleaved C5 domain of collagen alpha 3(VI) Cancer Res. 2004;64:817–820. doi: 10.1158/0008-5472.can-03-2408. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Wigelsworth DJ, Scobie HM, Young JA, Collier RJ. Crystal structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: An anthrax toxin receptor. Proc Natl Acad Sci U S A. 2004;101:6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation. Proc Natl Acad Sci U S A. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Bradley KA, Mogridge J, Jonah G, Rainey A, Batty S, et al. Binding of anthrax toxin to its receptor is similar to alpha integrin-ligand interactions. J Biol Chem. 2003;278:49342–49347. doi: 10.1074/jbc.M307900200. [DOI] [PubMed] [Google Scholar]
- Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, et al. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J Biol Chem. 2004;279:23349–23356. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- Wolfe JT, Krantz BA, Rainey GJ, Young JA, Collier RJ. Whole-cell voltage clamp measurements of anthrax toxin pore current. J Biol Chem. 2005;280:39417–39422. doi: 10.1074/jbc.M509049200. [DOI] [PubMed] [Google Scholar]
- Scobie HM, Thomas D, Marlett JM, Destito G, Wigelsworth DJ, et al. A soluble receptor decoy protects rats against anthrax lethal toxin challenge. J Infect Dis. 2005;192:1047–1051. doi: 10.1086/432731. [DOI] [PubMed] [Google Scholar]
- Benson EL, Huynh PD, Finkelstein A, Collier RJ. Identification of residues lining the anthrax protective antigen channel. Biochemistry. 1998;37:3941–3948. doi: 10.1021/bi972657b. [DOI] [PubMed] [Google Scholar]
- Wesche J, Elliott JL, Falnes PO, Olsnes S, Collier RJ. Characterization of membrane translocation by anthrax protective antigen. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- Milne JC, Blanke SR, Hanna PC, Collier RJ. Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol Microbiol. 1995;15:661–666. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences were aligned by the ClustalW method (MacVector). The actual hANTXR1 and hANTXR2 protein isoform sequences aligned are indicated above. The dark line above the sequence indicates the I domain region that binds PA. The residues of the MIDAS are indicated with green boxes. The residues implicated in PA contact in the hANTXR2-PA co-crystal structure are indicated with red boxes, and the residues important for binding PAD683N, as discussed in this report, are indicated with purple boxes. Asterisks (*) and periods (.) below the sequence designate conserved and similar residues, respectively, at that site.
(2.5 MB TIF)
(43 KB DOC)