Abstract
The nomadic Romani (gypsy) people are known for their deep-rooted traditions, but most of their history is recorded from external sources. We find evidence for a Romani genetic lineage in England long before their recorded arrival there. The most likely explanations are that either the historical record is wrong, or that early liaisons between Norse and Romani people during their coincident presence in ninth to tenth century Byzantium led to the spread of the haplotype to England.
Keywords: Romani, ancient DNA, historical record
1. Introduction
Ancient DNA (aDNA) has the potential to reveal changes in the genetic composition of populations over time, and thereby inform our perceptions of demographic history. When private alleles are found in regional populations, their occurrence elsewhere can aid our interpretation of individual movement and behaviour (e.g. Fabiani et al. 2003). In this study we amplified aDNA from 17 individuals from the Castle Mall archaeological site in Norwich, England, dated to approximately the tenth century. Among these we found an individual (young adult male) with a mitochondrial DNA haplotype that had previously only been identified in modern Romani populations.
Having originated in India (according to linguistics and genetic data), Romani people are known to have reached the Byzantine Empire (modern day Turkey and Greece) by the tenth century (Fraser 1992). The first record of Romani in the UK is dated to the early sixteenth century, based on official records (Fraser 1992), though some suggest their arrival could have been somewhat earlier (Jarman & Jarman 1992). Genetic studies of the Romani have shown a unique mitochondrial DNA (mtDNA) lineage, based on a transversion T→A at position 16 189 of the first hypervariable segment (HVS-1) of the control region (Gresham et al. 2001). This lineage comprises 5.5% of the 275 published Romani haplotypes (all from Bulgaria; Gresham et al. 2001) and it has not been found in any other human population sampled so far (out of a total of more than 10 000 mtDNA haplotypes for this sequence held in the GenBank sequence repository), except the Castle Mall individual reported on here.
2. Material and methods
Human skeletal remains exhumed from Castle Mall at Norwich were radiocarbon dated by liquid scintillation spectrometry to ca 930–1050 AD (Shepherd Popescu in press). A total of 118 dental samples (from 59 skeletons) were used for the DNA extractions. To ensure that the aDNA sequences obtained were authentic, we followed the most pertinent aspects of the criteria recommended by Cooper & Poinar (2000) for the analysis of a few relatively recent human specimens. aDNA extractions were carried out in a laboratory physically separated from the main building and exclusively dedicated to aDNA manipulation. PCR and post-PCR analyses were carried out in the main laboratory. In addition, a one-way (from the ancient laboratory to the PCR laboratory) procedure was always followed to avoid the imperceptible carrying of DNA aerosols on clothes or skin into the aDNA laboratory (MacHugh et al. 2000). To detect possible contamination, we included extraction, PCR and carrier negative controls every 10 samples. To trace observed contamination, DNA sequences from the authors and other laboratory personnel, as well as from any contaminated control, were recorded for comparisons.
Extracts were amplified several times; only independent extractions and amplifications (from different dental samples from the same skeleton) yielding identical sequences were accepted as real. Random lesions in the aDNA template would be expected to be non-reproducible from different extracts. In addition, two samples were replicated in independent laboratories (the Ancient Biomolecules Centre, Oxford and the Department of Ecology and Evolutionary Biology, Arizona), where amplification products were cloned and sequenced. Criteria suggesting evidence for DNA survival, such as the amplification of another species from the site or tests for biochemical preservation (e.g. amino acid racemization) were not undertaken, as internal and external replication should suffice to indicate the presence of DNA in the sample.
An extremely sensitive method was used for the DNA extraction, developed for this work and based on a protocol previously described by Schmerer and co-workers (Schmerer et al. 1999). Dental pulp or its remnants adhered to the wall of the pulp chamber were collected by drilling inside the tooth. Powdered dental material was incubated in a high concentration EDTA lysis buffer and 50 mg ml−1 proteinase K for 24 h at 55 °C with constant agitation. After incubation, an aliquot was extracted twice with phenol/chloroform/isoamylalcohol. A silica suspension was added, which in the presence of the appropriate chaotropic agent binds to the extracted DNA. DNA was eluted in alkaline buffer and stored at 4 °C prior to PCR amplification to reduce the effect of inhibitory compounds (Montiel et al. 1997). A fragment of 264 bp (including primers) of the mtDNA HVS-I was amplified using primers 16 099 (5′AACCGCTATGTATTTCGTAC3′) and 16 331 (5′TTTGACTGTAATGTGCTATGTA3′) (numbering according to Anderson et al. 1981). Longer HVS-1 fragments (500 bp) failed to amplify, and a large number of cycles (n=45) was needed to obtain amplification of the shorter fragment, as expected for the small and degraded number of templates present in the aDNA samples (see Rameckers et al. 1997). PCR products of the correct size were purified and DNA sequenced directly (using an Applied Biosystems 377 automated sequencer). As initial base pairs were often unreadable and had to be discarded, a final 207 bp sequence starting at 16 123 was used for the analysis.
Sequences were compared against all database samples using Blast and similar programs. For the two independent replicates, we carried out extraction, amplification, cloning and sequencing as by Gilbert et al. (2003).
Sequences were aligned using the Sequencher 3.0 software package (Ann Arbor, MI, USA, Gene Codes Corporation) and polymorphic positions were identified using Mega 2.1 (Kumar et al. 2001). A reduced median network (Bandelt et al. 1995) was constructed using Network 4.1 (www.fluxus-engineering.com) and restricted to just modern Romani and English database sequences (GenBank), and the ancient sequence. The network included only haplotypes represented by more than one person in the database, with the exception of the ancient sequence and the modern sequence most closely linked to it. To maximize the resolution, all of the segregating sites found along the 207 bp (including deletions) were used for the analysis. A genetic distance matrix for these haplotypes was constructed based on nucleotide differences and reduced to a two-dimensional space by means of a multidimensional scaling (MDS) analysis using the Spss 11.0 software (Chicago IL, USA, SPSS Inc.).
3. Results and Discussion
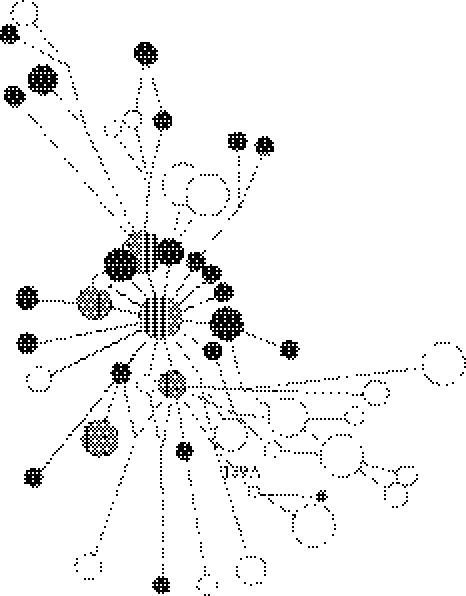
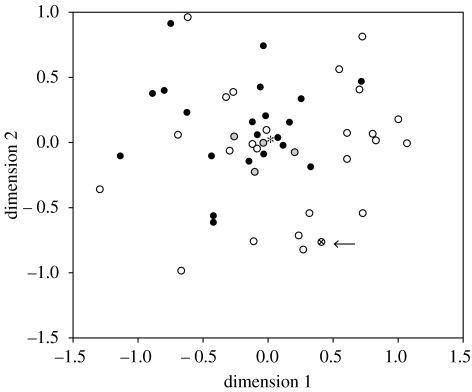
Of the 59 skeletons analysed from the Castle Mall site at Norwich, sequences were obtained from a total of 17 individuals. This study focuses on one individual, a male skeleton exhumed from grave 11 535, whose haplotype matched the rare modern lineage. Authentication of the ancient mtDNA sequence was done by replicated analyses of two different samples (and two extracts per sample) of the same individual following strict, established criteria (see §2) and all four replicate sequences matched. The ancient sample differs from the Cambridge reference sequence (Anderson et al. 1981) by the transversion at position 16 189, C→T transitions at 16 223 and 16 278, and a T→C transition at 16 271. The full haplotype (207 bp) matched a modern Romani haplotype at all positions with the exception of one transition, and fits well within a Romani clade based on modern lineages (figure 1). This is also supported by the MDS analysis (figure 2).
Figure 1.
Pruned spanning network (representing haplotypes found in two or more individuals, with the exception of the ancient British haplotype and the haplotype it connects to, both of which are unique in the database) showing the position of the ancient haplotype (black centre surrounded by white) relative to modern Romani (in white) and English (in black) haplotypes. Shared haplotypes are striped, and the position of the T→A transversion is shown.
Figure 2.
Multidimensional scaling (MDS) plot of genetic distances based on nucleotide differences for the English (black), Romani (white) and shared (grey) haplotypes. This is restricted to haplotypes found in two or more individuals (with the exception of the ancient British haplotype and the haplotype it connects to, both of which are unique in the database). The 54 dimensions of the genetic distance matrix were reduced to two dimensions, accounting for 94% of the dispersal. The ancient TA haplotype is indicated by an arrow, and the Cambridge reference sequence is shown as an asterisk.
In addition, an independent laboratory verified the sequence based on amplification from a third sample tooth from the same individual, both by direct sequencing and cloning (in two overlapping fragments: 16 055–16 218 and 16 209–16 356). For the first cloned fragment, all 3 clones showed the T→A transversion at 16 189. For the second fragment, all 5 clones for the ‘Romani’ sequence were consistent for variable sites 16 271 and 16 278, and 4 clones for site 16 223. Clones also showed other unique, non-reproducible variable sites, as was expected due to random lesions in the aDNA. If consistent variable sites were the result of non-random lesions, this should be evident by cloning amplicons from different individuals. However, none of the four variable sites observed in the ‘Romani’ haplotype were found in any of the 21 clones from a second cloned Norwich sample. Thus, we rule out the possibility that any of the variable sites, including the specific T→A transversion, are due to amplification or sequencing errors.
An independent T→A mutation at 16 189 in a British lineage is a possible alternative explanation, but very unlikely given the apparent lack of this mutation in modern Britain (see below), the low rate of transversion mutations in mtDNA, and the consistency of other sites in this sequence with the Romani lineage.
There are at least two other possible interpretations. The ancient ‘Romani’ haplotype may actually be present but undetected in modern European populations other than the Romani, however the probability is very low. Given 10 000 sequences, the frequency of the Romani haplotype would have to be less than 0.03% in non-Romani populations (at the 95% CI) in order for it to have not been detected so far (binomial test). Another possibility is that the haplotype was common in Saxon times, but since lost. However, this represents a period of only about 50 generations. It could have become extinct through genetic drift, but this process is slow in large populations, and Ne for European human populations is likely to have been in the thousands. It could have been lost through strong selection, but the loss of this haplotype by selection in all modern populations with the exception of the Romani seems a much less parsimonious explanation than the possibility that the Castle Mall individual shared ancestry with modern Romani.
If the rare TA haplotype found in ancient Britain instead suggests the presence of people of Romani ancestry in tenth century England, this is in surprising contradiction to historical evidence indicating that the Romani first left India—as mercenary soldiers or camp followers—at around AD 1000 (Hancock 2002). Some suggest that emigration from India could have been as early as the sixth century (Fraser 1992; Hancock 2002), and others have proposed much earlier routes via Egypt (see Kendrick 2000), but these theories are much less well supported.
One possible explanation would be if Romani women were enslaved by Vikings during trade expeditions to the Byzantine Empire, or formed liaisons with them during common association in Varangian army camps (in Byzantium) in the ninth and tenth centuries (Graham-Campbell 1994; Hancock 2002). These associations could also have been with Anglo Saxons, though known associations of Anglo Saxons with Varangian camps began only in the late eleventh century (Hancock 2002; Shepard 1973). Second generation Varangians are also known to have returned north (Hannestad 1970), and the mtDNA haplotype could have been introduced in this way. The gravesite at Norwich is typical of late Saxon, Christian sites with no grave goods and an east–west orientation, but this does not necessarily exclude Norse burial (Hadley 2002), and Viking artefacts were found nearby. However, the absence of the TA haplotype in modern Scandinavian or British populations suggests that if such associations happened, they were rare. Perhaps least likely is the independent arrival of Romani people in England, 500 years before the oldest known record, however census records are sparse from the Saxon and Medieval periods, and so this remains an open question.
Molecular genetic markers are valuable tools for interpreting the dispersion of human populations over evolutionary time-scales. However, the use of aDNA to clarify historical records from more recent times also has the potential to facilitate our understanding of population structure and historical patterns of mobility. This is especially true for nomadic peoples such as the Romani. Such information can inform and enhance our ability to interpret modern population genetic patterns.
Acknowledgments
We thank Liz Popescu for providing samples; Steven Lowe, Ken Lee and Ian Hancock for useful discussion; and Tom Gilbert for assistance with the replication of samples.
References
- Anderson S, et al. Sequence organisation of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J, Forster P, Sykes B.C, Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Poinar H.N. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- Fabiani A, Hoelzel A.R, Galimberti F, Muelbert M.M.C. Long-range paternal gene flow in the southern elephant seal. Science. 2003;299:676. doi: 10.1126/science.299.5607.676. [DOI] [PubMed] [Google Scholar]
- Fraser F.M. Blackwell; Oxford: 1992. The gypsies. [Google Scholar]
- Gilbert M.T, Willerslev E, Hansen A.J, Barnes I, Rudbeck L, Lynnerup N, Cooper A. Distribution patterns of postmortem damage in human mitochondrial DNA. Am. J. Hum. Genet. 2003;72:32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham-Campbell J, editor. Cultural atlas of the Viking world. Facts on File; New York: 1994. [Google Scholar]
- Gresham D, et al. Origins and divergence of the Roma (gypsies) Am. J. Hum. Genet. 2001;69:1314–1331. doi: 10.1086/324681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley D. Invisible Vikings. Br. Archaeol. 2002;64:16–20. [Google Scholar]
- Hancock I. University of Hertfordshire Press; Hatfield: 2002. We are the Romani people. [Google Scholar]
- Hannestad K, editor. Varangian problems. The eastern connections of the Nordic peoples in the Viking period and early middle ages. Munksgaard; Copenhagen: 1970. [Google Scholar]
- Jarman E, Jarman A.O.H. Cardiff University of Wales; 1992. The Welsh gypsies. [Google Scholar]
- Kendrick D. Romany origins and migration patterns. Int. J. Front. Missions. 2000;17:37–41. [Google Scholar]
- Kumar S, Tamura K, Jakobsen I.B, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- MacHugh D.E, Edwards C.J, Bailey J.F, Bancroft D.R, Bradley D. The extraction and analyses of ancient DNA from bone and teeth: a survey of current methodologies. Anc. Biomol. 2000;3:81–102. [Google Scholar]
- Montiel R, Malgosa A, Subirà E. Overcoming PCR inhibitors in ancient DNA extracts from teeth. Anc. Biomol. 1997;1:221–225. [Google Scholar]
- Rameckers J, Hummel S, Herrmann B. How many cycles does a PCR need? Determination of cycle numbers depending on the number of targets and reaction efficiency factor. Naturwissenschaften. 1997;84:259–262. doi: 10.1007/s001140050393. [DOI] [PubMed] [Google Scholar]
- Schmerer W, Hummel S, Herrmann B. Optimized DNA extraction to improve reproducibility of short tandem repeat genotyping with highly degraded DNA as target. Electrophoresis. 1999;20:1712–1716. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1712::AID-ELPS1712>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Shepard J. The English and Byzantium: a study of their role in the Byzantine army in the later eleventh century. Traditio: Stud. Anc. Mediaev. Hist. Thought Relig. 1973;29:53–92. [Google Scholar]
- Shepherd Popescu, E. In press. Norwich castle: excavations and historical survey 1987–98 Part 1. Anglo-Saxon to c.1345 Norfolk: NAU, East Anglian Archaeology.