Abstract
As once boldly stated, ‘bad taxonomy can kill’, highlighting the critical importance of accurate taxonomy for the conservation of endangered taxa. The concept continues to evolve almost 15 years later largely because most legal protections aimed at preserving biological diversity are based on formal taxonomic designations. In this paper we report unrecognized genetic divisions within the giant tortoises of the Galápagos. We found three distinct lineages among populations formerly considered a single taxon on the most populous and accessible island of Santa Cruz; their diagnosability, degree of genetic divergence and phylogenetic placement merit the recognition of at least one new taxon. These results demonstrate the fundamental importance of continuing taxonomic investigations to recognize biological diversity and designate units of conservation, even within long-studied organisms such as Galápagos tortoises, whose evolutionary heritage and contribution to human intellectual history warrant them special attention.
Keywords: Geochelone nigra (elephantopus), giant tortoises, microsatellites, conservation genetics, phylogeography, historical DNA
1. Introduction
The distinctiveness of giant tortoises (Geochelone nigra) throughout the Galápagos archipelago was an inspiration to Charles Darwin in developing his theory of natural selection as the mechanism of biological evolution. Fifteen formally described taxa of G. nigra are generally recognized, 11 of which are extant and threatened by human activities and introductions of non-native species (Pritchard 1996). The prevailing taxonomy is largely based on morphological differences among populations, primarily in carapace (shell) shape, which varies from domed to saddleback with intermediate forms also occurring (Fritts 1984). The taxonomic rank of populations that are often morphologically distinct on different islands and volcanoes has been contentious, especially as to whether such populations should be considered different species or subspecies (here referred to simply as taxa or lineages; Zug 1997). Our previous molecular studies have confirmed the distinctiveness of the extant named taxa (Caccone et al. 2002; Ciofi et al. 2002; Beheregaray et al. 2003; Beheregaray et al. 2004).
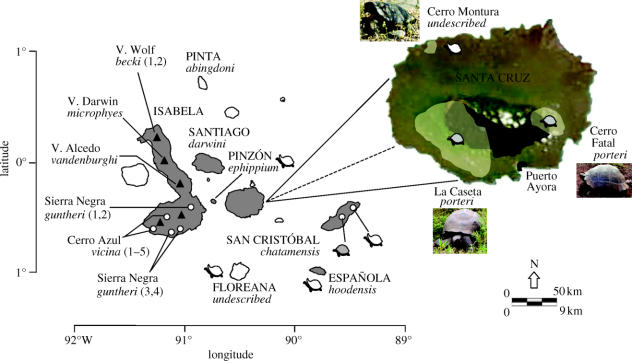
Santa Cruz is a moderately sized island central to the archipelago (figure 1; White et al. 1993) that supports one of the largest remaining populations of tortoises (ca 2000–4000 individuals). Paradoxically, this island also maintains the largest human population in Galápagos (currently greater than 20 000 and anticipated to double by 2013; MacFarland & Cifuentes 1995); thus, its unique biota is under heavy pressure from anthropogenic activities, including the conversion of substantial natural habitat for agriculture (figure 1). A single domed taxon of G. nigra has been described on Santa Cruz and given the name porteri (Fritts 1984). Two major populations of porteri occur on the island, each separated by urban and farm lands; one in the southwest Santa Rosa Tortoise Preserve (‘La Caseta’; figure 1) and the other to the east of the city of Puerto Ayora (‘Cerro Fatal’; figure 1). In addition, a very small, isolated population exhibiting the saddleback morphology has been documented in the northwest of Santa Cruz in an area called Cerro Montura (figure 1). While morphologically distinct from the domed porteri typical of Santa Cruz, it has never been assigned a formal Latinized name and is generally considered to be restricted to a few individuals in the wild, where only a single female and two males have been observed.
Figure 1.
Distribution maps of Galápagos tortoises throughout the archipelago and on Santa Cruz. Shaded islands indicate presence of extant tortoise populations and italized names represent current subspecific designations; island names indicated in all capitals, with distinct populations on Isabela specified by name; triangles represent volcanoes and circles indicate sampled populations throughout the archipelago. Shaded and unshaded tortoise caricatures signify ‘domed’ and ‘saddleback’ morphologies, respectively. The current distributions of tortoises (shaded yellow) and the agricultural corridor (shaded black) on Santa Cruz (Pritchard 1996) are plotted on a satellite map (MODOS Rapid Response Project, NASA/GSFC). Scale bar specified for the Galápagos archipelago (above) and Santa Cruz (below).
Previous studies (Caccone et al. 2002; Beheregaray et al. 2003) have hinted at a deep split among the extant lineages on Santa Cruz yet sampling limitations precluded a definitive evaluation of their genetic distinctiveness, taxonomic status and time of colonization. To explicitly address these questions, we conducted a comprehensive sampling of the Cerro Fatal and Cerro Montura populations on Santa Cruz to augment previous collections from La Caseta (Ciofi et al. 2002), and expanded character sampling by way of mitochondrial DNA (mtDNA) sequencing and microsatellite genotyping. Moreover, to specifically address the timing and origin of the Santa Cruz lineages, we have also included DNA sequence data from museum specimens of two extinct but biogeographically relevant taxa from the islands of Floreana and San Cristóbal (figure 1), the latter hypothesized as the source from which all other domed taxa may have been derived, including those currently extant on Santa Cruz.
2. Materials and methods
(a) Sample and data collection
Galápagos tortoise (G. nigra) blood samples were collected in three expeditions conducted over 7 years. All samples were obtained in accordance with local, national and international regulations. Population-level analyses were based on a total of 139 blood samples collected on Santa Cruz from the following populations: La Caseta (n=65), Cerro Fatal (n=71) and Cerro Montura (n=3). DNA was extracted using the Qiagen DNeasy Tissue Kit (Qiagen, Inc.) according to the manufacturer's protocol. DNA was sequenced for 697 bp of the mtDNA control region (CR) and genotyped at nine microsatellite loci following previously published conditions (Ciofi et al. 2002; Beheregaray et al. 2003). Higher-level analyses were based on consensus mtDNA sequences for extinct and extant lineages of Galápagos tortoises from four gene regions: control region (CR) (697 bp), 12s rDNA (12S; 425 bp), 16s rDNA (16S; 548 bp) and cytochrome b (cyt b; 416 bp). Bone samples were obtained from museum specimens representing the extinct Floreana population (Museum of Comparative Zoology No. R-46606) and extinct western, domed San Cristóbal population (California Academy of Sciences No. 8133). DNA was extracted from the museum samples in a dedicated facility for ancient DNA work according to a modified protocol (available from lead author), following all necessary precautions to prevent contamination by extant specimens. DNA was sequenced in 14 overlapping fragments not exceeding 200 bp in length across all four mtDNA gene regions (primer information available upon request). Sequences from the extinct lineages were combined with previously published consensus sequences of the extant G. nigra lineages (Caccone et al. 2002) and three out-group taxa (Geochelone chilensis, Geochelone carbonaria and Geochelone denticulata) for subsequent analyses. GenBank accession numbers for all DNA sequences used and generated over the course of this study are as follows: AY97476–AY098119, AF548216–AF548226 and AY956612–AY956623 (see Electronic Appendix A for details regarding population genetic and phylogenetic analyses).
3. Results and discussion
Levels of genetic variation at both mitochondrial and microsatellite loci are quite low in the Cerro Fatal population in contrast to the moderate to high levels exhibited in the Cerro Montura and La Caseta porteri populations (table 1). Overall, genetic variation among the three Santa Cruz populations is highly structured with most genetic variation being among, rather than within, populations (table 2). Genetic differentiation between populations was also revealed by highly significant fixation indices for all possible pairwise comparisons, observed for both the mitochondrial CR and microsatellite datasets (table 2). In fact, all three populations are reciprocally monophyletic as recovered from the mtDNA haplotype tree (data not shown) and population aggregation analysis revealed distinct aggregates among the domed porteri, with 31 fixed nucleotide differences across four mtDNA gene regions diagnosing Cerro Fatal porteri from La Caseta porteri (data not shown). From both a genealogical (Avise & Ball 1990) and phylogenetic species (Davis & Nixon 1992) perspective, the two domed lineages of porteri in Cerro Fatal and La Caseta represent distinct biological units. The presence of 100 private alleles between these two populations across nine microsatellite loci provides further evidence for this split, as do the results of assignment tests that identified 88 and 96% of La Caseta and Cerro Fatal individuals, respectively, as belonging to the populations in which they were collected. The three saddlebacked Cerro Montura individuals are also clearly genetically distinct from the two domed porteri on Santa Cruz, sharing all mtDNA haplotypes and the majority of microsatellite alleles (greater than 80%) with the saddleback ephippium population from the nearby island of Pinzón (figure 1). These individuals from north western Santa Cruz could represent the only remaining survivors of the source population from which the ephippium populations originated, or a recent secondary colonization from Pinzón (Fritts 1984).
Table 1.
Genetic divergence within Galápagos tortoises populations.
| population | n | mitochondrial DNAa | microsatellitesa | |||
|---|---|---|---|---|---|---|
| no. of haplotypes | haplotypic diversity, h | mean no. alleles/locus | HEb | HO | ||
| La Caseta | 65 | 12 | 0.80 (0.027)c | 15.8 | 0.71 | 0.80 |
| Cerro Fatal | 71 | 2 | 0.08 (0.043) | 5.0 | 0.55 | 0.56 |
| Cerro Montura | 3 | 2 | 0.67 (0.31) | 3.6 | 0.74 | 0.82 |
Results based on 697 base pairs of the mtDNA control region and genotypic data at nine microsatellite loci.
HE and HO are mean expected and observed heterozygosity.
Values in parentheses are the standard errors for h.
Table 2.
Genetic divergence among Galápagos tortoises populations.
| (a) analysis of molecular variance | ||||
|---|---|---|---|---|
| population comparisons | source of variationa | d.f. | % of variation | p-value |
| La Caseta | among | 2 | 91.21 | <0.0001 |
| Cerro Fatal | within | 137 | 9.79 | |
| Cerro Montura | total | 139 | ||
| La Caseta | among | 1 | 91.16 | <0.0001 |
| Cerro Fatal | within | 135 | 8.84 | |
| total | 136 | |||
| (b) fixation indicesb | |||
|---|---|---|---|
| population | La Caseta | Cerro Fatal | Cerro Montura |
| La Caseta | 0.149 | 0.107 | |
| Cerro Fatal | 0.912 | 0.279 | |
| Cerro Montura | 0.769 | 0.995 | |
Among populations, within populations or total.
Results based on mtDNA control region haplotypes (ϕst, below diagonal) and nine microsatellite loci (Θ, above diagonal). All pairwise comparisons were statistically significant (p<0.001).
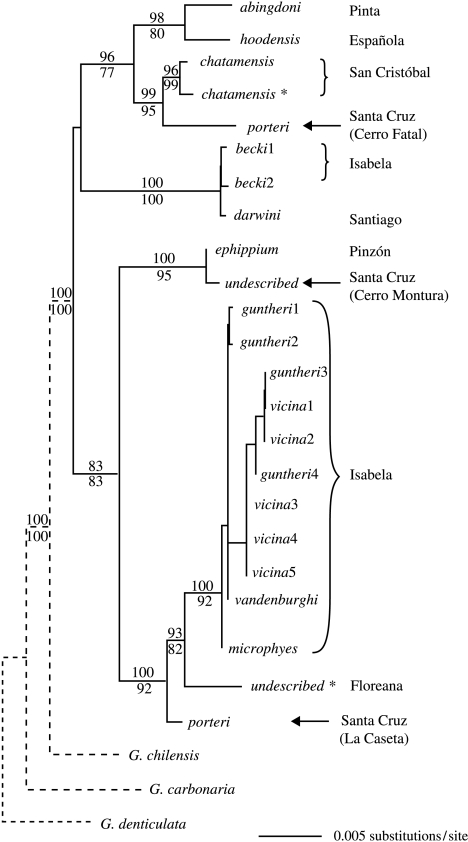
On a higher level, phylogenetic analysis reveals that the lineages on Santa Cruz are paraphyletic, all exhibiting closer affinities with taxa on other islands than with each other (figure 2). The Cerro Fatal porteri are most closely related to chatamensis on San Cristóbal, with pairwise genetic distances indicating a stronger association with the extinct domed population (figures 1 and 2). In contrast, La Caseta porteri is the basal lineage within a clade that includes the extinct Floreana taxon and several lineages from the younger, western island of Isabela (figures 1 and 2). Of all pairwise comparisons involving each taxon, La Caseta porteri and the extinct Floreana taxon reciprocally exhibit the shallowest sequence divergence. Lastly, the Cerro Montura individuals form a well-supported sister group with the ephippium taxon on Pinzón, consistent with both morphology and mtDNA haplotype data (figures 1 and 2). Overall, topological tests rejected hypotheses of monophyly (p<0.001) in the Santa Cruz lineages (Cerro Fatal, La Caseta and Cerro Montura) and in the domed porteri alone (Cerro Fatal and La Caseta).
Figure 2.
Bayesian phylogenetic tree of extant and extinct Galápagos tortoise taxa. Distinct haplogroups of becki, guntheri and vicina are indicated with numbers and follow figure 1; island of origin for each taxon is shown on the right; an asterisk (*) signifies an extinct taxon or population and arrows highlight the relative phylogenetic placement of the three populations of giant tortoises on Santa Cruz. Bayesian posterior probabilities and maximum likelihood bootstrap proportions (greater than 50%) are indicated above and below the branches, respectively. For illustration purposes, accurate branch lengths leading to out-group taxa are not shown (indicated by dashed line).
In addition to the distinct phylogenetic affinities of the three porteri lineages with taxa from different islands, application of a species-specific mtDNA CR rate (Beheregaray et al. 2004) suggests a temporal divide in their relative divergence times. The split between the Cerro Fatal porteri and San Cristóbal chatamensis lineages exhibits differing divergence times relative to the extinct, domed and extant saddlebacked chatamensis lineages, estimated at approximately 445 000 and 490 000 years ago (ya), respectively. Slightly deeper divergence times were reconstructed for La Caseta porteri and the extinct taxon on the southern island of Floreana, with a split dated between 584 000 and 926 000 ya. The timing of the saddleback lineages’ separation from Cerro Montura on Santa Cruz and ephippium on Pinzón was much more recent (0–261 000 ya).
Taken together, these findings suggest a significant revision of Galápagos tortoise taxonomy is in order. Of more immediate importance, the results of this molecular study unambiguously designate the La Caseta and Cerro Fatal porteri as distinct conservation units (Moritz 1994; Vogler & DeSalle 1994). This designation is especially pertinent for Cerro Fatal, as this population has suffered a dramatic decline in recent times due to heavy poaching and the conversion of their habitat to agricultural fields and lacks genetic variation compared with the fairly abundant and diverse population from La Caseta. From a broader perspective, despite years of study beginning with Darwin in 1835, modern DNA-based research coupled with thorough character and taxonomic sampling (extant and extinct) is still yielding insights into the taxonomy of the renowned giant tortoises of Galápagos. Of particular relevance are implications for conservation of the genetic, morphological and behavioural diversity of these extraordinary organisms, whose historical contribution to human intellectual history warrant them special attention.
Acknowledgments
T. Fritts first encouraged us to examine the Cerro Fatal population to determine its genetic affinities. We thank L. Behergaray and C. Ciofi for their initial studies and insights. H. Snell, W. Tapia and other members of the Charles Darwin Research Station and Galapagos National Park were crucial in providing logistical support for fieldwork and M. Milinkovitch, G. Gentile, N. Karraker, C. Martinez and B. Guerrero assisted in sample collection. J. Vindum and J. Rosado offered invaluable assistance regarding sample collection from the museum specimens. We thank G. Amato, S. Steinfartz and M. Olson for comments on the manuscript. Financial support was provided by the Yale Institute for Biospherics Studies through their ECOSAVE program (directed by Elisabeth Vrba) and a National Geographic Research grant (no. 6800-00).
Supplementary Material
References
- Avise J.C, Ball R.M. Principles of genealogical concordance in species concepts and biological taxonomy. Oxf. Surv. Evol. Biol. 1990;7:45–67. [Google Scholar]
- Beheregaray L.B, Ciofi C, Caccone A, Gibbs J.P, Powell J.R. Genetic divergence, phylogeography and conservation units of giant tortoises from Santa Cruz and Pinzón, Galápagos Islands. Conserv. Genet. 2003;4:31–46. [Google Scholar]
- Beheregaray L.B, et al. Giant tortoises are not so slow: rapid diversification and biogeographic consensus in the Galápagos. Proc. Natl Acad. Sci. USA. 2004;101:6514–6519. doi: 10.1073/pnas.0400393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccone A, et al. Phylogeography and history of giant Galápagos tortoises. Evolution. 2002;56:2052–2066. doi: 10.1111/j.0014-3820.2002.tb00131.x. [DOI] [PubMed] [Google Scholar]
- Ciofi C, Milinkovitch M.C, Gibbs J.P, Caccone A, Powell J.R. Microsatellite analysis of genetic divergence among populations of giant Galápagos tortoises. Mol. Ecol. 2002;11:2265–2283. doi: 10.1046/j.1365-294x.2002.01617.x. [DOI] [PubMed] [Google Scholar]
- Davis J.I, Nixon K.C. Populations, genetic variation, and the delimitation of phylogenetic species. Syst. Biol. 1992;41:421–435. [Google Scholar]
- Fritts T.H. Evolutionary divergence of giant tortoises in Galápagos. Biol. J. Linn. Soc. 1984;21:165–176. [Google Scholar]
- MacFarland C, Cifuentes M. In: Case study: Galápagos, Ecuador. In Human population, biodiversity and protected areas: science and policy issues. Dompka V, editor. American Association for the Advancement of Science; Washington, DC: 1995. pp. 135–188. [Google Scholar]
- Moritz C. Defining evolutionarily-significant-units for conservation. Trends Ecol. Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Pritchard P.C.H. The Galápagos tortoises—nomenclatural and survival status. Chelonian Res. Monogr. 1996;1:1–85. [Google Scholar]
- Vogler A.P, DeSalle R. Diagnosing units of conservation for management. Conserv. Biol. 1994;8:354–363. [Google Scholar]
- White W.M, McBirney A.R, Duncan R.A. Petrology and geochemistry of the Galápagos Islands—portrait of a pathological mantle plume. J. Geophys. Res. 1993;98:19 533–19 563. [Google Scholar]
- Zug G.R. Galápagos tortoise nomenclature: still unresolved. Chelonian Conserv. Biol. 1997;2:618–619. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.