Abstract
It is well known theoretically that gene flow can impede genetic differentiation between populations. In this study, we show that in a highly mobile bird species, where dispersal is well documented, there is a strong genetic and morphological differentiation over a very short geographical scale (less than 5 km). Allocation tests revealed that birds caught in one area were assigned genetically to the same area with a very high probability, in spite of current gene flow. Populations were also morphologically differentiated. The results suggest that the relationship between gene flow and differentiation can be rather complicated and non-intuitive.
Keywords: gene flow, genetic differentiation, morphological differentiation, dispersal, finches
1. Introduction
Populations are unlikely to adapt locally if there is strong gene flow from other populations with different environmental conditions, which creates a conflict between gene flow and local differentiation (Mayr 1963). Despite this, population divergence with gene flow has been observed in several taxa. These cases share common features. One set of studies show divergence at small spatial scales, where movements are not necessarily restricted by physical barriers (Blondel et al. 1999; Postma & Van Noordwijk 2005; Garant et al. 2005). These populations can broadly be described as parapatric but at different spatial scales. Another set of studies concern populations that spend most of their time in allopatry (ecotypes), but become sympatric during breeding (e.g. Skulason et al. 1999), hence maximizing probabilities for gene flow. However, in both of these cases, selection within habitats can overcome gene flow if there is a long period of selection relative to gene flow. This indicates that the timing between selection and gene flow can be important in creating differences in spite of gene flow. Based on this, we should hypothesize that populations that face diversifying selection at the time of gene flow should not be able to differentiate to the same extent.
The citril finch (Serinus citrinella) is a small (12 g) granivorous non-territorial Cardueline finch breeding at high densities in the Pyrenees. In this area, the interaction between a Mediterranean influence and high altitudes creates a significant contrast between north- and south- facing slopes, so that patches of highly differentiated habitats appear at very close proximity. Populations leave the breeding areas in winter and gather at overwintering sites, where they are found together. In spring, they leave the overwintering areas and again settle in the different subalpine habitats. We studied two populations that differ significantly in survival rate, breeding success and body mass, and where there is also substantial dispersal between the populations (Senar et al. 2002). This means that the citril finch populations spend a substantial part of the year in sympatry, but diverge only in terms of breeding habitat choice. The citril finch is, therefore, an ideal model species for a study of divergence with gene flow, since the periods of selection and gene flow (breeding) are the same.
2. Material and methods
We studied two populations in different habitats in the Spanish Pyrenees, both situated at high altitudes (2000 m), but on two sides of a mountain slope differing in direction and vegetation structure (Senar et al. 2002). La Vansa (42°12′ N, 1°35′ E) faces north and is moist and cool, and has a high abundance of mountain pines (Pinus uncinata), which is a main food source for citril finches. La Bofia (42°10′ N, 1°32′ E) is situated 5 km from La Vansa and faces south, is drier and sunnier and has a lower abundance of pines. Citril finches were trapped with mist nets at drinking vessels and individually marked. We recorded sex and age, and measured wing and tarsus lengths (Borràs et al. 1998). Morphological analyses are based on data obtained from 1996 to 2003 (Bofia: n=798; Vansa: n=1.509). We extracted DNA from blood samples from birds captured at random during 1998 in the field in the two locations (n=19 from each population) by means of Chelex extraction. We only used blood from breeding adult birds, therefore avoiding catching family groups.
For microsatellite analysis, we used the following primers: Lox1, HrU2, HrU7, Pocc6, Ppi2, Pca2, Pca9 (Primmer et al. 1996; Piertney et al. 1999; Dawson et al. 2000). These primers were amplified using a PCR-cycle of 94 °C for 2 min, followed by 30×(94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s), and 72 °C for 10 min using standard PCR conditions.
The microsatellite data were analysed using the GenePop (Raymond & Rousset 1997) and Arlequin software (Schneider et al. 2000) for estimation of heterozygosity, test of Hardy–Weinberg (HW) equilibrium, FST-values and test for genetic differentiation. We performed several different multilocus assignment tests, which all differ in terms of method and assumptions. We did this to make sure that our conclusions are robust for details regarding algorithms and assumptions. We used three different approaches based on an extensive resampling of individuals and a calculation of genotype likelihoods from these resampled datasets, using GeneClass2 (Paetkau et al. 2004). We also used the Bayesian clustering method developed by Pritchard et al. (2000) and implemented in the software STRUCTURE. Briefly, this method assigns individual genotypes to different populations based on the allele frequency distributions in each population and returns probabilities of the assignment of a genotype to the different populations. We also used the software Doh (http://www2.ualberta.ca/jbrzusto/Doh.php) that calculates an assignment index with the highest probability of an individual's genotype in any population. Finally, we used MIGRATE 1.7.6 (Beerli & Felsenstein 2001) to estimate M=m/μ (where m is the migration rate), simultaneously, with effective population size (Neμ). This program was run 10 times to secure accurate estimates.
3. Results
The number of alleles differed between different loci and between populations, as did the frequencies of alleles. In La Vansa, three loci (Lox1, HrU7, HrU2) differed from HW expectations, which show a heterozygote deficiency. In La Bofia, two loci (HrU7, HrU2) differed from HW expectations; in all cases, we found a heterozygote deficiency. La Vansa had a slightly higher level of heterozygosity (mean±s.e.m.=0.39±0.067) than La Bofia (mean±s.e.m.=0.27±0.074), but the difference was not significant (p>0.24, Mann–Whitney U-test).
Rates of gene flow were low between the two populations based on simulations using BayesAss and Migrate. The gene flow from La Bofia to La Vansa was 0.023 (s.e.=0.039) and from La Vansa to La Bofia was 0.013 (s.e.=0.020). The estimates of effective population sizes were highly consistent among runs in Migrate (La Vansa Neμ=0.224 (CV=0.11); La Bofia Neμ=0.305 (CV=0.13); mean values over 10 runs), with La Bofia having the larger effective population size in all runs. Taken together this means that gene flow (Nem) can be estimated to be around five to ten individuals every generation in each direction, assuming a mutation rate of 10−3.
Most of the genetic variation was found within individuals (62.9%), but 9.4% was found among populations, which was highly significant (locus-by-locus AMOVA, p=0.002), corresponding to an FST of 0.093 (RST=0.043). An exact test of population differentiation using GENEPOP gave the same result (combined =52.89, p<0.001). The different population assignment programs all classified individuals to their original population to a very large extent, with the number of correctly classified individuals ranging between 73.6 and 78.9%. The results from STRUCTURE show that La Vansa birds were assigned to La Vansa with a very high probability, whereas La Bofia birds were assigned to La Bofia with a very high probability (figure 1).
Figure 1.
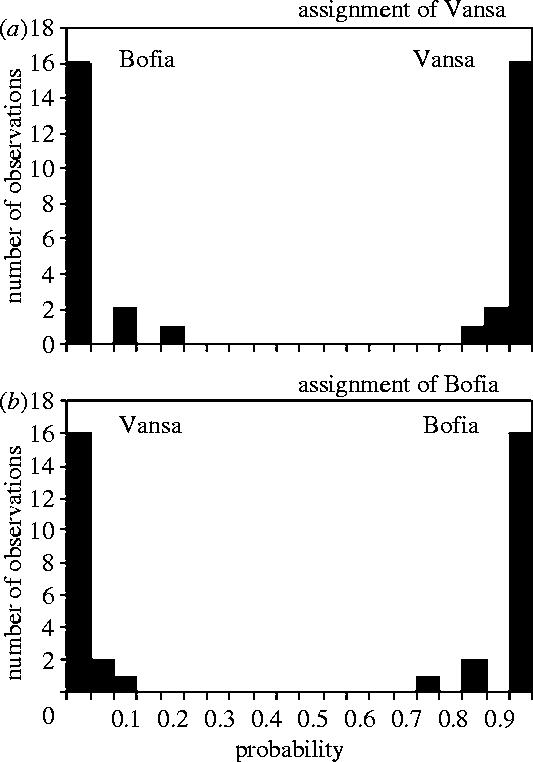
The probability of assigning an individual captured in one locality to each of the two localities. (a) assignment to La Vansa. (b) Assignment to La Bofia. The text in the figures refers to where the individuals were captured.
Citril finches differed significantly in morphology between the localities when sex, age and year had been accounted for in a factorial ANOVA (Wilks' lambda=0.96, d.f.=2,925, p<0.001). Birds in Bofia had longer tarsus and shorter wings than birds in Vansa (wing length: F1,2243=44.92, p<0.001, tarsus length: F1,2243=17.97, p<0.001; figure 2).
Figure 2.
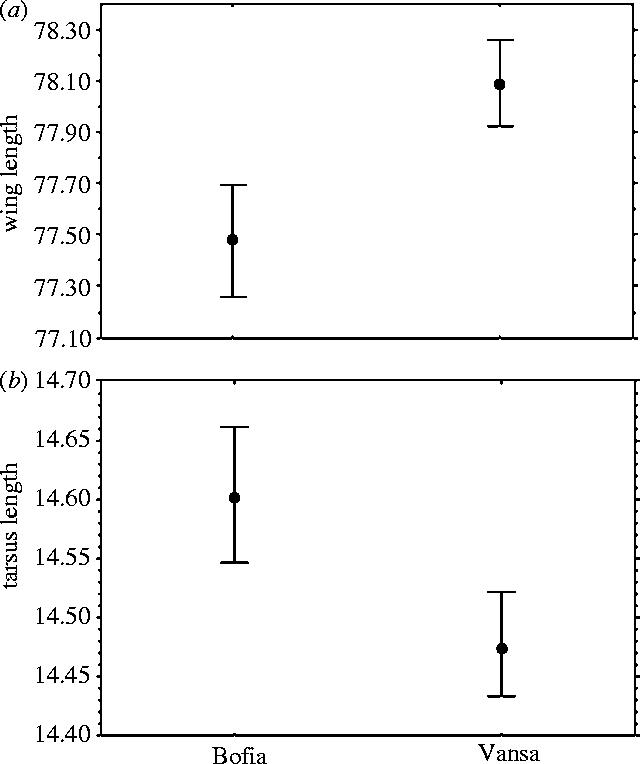
(a) Wing and (b) tarsus length variation (mean±s.e.) of citril finches comparing Bofia versus Vansa localities. Values have been standardized for sex, age and yearly variations with factorial ANOVA.
4. Discussion
The results show that there was significant and substantial genetic differentiation between the La Vansa and La Bofia. This result was based on a variety of methods and we therefore regard the result as being robust. This shows that despite the short distance, the well-documented movements and gene flow between the localities (Senar et al. 2002), strong genetic differences can still occur. In addition to differences in allele frequencies, we also found differences in morphology. This is consistent with differences in the feeding ecology and habitat structure between the two localities: La Vansa birds, which rely more often on pine seed obtained directly from cones in trees in hanging positions, had shorter tarsus lengths than La Bofia birds, which rely more on grasses and, hence, ground feeding (Senar et al. 2002). This is in accordance with the general view that ground-feeding species have longer tarsus than clinging and hanging ones (Newton 1967; Moreno & Carrascal 1993). The shorter wings of La Bofia citril finches may be related to a higher presence of aerial predators in this area (A. Borcas 2005, personal observation), as shorter wings are more manoeuvrable and, hence, may be more adaptative (Alatalo et al. 1984; Winkler & Leisler 1985; Rayner 1988). We have to point out that, although the differences in biometry between localities were small, differences between localities were in the opposite directions: wing length was larger in La Vansa, while tarsus length was larger in La Bofia.
These results suggest that even when selection and gene flow occur at the same time, morphological and genetic divergence can still occur. The degree of genetic divergence was high and the results from the assignment tests show that individuals could be accurately assigned genetically to the population where they were caught. The mechanism behind this pattern is not currently known, but two non-mutually exclusive factors can be imagined. First, there might be selection acting on individuals with the ‘wrong’ genotype. This assumes that there is a connection between the diversity at the microsatellite level and phenotype. We know that there are morphological differences (Senar et al. 2002), but whether these are based on genetic differences, or are only a result of plasticity, is not known. Second, we might have phenotype-dependent dispersal, so that individuals facing the wrong environment are more likely to disperse than others, which then might act to promote the differences (Postma & Van Noordwijk 2005). Again, this assumes genetic differences at a large scale and not only in a few microsatellite loci.
Acknowledgments
We thank Paula Dias and Ted Morrow for comments, and Peter Beerli for considerable support concerning the use of Migrate. We are grateful to Xevi Colomer, José Molina and Daniel Mañas for field assistance and Jordi García-Petit (Departament Medi Ambient, Generalitat Catalunya) for support and facilities. We thank M. Hultqvist for doing the lab-work. M.B. was supported by grants from the Swedish Research Council and the Nordic Centre of Excellence in Population Ecology and J.C.S. by BOS 2003-09589 grant, Ministerio de Ciéncia y Tecnología.
References
- Alatalo R.V, Gustafsson L, Lundberg A. Why do young passerine birds have shorter wings than older birds? Ibis. 1984;126:410–415. [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl Acad. Sci. USA. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. doi:10.1073/pnas.081068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel J, Dias P.C, Perret P, Maistre M, Lambrechts M.M. Selection-based biodiversity at a small spatial scale in a low-dispersing insular bird. Science. 1999;285:1399–1402. doi: 10.1126/science.285.5432.1399. doi:10.1126/science.285.5432.1399 [DOI] [PubMed] [Google Scholar]
- Borràs A, Cabrera J, Cabrera T, Senar J.C. Sex and age related biometrical patterns in Pyrenean Citril Finches (Serinus citrinella) Vogelwarte. 1998;39:196–202. [Google Scholar]
- Dawson D.A, Hanotte O, Greig C, Stewart I.R, Burke T. Polymorphic microsatellites in the blue tit Parus caeruleus and their cross-species utility in 20 songbird families. Mol. Ecol. 2000;9:1941–1944. doi: 10.1046/j.1365-294x.2000.01094-14.x. doi:10.1046/j.1365-294x.2000.01094-14.x [DOI] [PubMed] [Google Scholar]
- Garant D, Kruuk L.E.B, Wilkin T.A, McCleery R.H, Sheldon B.C. Evolution driven by differential dispersal within a wild bird population. Nature. 2005;433:60–65. doi: 10.1038/nature03051. doi:10.1038/nature03051 [DOI] [PubMed] [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1963. Animal species and evolution. [Google Scholar]
- Moreno E, Carrascal L.M. Leg morphology and feeding postures in four Parus species. An experimental ecomorphological approach. Ecology. 1993;74:2037–2044. [Google Scholar]
- Newton I. The adaptive radiation and feeding ecology of some British finches. Ibis. 1967;109:33–98. [Google Scholar]
- Paetkau D, Slade R, Burdens M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol. Ecol. 2004;13:55–65. doi: 10.1046/j.1365-294x.2004.02008.x. doi:10.1046/j.1365-294X.2004.02008.x [DOI] [PubMed] [Google Scholar]
- Piertney S.B, Marquiss M, Summers R. Characterisation of tetranucleotide microsatellites in the Scottish crossbill (Loxia scotica) Mol. Ecol. 1999;7:1247–1263. [PubMed] [Google Scholar]
- Postma E, Van Noordwijk A.J. Gene flow maintains a large genetic difference in clutch size at a small spatial scale. Nature. 2005;433:65–68. doi: 10.1038/nature03083. doi:10.1038/nature03083 [DOI] [PubMed] [Google Scholar]
- Primmer C.R, Møller A.P, Ellegren H. A wide-range survey of cross-species microsatellite amplification in birds. Mol. Ecol. 1996;5:365–378. doi: 10.1111/j.1365-294x.1996.tb00327.x. doi:10.1046/j.1365-294X.1996.00092.x [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (v. 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 1997;86:248–249. [Google Scholar]
- Rayner J.M.V. Form and function in avian flight. Curr. Ornithol. 1988;5:1–66. [Google Scholar]
- Schneider, S., Roessli, D. & Excoffier, L. 2000 Arlequin v. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- Senar J.C, Conroy M.J, Borras A. Asymmetric exchange between populations differing in habitat quality: a metapopulation study on the citril Finch. J. Appl. Stat. 2002;29:425–441. doi:10.1080/02664760120108791 [Google Scholar]
- Skulason S, Snorrason S.S, Jonson B. Sympatric morphs, populations and speciation in freshwater fish with emphasis on arctic charr. In: Magurran A.E, May R.M, editors. Evolution of biological diversity. Oxford University Press; New York: 1999. pp. 70–92. [Google Scholar]
- Winkler H, Leisler B. Morphological aspects of habitat selection in birds. In: Cody M.L, editor. Habitat selection in birds. Academic Press; Orlando, FL: 1985. pp. 415–434. [Google Scholar]


