Abstract
Hantaviruses cause two diseases with prominent vascular permeability defects, hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. All hantaviruses infect human endothelial cells, although it is unclear what differentiates pathogenic from nonpathogenic hantaviruses. We observed dramatic differences in interferon-specific transcriptional responses between pathogenic and nonpathogenic hantaviruses at 1 day postinfection, suggesting that hantavirus pathogenesis may in part be determined by viral regulation of cellular interferon responses. In contrast to pathogenic NY-1 virus (NY-1V) and Hantaan virus (HTNV), nonpathogenic Prospect Hill virus (PHV) elicits early interferon responses following infection of human endothelial cells. We determined that PHV replication is blocked in human endothelial cells and that RNA and protein synthesis by PHV, but not NY-1V or HTNV, is inhibited at 2 to 4 days postinfection. The addition of antibodies to beta interferon (IFN-β) blocked interferon-directed MxA induction by >90% and demonstrated that hantavirus infection induces the secretion of IFN-β from endothelial cells. Coinfecting endothelial cells with NY-1V and PHV resulted in a 60% decrease in the induction of interferon-responsive MxA transcripts by PHV and further suggested the potential for NY-1V to regulate early IFN responses. Expression of the NY-1V G1 cytoplasmic tail inhibited by >90% RIG-I- and downstream TBK-1-directed transcription from interferon-stimulated response elements or β-interferon promoters in a dose-dependent manner. In contrast, expression of the NY-1V nucleocapsid or PHV G1 tail had no effect on RIG-I- or TBK-1-directed transcriptional responses. Further, neither the NY-1V nor PHV G1 tails inhibited transcriptional responses directed by a constitutively active form of interferon regulatory factor 3 (IRF-3 5D), and IRF-3 is a direct target of TBK-1 phosphorylation. These findings indicate that the pathogenic NY-1V G1 protein regulates cellular IFN responses upstream of IRF-3 phosphorylation at the level of the TBK-1 complex. These findings further suggest that the G1 cytoplasmic tail contains a virulence element which determines the ability of hantaviruses to bypass innate cellular immune responses and delineates a mechanism for pathogenic hantaviruses to successfully replicate within human endothelial cells.
Hantaviruses are spread to humans from small mammal hosts and cause two diseases with acute vascular permeability defects, hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome (HPS) (7, 35, 48, 66). Hantaviruses predominantly infect vascular endothelial cells and cause acute thrombocytopenia, providing a focus for hantavirus-directed changes in vascular permeability and viral pathogenesis (10, 35, 38, 66, 67). The use of αvβ3 integrins on endothelial cells is a common feature of pathogenic HPS (NY-1 virus [NY-1V] and Sin Nombre virus) and HFRS (Hantaan virus [HTNV], Puumala virus, and Seoul virus) causing hantaviruses and is linked to vascular diseases caused by the absence of β3 integrin function (15, 16, 43). Pathogenic hantaviruses use αvβ3 for cellular entry and also dysregulate αvβ3 functions which maintain vascular integrity. In contrast to pathogenic hantaviruses, α5β1 integrins are used by Prospect Hill virus (PHV) and Tula virus, which are not associated with any human disease.
The ability of both pathogenic and nonpathogenic hantaviruses to infect human endothelial cells indicates that cellular entry is not the only determinant of hantavirus pathogenesis. Hantavirus infection of endothelial cells is not lytic, and hantaviruses cause little or no damage to the vasculature (30, 35, 38, 42, 58, 64, 66). Hantaviruses can be passaged with and persistently infect cells in vitro, and hantavirus infection of host endothelial cells results in a persistent infection of the animal in the absence of disease (48). The ability of pathogenic and nonpathogenic hantaviruses to infect human endothelial cells suggests that the regulation of intracellular responses and pathways is likely to contribute to the pathogenic potential of specific hantaviruses.
Viral infections produce small amounts of double-stranded RNA (dsRNA) which trigger the induction of type I interferon (alpha/beta interferon [IFN-α/β]) and direct an innate cellular defense program that limits the spread of invading pathogens (20, 54, 60). Viral dsRNA is detected by a recently discovered RNA helicase, RIG-I (retinoic acid-inducible gene I), which activates IFN signaling pathways (65). RIG-I directs IFN signaling responses through protein-protein interactions of its caspase recruitment-like domains (CARDs) with MAVS, resulting in the activation of TBK-1 (TANK binding kinase 1) and IKKɛ (28, 34, 51, 62). TBK-1 interacts with TANK and recruits tumor necrosis factor receptor-associated factor scaffolding proteins to TBK-1 complexes (40, 46). Activated TBK-1 directs interferon regulatory factor 3 (IRF-3) phosphorylation and NF-κB activation, both of which are required for IFN-β transcription (12, 23). IFN-β is secreted from infected cells and directs autocrine and paracrine signaling responses by binding to cellular IFN receptors. In response to IFN, IFN receptors activate JAK/STAT signaling pathways and direct transcription of interferon-stimulated genes (ISGs), which amplify the IFN response and disrupt viral transcription and translation (8, 9, 54).
Regulating IFN responses is fundamental to viral success and may further determine the ability of viruses to replicate in specific hosts, tissues, or cells. To negotiate cellular defenses, viruses have developed mechanisms that block dsRNA recognition, regulate cell signaling pathways that induce interferon, or compensate for ISG functions (5, 22, 27). However, hantaviruses have relatively little genetic capacity for regulating cellular IFN responses. Hantaviruses are enveloped negative-stranded RNA viruses with a tripartite genome and replicate cytoplasmically (48). Hantaviruses encode four proteins: a polymerase, a nucleocapsid protein, and two virion surface glycoproteins, G1 and G2, which are synthesized as a polyprotein and cotranslationally cleaved during translocation. G1 and G2 form heterodimers which assemble into higher-order oligomers and comprise the highly structured virion surface of hantavirus particles. G1 and G2 oligomers are trafficked to the cis-Golgi, and hantaviruses bud into the lumen of the cis-Golgi. G1 and G2 are not posttranslationally modified in the Golgi, and virion release is consistent with a novel vesicular secretory process (25, 36, 37, 47).
There is little information on the regulation of endothelial cell signaling and cellular defense responses by hantaviruses. Hantaviruses lack the nonstructural proteins of other bunyaviruses which reportedly facilitate viral replication and regulate cellular interferon responses. Hantaviruses have three cytoplasmic proteins, the polymerase, the nucleocapsid protein, and the G1 cytoplasmic tail, that have the potential to regulate innate cellular responses (50, 53). The hantavirus G1 protein contains a large 142-residue cytoplasmic tail which reportedly binds cellular kinases and is ubiquitinated and degraded by the proteasome (17, 18). However, a role for G1 tail interactions in the regulation of cellular IFN responses has not been established.
The ability of hantaviruses to enter endothelial cells and establish persistence in animals suggests that all hantaviruses are able to regulate innate endothelial cell defenses within their hosts. The dramatic induction of IFN responses by PHV, but not pathogenic NY-1V (HPS) or HTNV (HFRS), was reported 1 day postinfection (p.i.) of human endothelial cells, using DNA arrays (19, 29). These findings suggested that differences in hantavirus-directed endothelial cell responses might determine the pathogenic potential of hantaviruses at a postentry step. The effect of IFN responses directed by PHV has not been further evaluated, and it is unclear whether IFN responses limit PHV infection of human endothelial cells.
Here, we report that PHV replication within human endothelial cells is blocked and that PHV protein and RNA synthesis are inversely related to PHV-induced IFN responses 2 to 5 days p.i. In contrast, HTNV and NY-1V productively replicate within human endothelial cells and protein and RNA synthesis by these pathogenic hantaviruses are not restricted at 1 to 5 days p.i. Coinfection of cells with PHV and NY-1V reduced PHV-directed IFN responses, and cells infected with pathogenic hantaviruses developed resistance to exogenously applied IFN from 12 to 24 h p.i. Expression of the NY-1V, but not the PHV, G1 cytoplasmic tail inhibited RIG-I- and TBK-1-directed IFN responses. The inability of the NY-1V G1 tail to inhibit IRF-3 5D-directed transcription indicates that the G1 tail blocks IFN pathway activation upstream of IRF-3 phosphorylation at the level of the TBK-1 complex. Our findings indicate that the pathogenic NY-1V G1 cytoplasmic tail regulates IFN signaling pathway activation, which permits the productive replication of NY-1V within human endothelial cells.
MATERIALS AND METHODS
Cells and virus.
Biosafety level 3 facilities were used throughout these experiments for hantavirus cultivation. Vero E6 cells were grown in Dulbecco's minimal essential medium, 10% fetal calf serum (FCS), l-glutamine, penicillin, and streptomycin (Gibco). Human umbilical vein endothelial cells (HUVECs) were purchased from Clonetics (Walkersville, MD). Endothelial cells were grown in endothelial cell basal medium 2 supplemented with human recombinant epidermal growth factor (10 ng/ml), hydrocortisone (1 μg/ml), gentamicin (50 μg/ml), amphotericin B (50 μg/ml), 0.1% endothelial growth factor, and 2% FCS (Clonetics) (16). HUVECs were used for three to six passages. NY-1V, HTNV 76-118, and PHV were mycoplasma free and cultivated in Vero E6 cells as previously described (48).
Hantavirus infection.
Cells were infected in six-well plates with viral stocks containing approximately 5 × 105 focus-forming units (FFU) per ml, as determined by a focus assay with Vero E6 cells (16). Virus was adsorbed to cells at a multiplicity of infection (MOI) of 1 for 1 h at 37°C as indicated. Virus was removed, monolayers were washed with media, and cells were maintained in complete media with 2% FCS. At various times p.i., the titer of virus in cell supernatants was determined by titration on Vero E6 cells as previously described (16). In some experiments, 1,000 IU/ml human IFN-α (Sigma) was added 24 h prior to, or at various times after, mock infection or infection with NY-1V, HTNV, or PHV (MOI of 1). In other experiments, cells were treated with 500 U/ml of neutralizing antibody to IFN-α or IFN-β, as indicated, following adsorption. These experiments were performed at least five times with similar results.
Immunoperoxidase staining of hantavirus-infected cells.
Immunoperoxidase staining of the hantavirus nucleocapsid protein within infected cells has previously been described (14). Briefly, cell monolayers were fixed with 100% methanol and incubated with a rabbit polyclonal anti-nucleocapsid serum (1:2,000) made to the recombinant nucleocapsid protein of NY-1V. Monolayers were washed with phosphate-buffered saline (PBS) and incubated with goat anti-rabbit horseradish peroxidase (HRP) conjugate (Kirkegaarde and Perry Laboratories). Monolayers were washed with PBS and stained with 3-amino-9-ethylcarbazole (0.026%) in 0.1 M sodium acetate, pH 5.2, and 0.03% H2O2 for 5 to 30 min (14). Monolayers were washed with distilled water, and the number of nucleocapsid protein-containing infected cells present in duplicate wells was quantified and compared to that of the controls.
Real-time PCR analysis.
Total RNA was extracted from cells using the RNeasy kit (QIAGEN, Chatsworth, CA), according to the manufacturer's protocol. RNA (1 μg) was reverse transcribed using oligo-p(dT)15 or p(dN)6 primers and the First Strand cDNA synthesis kit (Roche) according to the company's protocol. Primers were designed using Vector NTI Suite so that primer pairs were 100 to 150 nucleotides apart and had similar GC contents and melting temperatures. Specific primers were made to S segments from NY-1V, HTNV, and PHV as well as GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and MxA at the following nucleotide positions: NY-1V forward, positions 126 to 145; NY-1V reverse, positions 257 to 277 (52); HTNV forward, positions 798 to 817; HTNV reverse, positions 914 to 933 (49); PHV forward, positions 184 to 203; PHV reverse, positions 307 to 326 (39); GAPDH forward, positions 4357 to 4375 (GGAAGCTCACTGGCATGGC); GAPDH reverse, positions 4408 to 4427 (TAGACGGCAGGTCAGGTCCA) (33); MxA forward, positions 379 to 397 (TGATCCAGCTGCTGCATCCC); and MxA reverse, positions 477 to 496 (GGCGCACCTTCTCCTCATAC) (1). Real-time PCR was performed with cDNA templates in a LightCycler reaction mixture: FastStart DNA Master SYBR green I (Roche), 4 mM MgCl2, and 0.5 μM of each primer pair. LightCycler PCR conditions were as follows: 95°C for 5 min, followed by 30 amplification cycles of 95°C for 15 seconds, 62°C for 5 seconds, and 72°C for 5 seconds. Melting curve analysis was used to confirm PCR product identity. The results were compared to those of controls lacking cDNA template, and GAPDH primer-directed real-time PCRs were used to normalize sample cDNA levels (19). Experiments were performed three times with similar results.
For ISG56 analysis, HUVECs were infected with PHV or NY-1V and RNA was extracted as described above. ISG56 mRNA levels were determined relative to those of mock-infected controls, using TaqMan primers for ISG56 (Applied Biosystems). Real-time PCR was performed using an Applied Biosystems 7300 real-time PCR machine. Thermocycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, 95°C for 15 sec, and 60°C for 1 min for a total of 40 cycles. All reactions were normalized to TaqMan-derived GAPDH mRNA levels.
Western blot analysis.
Western blot analysis was performed as previously described (17, 18). Briefly, infected HUVECs (1 × 105) were lysed as previously described at various times p.i., protein levels were determined by a bicinchoninic acid assay (Pierce), and 2.5 to 5 μg of protein was separated by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Proteins were transferred to nitrocellulose using a Novex XCell electroblotter and blocked with 5% bovine serum albumin, and nucleocapsid proteins were detected using rabbit anti-nucleocapsid sera (1:10,000) followed by anti-rabbit HRP-conjugated antibody (1:5,000) (Amersham). G1 cytoplasmic tail and N protein expression was evaluated following transfection of COS7 cells that were treated with MG132 (50 μM) at 40 h posttransfection. Cells were lysed in 2× sample buffer (100 mM Tris-Cl, pH 6.8, 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol) and subjected to Western blot analysis at 48 h posttransfection as described above, using a monoclonal anti-GAL4 antibody (1:2,000) from Santa Cruz Biotechnology (GAL4 [RK5C1], sc-510). Tubulin was detected using a monoclonal anti-tubulin antibody (1:2,000) from Sigma (anti-β-tubulin clone TUB 2.1, T-4026), and Western blots were developed by using ECL reagent (Amersham) and exposing the blots to XAR-5 film (Kodak). Western blotting was performed three times with similar results.
Hantavirus protein expression plasmids and transfections.
Constructs expressing the NY-1V G1 cytoplasmic tail (pBIND-NY1G1cyto), the PHV G1 cytoplasmic tail (pBIND-PHVG1cyto), and the NY-1V nucleocapsid protein (pBIND-NY1-S) were generated by C-terminally fusing coding regions to a GAL4 tag as previously reported (17). Briefly, coding regions were amplified with PCR primers containing BamHI and XbaI restriction sites and ligated directionally into the pBIND expression vector (Promega). All transfections were performed in subconfluent monolayers of HEK 293 cells (∼1 × 106) in six-well plates (Corning, Inc.), using the calcium phosphate method as previously described (17). Cells were transfected with constant amounts of plasmid DNA for 16 h, washed in 2 ml of PBS (1 min), and grown in complete medium (Dulbecco's minimal essential medium with 10% FCS, penicillin, and streptomycin) for 48 h before analysis.
Luciferase assays.
HEK 293 cells were cotransfected with 0.5 μg pISRE-Luc (Clontech Laboratories, Inc.) containing five copies of the interferon-stimulated response element (ISRE) binding site or 0.5 μg of the IFN-β promoter luciferase reporter as indicated, as well as 250 ng of PRL null (Promega), a constitutively active Renilla luciferase expression vector. Cells were cotransfected with 250 to 500 ng of a plasmid expressing myc-tagged TBK-1 (pC-TBK-1), generously provided by Joel Pomerantz and David Baltimore (California Institute of Technology, Pasadena, CA) (40), or indicated amounts of a construct expressing N-terminal CARDs of RIG-I [(N)RIG, residues 1 to 284; a gift from Michael Gale, University of Texas Southwest Medical Center, Dallas, TX] (55) or a constitutively active form of IRF-3 (IRF-3 5D) (31, 32) with an N-terminal green fluorescent protein tag (a gift from John Hiscott, McGill University, Montreal, Canada). Cells were transfected with the indicated amounts of expression vector, and the total amount of plasmid DNA transfected was kept constant using empty pBIND vector. Cells were lysed at 48 h posttransfection in 1× passive lysis buffer (Promega) by incubation at 25°C (15 min) with gentle agitation. All luciferase assays were performed at least three times with similar results using the Dual-Luciferase assay system (Promega) and a Turner Designs TD 20/20 luminometer.
RESULTS
PHV replication is inhibited in endothelial cells.
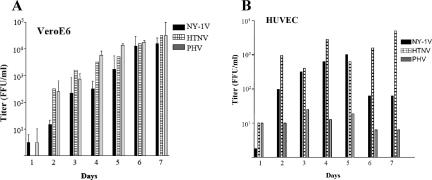
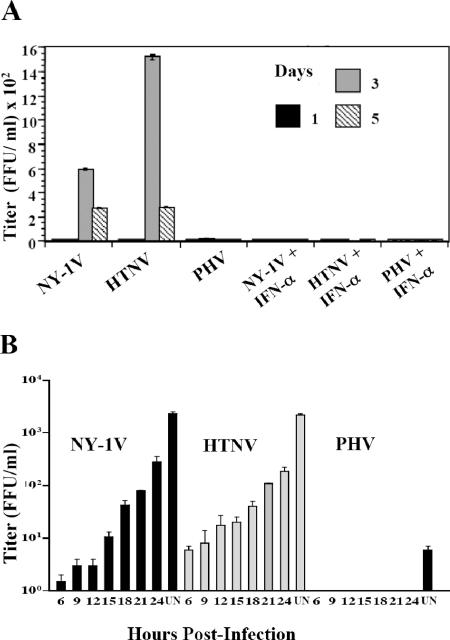
Pathogenic and nonpathogenic hantaviruses reportedly infect human endothelial cells (64). However, recent findings suggest that fundamental differences in hantavirus-elicited interferon responses might limit replication of some nonpathogenic hantaviruses. Vero E6 cells, which are used for the growth of all hantaviruses, lack the type I interferon locus, further suggesting that interferon responses of endothelial cells may restrict replication of some hantaviruses. Since there is little information on the replication of nonpathogenic hantaviruses in human endothelial cells, we compared the abilities of nonpathogenic (PHV) and pathogenic (HTNV and NY-1V) hantaviruses to replicate in Vero E6 cells and HUVECs. Cells were infected at an MOI of 1, and virus present in cell supernatants was titered at various times postinfection (Fig. 1A). PHV, NY-1V, and HTNV replicated in Vero E6 cells, resulting in titers of 1 × 103 to 1 × 104 FFU/ml 3 to 4 days p.i. and reaching higher titers at 6 to 7 days p.i. (Fig. 1A). NY-1V and HTNV similarly reached titers of 1 × 103 to 1 × 104 FFU/ml at 3 to 4 days postinfection of HUVECs (Fig. 1B). In contrast, there was little PHV replication within HUVECs at 1 to 7 days postinfection, with maximal titers of 5 to 50 FFU/ml in supernatants (Fig. 1B). These findings suggest that PHV replication is inhibited within human endothelial cells.
FIG. 1.
Hantavirus replication in HUVECs and Vero E6 cells. Vero E6 cells (A) and HUVECs (B) were infected with NY-1V, HTNV, or PHV at an MOI of 1. Viral titers in the supernatant of infected cells were analyzed 1 to 7 days postinfection by an infectious focus assay (16). Days postinfection are indicated on the x axis, and titers are represented as FFU/ml.
PHV transcription and protein synthesis are altered in endothelial cells.
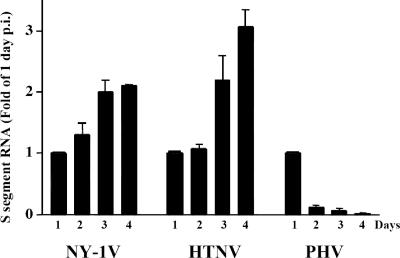
In order to determine why PHV replication is blocked in endothelial cells, we comparatively evaluated hantavirus transcription levels within HUVECs. Hantavirus mRNA levels were quantitated using real-time PCR and standardized to cellular GAPDH mRNA levels (Fig. 2). As previously reported, S segment mRNA levels within NY-1V-, HTNV-, and PHV-infected endothelial cells were similar 1 day p.i (19), and this similarity is reflected in comparable N protein levels within infected cells (Fig. 3). NY-1V and HTNV S segment mRNAs increased to maximal level 3 to 4 days p.i., while peak PHV S segment mRNA levels were observed 1 day p.i. and decreased 10- to 20-fold 2 to 4 days p.i. (Fig. 2).
FIG. 2.
Kinetics of S segment RNA synthesis during hantavirus infection. HUVECs were mock infected or infected with NY-1V, HTNV, or PHV (MOI of 1). S segment RNA levels were determined by quantitative real-time PCR using hantavirus S segment-specific primers 1 to 4 days postinfection, and responses from duplicates were normalized to GAPDH mRNA levels. Experiments were performed twice with similar results, and mRNA levels are expressed as the change (n-fold) from 1-day levels.
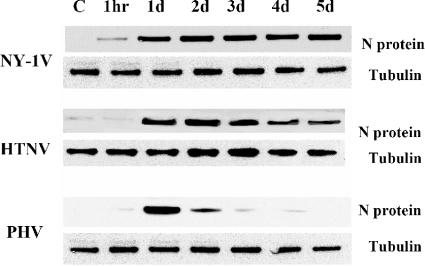
FIG. 3.
Western blot analysis of N protein expression. HUVECs were infected with NY-1V, HTNV, or PHV (MOI of 1) or mock infected (C). Cells were lysed at 1 hour or 1 to 5 days postinfection as indicated. Total protein levels were determined, and an equivalent amount of whole-cell lysate was separated by 10% SDS-polyacrylamide gel electrophoresis. Proteins were detected by Western blotting using anti-nucleocapsid polyclonal rabbit antibody or anti-tubulin monoclonal antibody (Sigma), species-specific secondary antibodies (HRP conjugated), and enhanced chemiluminescence (Amersham).
Immunoperoxidase staining of nucleocapsid protein in infected monolayers indicated that nearly every cell contained clearly detectable levels of N protein at 1 day postinfection with PHV, NY-1V, and HTNV. Interestingly, we visually noted that nucleocapsid protein levels within PHV-, but not HTNV- or NY-1V-, infected cells declined 1 to 4 days postinfection. In order to demonstrate differences in nucleocapsid protein levels, we evaluated nucleocapsid protein expression in NY-1V-, HTNV-, and PHV-infected endothelial cells by Western blot analysis 1 to 5 days p.i. Comparable levels of nucleocapsid protein were observed 1 day postinfection with NY-1V, HTNV, and PHV, and nucleocapsid protein levels persisted within NY-1V- and HTNV-infected cells from 1 to 5 days postinfection (Fig. 3). In contrast, nucleocapsid protein levels in PHV-infected cells decreased dramatically 2 to 4 days p.i., with no apparent nucleocapsid protein present 5 days p.i. (Fig. 3). Compared with pathogenic NY-1V and HTNV, our findings demonstrate that PHV mRNA and nucleocapsid protein synthesis are dramatically reduced 2 to 5 days postinfection of human endothelial cells.
Induction of ISG56 and MxA following hantavirus infection.
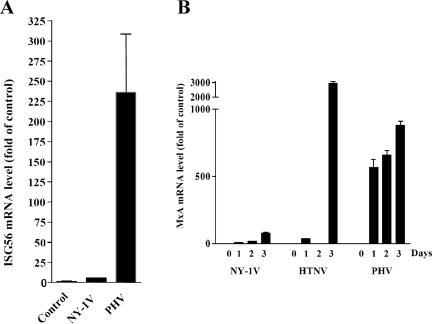
IFNs and ISGs are known to regulate viral transcription and replication (22). Using real-time PCR, we studied the transcription of ISG56 and MxA, which are induced by IFN secretion and binding to IFN receptors. One day postinfection, we found that PHV infection of human endothelial cells directed a >225-fold increase in ISG56 mRNA compared to a 6-fold increase in ISG56 mRNA by NY-1V (Fig. 4A). PHV also directed a 539-fold increase in MxA, while pathogenic NY-1V or HTNV induced a substantially smaller 9- or 31-fold increase in MxA, respectively, (Fig. 4B). By 3 days p.i. all three hantaviruses induced MxA to high levels, although NY-1V directed an approximately 30-fold-lower MxA response than HTNV or PHV. These studies demonstrate that nonpathogenic PHV and pathogenic NY-1V and HTNV differentially direct IFN responses 1 day postinfection of human endothelial cells. High-level IFN responses induced by PHV at 1 day postinfection are consistent with reduced PHV transcription, protein synthesis, and replication in human endothelial cells. These findings further suggest that NY-1V and HTNV may regulate early IFN responses during infection.
FIG. 4.
Kinetics of MxA and ISG56 mRNA induction during hantavirus infection. HUVECs were infected with NY-1V or PHV (MOI of 1) or mock infected. One day postinfection, ISG56 (A) and MxA (B) mRNA levels were determined, relative to those of mock-infected controls, using quantitative real-time PCR and normalized to GAPDH mRNA levels. MxA mRNA levels were quantified 0, 1, 2, and 3 days postinfection.
Hantaviruses direct the secretion of IFN-β from endothelial cells.
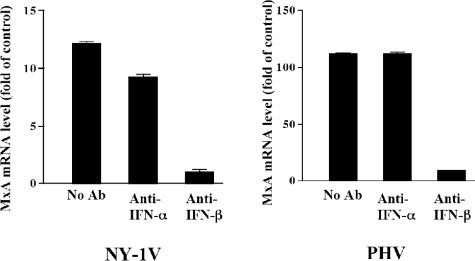
Minute amounts of secreted interferon (type I, alpha or beta) direct the autocrine or paracrine activation of IFN receptors, which amplify IFN signals and induce the high-level transcription of a large number of ISGs, including MxA (3, 44, 45). In order to demonstrate that hantaviruses direct MxA responses through secreted IFNs, we added function-blocking antibodies to IFN-α or IFN-β to the media of infected cells and assayed MxA induction 1 day after hantavirus infection. PHV directed a 10-fold-higher level of MxA induction than NY-1V 1 day postinfection (Fig. 5). When neutralizing antibodies to IFN-β, but not IFN-α, were added to the media, MxA induction was blocked following infection by NY-1V or PHV. This demonstrates that hantavirus infection directs the secretion of IFN-β from human endothelial cells, which in turn directs MxA transcription (Fig. 5).
FIG. 5.
Antibody to IFN-β inhibits hantavirus-directed MxA induction. HUVECs were infected at an MOI of 1 with NY-1V or PHV or mock infected. Following adsorption, anti-IFN-α or anti-IFN-β neutralizing antibodies were added to the media as indicated and MxA mRNA levels were quantified by real-time PCR as described above. MxA levels are shown as the increase (n-fold) compared to those of mock-infected controls. The scales of the y axes for NY-1V and PHV differ 10-fold. Ab, antibody.
Hantavirus replication is IFN sensitive.
In order to determine whether hantaviruses are sensitive to type I IFN, human endothelial cells were pretreated with IFN-α (1,000 IU/ml) 1 day prior to infection with HTNV, NY-1V, or PHV. IFN pretreatment completely inhibited the replication of NY-1V, HTNV, and PHV, while PHV replication in human endothelial cells was restricted even without added IFN (Fig. 6A). These findings indicate that pathogenic hantaviruses are acutely sensitive to the prior addition of IFN and suggest that pathogenic hantaviruses need to regulate IFN responses to successfully replicate within endothelial cells.
FIG. 6.
Hantavirus replication in the presence of type I interferon. (A) Vero E6 cells were pretreated in duplicate with 1,000 IU/ml IFN-α or left untreated for 24 h prior to infection with NY-1V, HTNV, or PHV (MOI of 1) or mock infection. Hantavirus titers were determined using a focus assay 0, 1, 3, and 5 days postinfection as previously described (16). (B) HUVECs were infected as for panel A prior to the addition of 1,000 IU/ml of IFN-α 6 to 24 h postinfection or were left untreated (UN) as indicated. Three days postinfection, cell supernatants were titered on Vero E6 cells by using a focus assay (16).
Pathogenic hantaviruses develop resistance to IFN addition during infection.
To determine whether pathogenic hantaviruses develop resistance to IFN during the course of infection, we added IFN to cells at various times following hantavirus infection (6 to 24 h) and monitored viral titers at 3 days postinfection. PHV replication was not detectable in endothelial cells with or without IFN treatment, and the addition of IFN 6 to 12 h after NY-1V or HTNV infection blocked replication almost entirely (Fig. 6B). In contrast, when IFN was added between 15 and 24 h p.i. we observed a gradual increase in NY-1V and HTNV titers (Fig. 6B), demonstrating the progressive insensitivity of NY-1V and HTNV replication to IFN addition. Collectively, our findings are consistent with the ability of pathogenic hantaviruses to synthesize products which regulate early IFN responses and, at later times postinfection, block the effects of IFN-induced ISGs that limit viral replication.
NY-1V coinfection of human endothelial cells attenuates PHV-directed MxA induction.
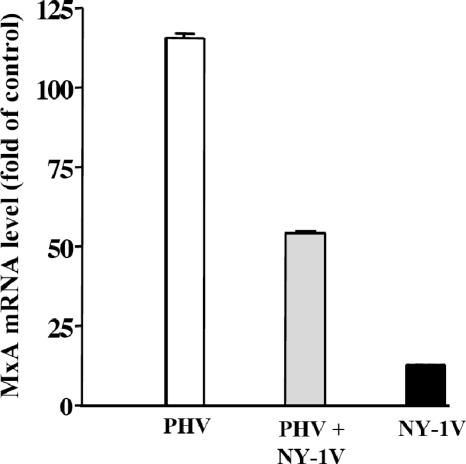
The potential for pathogenic hantaviruses to regulate IFN responses suggested that NY-1V might suppress IFN responses directed by PHV (19). In order to test this, we compared the induction of MxA during NY-1V and PHV coinfection with that of MxA during infection with each virus alone by using real-time PCR. We found that PHV induced MxA over 100-fold and at least 10-fold more than NY-1V infection alone (Fig. 7). In contrast, coinfecting endothelial cells with PHV and NY-1V resulted in a 66% reduction in PHV-directed MxA transcription (Fig. 7). Although these findings suggest that NY-1V might repress early IFN responses directed by PHV, this experiment does not exclude the possibility that NY-1V interferes with PHV transcription.
FIG. 7.
NY-1V coinfection attenuates PHV-directed MxA induction. HUVECs were mock infected, infected with NY-1V or PHV (MOI of 0.5), or infected with NY-1V and PHV (MOI of 0.5 each) in duplicate. One day postinfection, MxA mRNA levels were quantified by real-time PCR and standardized to GAPDH mRNA levels as described above, and they are reported as the increase (n-fold) in mRNA levels compared to those of mock-infected controls.
Hantavirus G1 protein blocks cellular IFN responses.
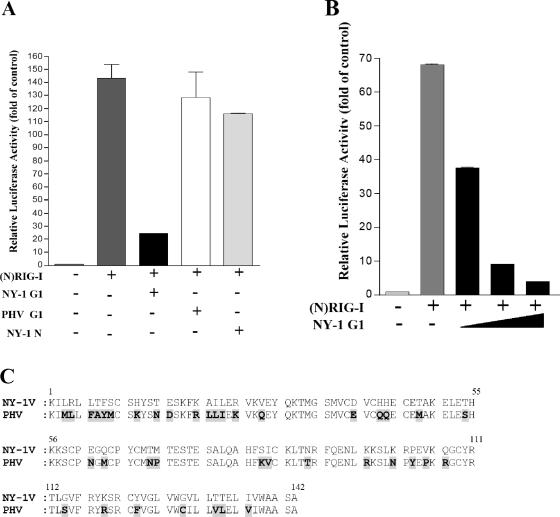
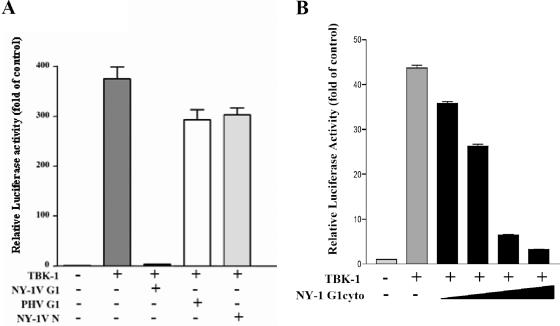
In order to determine whether pathogenic hantavirus proteins are capable of regulating IFN signaling pathways, we compared the abilities of the hantavirus N and G1 cytoplasmic tail proteins to regulate IFN pathway activation directed by RIG-I and TBK-1. We cotransfected cells with a constitutively active RIG-I construct containing only the N-terminal CARD and a luciferase reporter driven by an ISRE. We evaluated the ability of NY-1V nucleocapsid or G1 cytoplasmic tail proteins to regulate RIG-I-directed ISRE activation. RIG-I induced ISRE transcription approximately 130-fold compared to controls, and expression of the NY-1V N protein or the PHV G1 cytoplasmic tail had little effect on RIG-I-directed ISRE transcription (Fig. 8A). In contrast, expressing the NY-1V G1 cytoplasmic tail inhibited RIG-I-directed ISRE transcription (Fig. 8A) and cotransfecting increasing amounts of the NY-1V G1 tail expression plasmid resulted in a concomitant decrease in RIG-I-directed ISRE transcription (>90%) (Fig. 8B). These results indicate that the NY-1V G1 cytoplasmic tail blocks RIG-I-directed transcriptional responses from ISRE-containing promoters.
FIG. 8.
NY-1V G1 cytoplasmic tail inhibits RIG-I-directed ISRE activation. (A) HEK 293 cells were transfected with an ISRE-driven luciferase reporter construct with or without cotransfection of the (N)RIG-I expression plasmid (500 ng). Cells were cotransfected with plasmids expressing the NY-1V or PHV G1 cytoplasmic tail or the NY-1V N protein (2 μg). (B) Cells were cotransfected with the (N)RIG-I expression vector (250 ng) and increasing amounts of plasmid expressing the NY-1V G1 cytoplasmic tail (0.5, 1, or 2 μg) or the control empty vector in order to transfect cells with constant amounts of total DNA. Two days posttransfection, cells were lysed, luciferase activity was assayed, and the increase (n-fold) in luciferase activity levels compared to those of controls which were not transfected with (N)RIG-I was reported after being normalized to Renilla luciferase levels. (C) Amino acid alignment of 142-residue G1 tail sequences from NY-1V (G1, positions 510 to 652) and PHV (G1, positions 513 to 655). PHV residues which differ from NY-1V are highlighted and bolded.
Sequence differences within the G1 tails of NY-1V and PHV.
An analysis of NY-1V and PHV G1 cytoplasmic tails revealed the presence of 41 different residues in their 142-amino-acid cytoplasmic tails (27% divergence) (Fig. 8C). There are notable changes within the G1 tails which include charge changes, proline insertions, tyrosine differences, and the presence of an additional cysteine at residue 128 within the PHV G1 tail. However, the broad divergence of PHV and NY-1V G1 tails does not delineate a specific residue change or domain that might account for the differential regulation of RIG-I-directed responses.
NY-1V G1 tail regulates TBK-1-directed transcription from ISRE and IFN-β promoters.
TBK-1 is a downstream effector of RIG-I activation, and TBK-1 activation is required for ISRE and IFN-β transcriptional responses. To determine whether the G1 cytoplasmic tail inhibits IFN signaling upstream or downstream of TBK-1, we determined whether hantavirus proteins regulate TBK-1-directed transcriptional responses. We determined the effect of the NY-1V G1 cytoplasmic tail on transcriptional responses directed by the complete IFN-β promoter. TBK-1 directed transcription from the IFN-β promoter >40-fold, while coexpression of the NY-1V G1 cytoplasmic tail inhibited TBK-1-directed IFN-β transcriptional responses by >95% (Fig. 9A). Transfecting identical amounts of NY-1V N or PHV G1 cytoplasmic tail expression plasmids had little effect on TBK-1-directed IFN-β transcription (Fig. 9A). Transfecting cells with increasing amounts of NY-1V G1 cytoplasmic tail resulted in a dose-dependent decrease in TBK-1-directed transcription from the IFN-β promoter (Fig. 9B).
FIG. 9.
Hantavirus G1 cytoplasmic tail disrupts IFN-β promoter activation. (A) Duplicate wells of HEK 293 cells were transfected with an IFN-β promoter-driven luciferase reporter with or without a TBK-1 expression vector (500 ng). Cells were cotransfected with equal amounts of NY-1V or PHV G1 cytoplasmic (cyto) tail or NY-1V N protein expression vector (4 μg) as indicated. Luciferase activity was determined 48 h posttransfection and normalized to Renilla luciferase activity. Luciferase activity is reported as the increase (n-fold) compared to that of controls not transfected with TBK-1. (B) Cells were transfected with the TBK-1 expression vector as described above (250 ng), along with increasing amounts of plasmid expressing the NY-1V G1 cytoplasmic tail (0.5, 1, 2, or 4 μg) or the control empty vector in order to transfect cells with a constant amount of DNA.
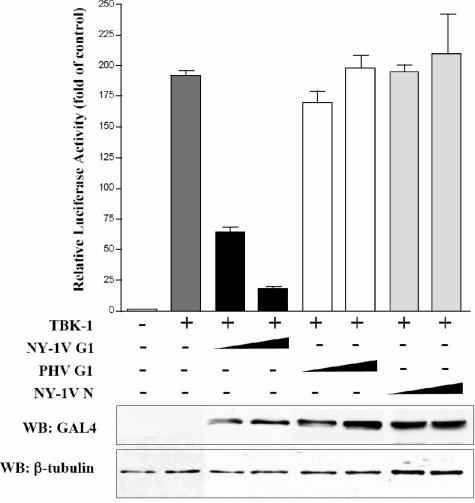
We similarly found that TBK-1 activated ISRE transcription approximately 190-fold compared to controls and that coexpression of the NY-1V G1 cytoplasmic tail resulted in a >90% reduction in TBK-1-directed ISRE transcription (Fig. 10). In contrast, coexpression of either the NY-1V N protein or the PHV G1 cytoplasmic tail had little or no effect on TBK-1-directed ISRE transcriptional responses (Fig. 10). Similar to RIG-I, cotransfecting cells with increasing amounts of the NY-1V G1 cytoplasmic tail resulted in a dose-dependent inhibition of TBK-1-directed transcription (Fig. 10). These findings indicate that expression of the NY-1V G1 cytoplasmic tail blocks transcription from ISRE promoters at or downstream of the TBK-1 complex.
FIG. 10.
NY-1V G1 cytoplasmic tail blocks TBK-1-directed ISRE activation. ISRE luciferase reporters were transfected into HEK 293 cells with or without TBK-1 expression plasmids (500 ng). Cells were cotransfected with increasing amounts of NY-1V G1 cytoplasmic tail, PHV G1 cytoplasmic tail, or NY-1V N protein expression vector (1 or 2 μg) or empty vector in order to transfect cells with a constant amount of DNA. Luciferase reporter activity was assayed as for Fig. 8 48 h posttransfection and normalized to Renilla luciferase activity, and it is reported as the increase (n-fold) compared to that of controls lacking TBK-1 activation. Protein expression levels of cells were similarly evaluated by Western blotting (WB) 48 h p.i. from parallel, comparably transfected monolayers using an anti-GAL4 antibody, and β-tubulin levels were analyzed as internal controls by Western blotting.
NY-1V G1 tail regulates transcription upstream of IRF-3 phosphorylation.
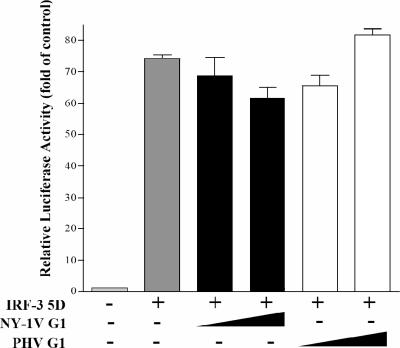
Activated TBK-1 phosphorylates IRF-3, and phosphorylated IRF-3 dimerizes and translocates to the nucleus, where it directs transcriptional responses. To determine whether the NY-1V G1 tail regulates IFN responses upstream or downstream of IRF-3 phosphorylation, we induced ISRE transcription using a constitutively active phosphomimetic form of IRF-3 (IRF-3 5D) (32). IRF-3 5D expression resulted in a >70-fold induction of the ISRE reporter, and coexpression of either the NY-1V or PHV G1 tail failed to inhibit IRF-3 5D-directed transcriptional responses (Fig. 11). These findings indicate that the NY-1V G1 tail is incapable of inhibiting phospho-IRF-3-directed transcriptional responses and suggest that G1 regulates IFN transcriptional responses upstream of IRF-3 phosphorylation at the level of the TBK-1 complex. This suggests a pathway-specific mechanism for the pathogenic NY-1 hantavirus to regulate cellular IFN signaling pathways and prevent early IFN responses that limit the replication of PHV within human endothelial cells.
FIG. 11.
NY-1V G1 tail inhibits transcription upstream of IRF-3 phosphorylation. ISRE luciferase reporters were transfected into HEK 293 cells with or without IRF-3 5D expression plasmids (500 ng). Cells were cotransfected with the NY-1V or PHV G1 cytoplasmic tail or NY-1V N protein expression vector as indicated (2 μg). Luciferase reporter activity was assayed as for Fig. 8 48 h posttransfection and is reported as the increase (n-fold) compared to that of controls lacking IRF-3 5D.
DISCUSSION
Hantaviruses predominantly infect endothelial cells and cause two diseases with defects in vascular endothelial barrier functions (6, 24). There are a few hantaviruses which are not associated with any human disease, although there is little information about the replication of nonpathogenic hantaviruses in human endothelial cells (57, 63). The cellular entry of pathogenic and nonpathogenic hantaviruses reportedly requires different integrin receptors with differing roles in vascular permeability; however, nonpathogenic hantaviruses still enter human endothelial cells (15, 64). This suggests that at one level, intracellular processes are likely to determine the pathogenic potential of specific hantaviruses. In this report, we demonstrate that, in contrast to pathogenic hantaviruses, the nonpathogenic hantavirus PHV induces early IFN responses and that PHV replication is inhibited in human endothelial cells. Our findings indicate that expression of the G1 cytoplasmic tail from pathogenic NY-1V, but not PHV, regulates the induction of cellular IFN responses. These findings demonstrate that a specific hantavirus protein is capable of regulating innate immune responses and suggest that IFN regulation is a primary determinant of hantavirus pathogenesis.
DNA array data suggested that PHV infection of human endothelial cells is unique from that of HTNV or NY-1V (19). PHV directs strong IFN responses within endothelial cells 1 day postinfection which are absent following infection by HTNV or NY-1V (19). This prompted us to compare pathogenic and nonpathogenic hantavirus RNA syntheses and replications within human endothelial cells. Our findings demonstrate that PHV RNA and protein synthesis are restricted primarily to the first day of hantavirus infection and decrease dramatically 2 to 5 days postinfection of human endothelial cells. Our results also demonstrate that PHV replication is nearly completely blocked following infection of human endothelial cells. This is in contrast to pathogenic HTNV and NY-1V, which continue to synthesize viral mRNA and protein 1 to 5 days postinfection and successfully replicate within human endothelial cells. Consistent with a role for IFN in regulating PHV replication within human endothelial cells, PHV replicates successfully in Vero E6 cells which lack a type I IFN genetic locus (11, 59). The successful replication of pathogenic hantaviruses in endothelial cells, the absence of early ISG induction by HTNV or NY-1V, and the inhibition of PHV-directed ISG responses by NY-1V coinfection suggest that pathogenic hantaviruses regulate early IFN responses following infection. The findings presented here suggest that early PHV-directed interferon responses restrict PHV's ability to replicate within endothelial cells and provide a strong rationale for why PHV is not a human pathogen.
The ability of type I interferon to protect endothelial cells from HTNV infection has been reported previously (38). Pretreating endothelial cells with type I interferon also inhibits NY-1V and PHV replication, demonstrating that pathogenic and nonpathogenic hantaviruses are sensitive to the effects of interferon added prior to infection. In fact, the addition of IFN up to 12 h postinfection still blocked NY-1V and HTNV replication and suggested a rationale for why hantaviruses need to regulate the early induction of IFN responses by endothelial cells in order to replicate. Interestingly, the addition of IFN at 15 to 24 h postinfection coincides with an increase in NY-1V and HTNV replication, suggesting that a viral product is synthesized 12 to 24 h postinfection which compensates for IFN-induced responses. These findings suggest that pathogenic hantaviruses have at least two means of regulating innate cellular responses, one which inhibits the early induction of IFN and a second which permits hantavirus replication in the presence of added IFN or IFN induced at later times postinfection. Currently, there is no information on hantavirus proteins that might temper the effects of IFN on hantavirus replication or regulate the function of ISGs.
MxA is an ISG that reportedly blocks Puumala virus, Tula virus, and HTNV replication when constitutively expressed in cells prior to infection (13, 21, 26), and we have demonstrated an inverse correlation between the induction of MxA and PHV replication. The early high-level MxA responses directed by PHV, but not NY-1V or HTNV, suggest that MxA alone could be responsible for limiting PHV transcription and replication within endothelial cells. High levels of MxA are also induced by pathogenic hantaviruses 2 to 4 days postinfection of human endothelial cells, but MxA induction at this point appears to have little effect on NY-1V or HTNV replication. These findings are consistent with clinical data indicating that only early IFN treatment is effective against hantavirus disease (2). Collectively, these data suggest that hantaviruses are unable to compensate for preinduced ISGs or even preexisting MxA protein (26) but that during the course of infection, hantaviruses produce a compensatory product which counters the effects of MxA and presumably other ISGs that otherwise limit hantavirus replication. The means by which pathogenic hantaviruses replicate in the presence of high levels of MxA and other ISGs at late times postinfection remain to be investigated.
There are several papers in the literature which indicate that hantaviruses induce IFN-β at late times postinfection, and the absence of early IFN responses by some pathogenic hantaviruses has been noted (29, 38). There are also conflicting reports suggesting that type I interferons are not induced by hantaviruses, that early IFN responses are elicited by some pathogenic hantaviruses, and that inactivated hantaviruses induce early IFN responses (41, 56). However, IFN transcription is transient and IFN enzyme-linked immunosorbent assays are of low sensitivity relative to assays of IFN function which are amplified by IFN receptor-directed ISG responses. We and others have noted that IFN-β is induced by hantavirus infection of human endothelial cells, and antibodies to IFN-β have previously been shown to enhance HTNV replication (38). Here, we demonstrate that antibodies to IFN-β inhibit the induction of ISGs following hantavirus infection and thus hantaviruses induce the secretion of IFN-β that directs ISG responses (38). Although reported, no explanation has been provided for how or why inactivated hantaviruses might play a role in inducing IFN responses in endothelial cells or the means by which this response might be elicited in an in vivo setting (40). Since viruses are clearly sensitive to the induction or addition of IFN at early points postinfection, it is also unclear how pathogenic hantaviruses might compensate for inactivated virus induction of early IFN responses.
Hantaviruses replicate in endothelial cells and establish persistent infections of their small mammal hosts in the absence of pathogenesis (48). This indicates that hantavirus entry and replication within endothelial cells are not by themselves sufficient for hantavirus pathogenesis and suggests that all hantaviruses are capable of inhibiting innate cellular responses that would otherwise limit viral replication within their hosts. A subset of hantaviruses appear to be capable of blocking early IFN responses in human endothelial cells as well as the function of ISGs induced at later times, and this appears to be a prerequisite for hantavirus pathogenesis. Differences in human and host IFN pathway proteins that might differentiate these regulatory responses are currently unknown. However, delineating pathway-specific proteins and ISGs regulated by pathogenic hantaviruses may lead to the discovery of specific viral protein interactions which differentiate hantavirus replication in host and human cells.
Other bunyaviruses have been shown to inhibit the induction of cellular interferon responses. The Bunyamwera virus encodes a nonstructural protein, NSs, which blocks the induction of IFN-β (4), and an NSs-deficient Bunyamwera virus mutant induces IFN-β and replicates poorly in IFN-competent cell lines (61). However, hantaviruses do not encode known NSs proteins, suggesting that hantaviruses employ a unique mechanism of suppressing IFN activation. Our findings show that the G1 cytoplasmic tail of the pathogenic NY-1V, but not PHV, suppresses ISRE and IFN-β promoter activation in response to RIG-I- or TBK-1-directed IFN pathway activation. These results indicate that the NY-1V G1 cytoplasmic tail inhibits IFN induction and functions in the regulation of IFN responses in the absence of NSs proteins.
TBK-1/IKKɛ activates cytoplasmic forms of IRF-3 and NF-κB, and activation permits their nuclear translocation and transcriptional responses. Activation of the IFN-β promoter is directed by an IRF-3/NF-κB/CBP-p300 transcription complex, while ISRE promoters are activated by cellular IRFs but do not require NF-κB activation. Our findings indicate that the G1 cytoplasmic tail inhibits both ISRE- and IFN-β-directed transcriptional responses, consistent with G1 inhibiting TBK-1/IRF-3 but not TBK-1/NF-κB, responses that are required for the activation of both promoters. However, the NY-1V G1 tail had no effect on transcription directed by a constitutively active phosphomimetic IRF-3, suggesting that G1 acts upstream of IRF-3 phosphorylation at the level of the TBK-1 complex.
Previous studies have demonstrated that the NY-1V G1 cytoplasmic tail binds cellular Src and Syk family kinases and is ubiquitinated and degraded (17). Although it is unclear which proteins within the TBK-1 complex are bound by G1 or whether G1 directs the degradation of specific TBK-1 complex regulatory proteins, these possibilities provide plausible mechanisms for the NY-1V G1 tail to regulate TBK-1 complex-directed IFN signaling responses.
In order to successfully replicate, hantaviruses must evade host cell defenses, and here, we demonstrate that the NY-1V G1 cytoplasmic tail blocks RIG-I- and TBK-1-directed IFN transcription upstream of IRF-3 phosphorylation. We further demonstrate that the G1 protein of the nonpathogenic hantavirus PHV fails to regulate TBK-1-directed IFN responses, and these findings correlate with failed PHV replication within human endothelial cells. These findings establish fundamental differences between pathogenic and nonpathogenic hantaviruses and suggest that at one level, the G1 tail is a virulence element which determines the success of a hantavirus infection within human endothelial cells. Our results also point to a second level of hantavirus regulation of innate cellular responses and suggest that hantavirus-derived products prevent the inhibitory function of IFN-directed ISGs that otherwise limit viral replication at late times postinfection. The combination of the early regulation of IFN induction and the late regulation of IFN-directed effectors provides a mechanism for hantaviruses to bypass innate cellular responses and permits hantaviruses to effect subsequent pathogenic programs.
Acknowledgments
We thank Adrish Sen for helpful discussions and critical reading of the manuscript, Matt Weintraub for technical assistance, and Timothy Pepini for helpful discussions. We are very grateful for plasmids provided by Joel Pomerantz, David Baltimore, and Michael Gale for these experiments.
This work was supported by National Institutes of Health grants R01AI47873 and PO1AI055621 and by a Veterans Affairs Merit Award.
REFERENCES
- 1.Aebi, M., J. Fah, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, J., K. Zhu, and G. Zhou. 1997. [The therapeutic effect of purified human leucocytic interferon-alpha on hemorrhagic fever with renal syndrome]. Zhonghua Nei Ke Za Zhi 36:90-93. (In Chinese.) [PubMed] [Google Scholar]
- 3.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 4.Bridgen, A., F. Weber, J. K. Fazakerley, and R. M. Elliott. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. USA 98:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conzelmann, K.-K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosgriff, T. M. 1991. Mechanisms of disease in Hantavirus infection: pathophysiology of hemorrhagic fever with renal syndrome. Rev. Infect. Dis. 13:97-107. [DOI] [PubMed] [Google Scholar]
- 7.Cosgriff, T. M., and R. M. Lewis. 1991. Mechanisms of disease in hemorrhagic fever with renal syndrome. Kidney Int. Suppl. 35:S72-S9. [PubMed] [Google Scholar]
- 8.Daly, C., and N. C. Reich. 1995. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J. Biol. Chem. 270:23739-23746. [DOI] [PubMed] [Google Scholar]
- 9.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 10.Duchin, J. S., F. T. Koster, C. J. Peters, G. L. Simpson, B. Tempest, S. R. Zaki, T. G. Ksiazek, P. E. Rollin, S. Nichol, E. T. Umland, R. L. Moolenaar, S. E. Reef, K. B. Nolte, M. M. Gallaher, J. C. Butler, and R. F. Breiman. 1994. Hantavirus pulmonary syndrome: a clinical description of 17 patients with a newly recognized disease. The Hantavirus Study Group. N. Engl. J. Med. 330:949-955. [DOI] [PubMed] [Google Scholar]
- 11.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 13.Frese, M., G. Kochs, H. Feldmann, C. Hertkorn, and O. Haller. 1996. Inhibition of bunyaviruses, phleboviruses, and hantaviruses by human MxA protein. J. Virol. 70:915-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilovskaya, I., R. LaMonica, M. E. Fay, B. Hjelle, C. Schmaljohn, R. Shaw, and E. R. Mackow. 1999. New York 1 and Sin Nombre viruses are serotypically distinct viruses associated with hantavirus pulmonary syndrome. J. Clin. Microbiol. 37:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavrilovskaya, I. N., E. J. Brown, M. H. Ginsberg, and E. R. Mackow. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by β3 integrins. J. Virol. 73:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. Beta3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geimonen, E., I. Fernandez, I. N. Gavrilovskaya, and E. R. Mackow. 2003. Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail. J. Virol. 77:10760-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geimonen, E., R. LaMonica, K. Springer, Y. Farooqui, I. N. Gavrilovskaya, and E. R. Mackow. 2003. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. J. Virol. 77:1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geimonen, E., S. Neff, T. Raymond, S. S. Kocer, I. N. Gavrilovskaya, and E. R. Mackow. 2002. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. USA 99:13837-13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 21.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. Off. Int. Epizoot. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 22.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26:396-401. [DOI] [PubMed] [Google Scholar]
- 23.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010:237-248. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, J. M., C. J. Peters, M. L. Cohen, and B. W. Mahy. 1993. Hantavirus pulmonary syndrome: an emerging infectious disease. Science 262:850-851. [DOI] [PubMed] [Google Scholar]
- 25.Kamrud, K. I., and C. S. Schmaljohn. 1994. Expression strategy of the M genome segment of Hantaan virus. Virus Res. 31:109-121. [DOI] [PubMed] [Google Scholar]
- 26.Kanerva, M., K. Melen, A. Vaheri, and I. Julkunen. 1996. Inhibition of Puumala and Tula hantaviruses in Vero cells by MxA protein. Virology 224:55-62. [DOI] [PubMed] [Google Scholar]
- 27.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 28.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 29.Kraus, A. A., M. J. Raftery, T. Giese, R. Ulrich, R. Zawatzky, S. Hippenstiel, N. Suttorp, D. H. Kruger, and G. Schonrich. 2004. Differential antiviral response of endothelial cells after infection with pathogenic and nonpathogenic hantaviruses. J. Virol. 78:6143-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, H. W. 1982. Hemorrhagic fever with renal syndrome (HFRS). Scand. J. Infect. Dis. Suppl. 36:82-85. [PubMed] [Google Scholar]
- 31.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19:959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medhurst, A. D., D. C. Harrison, S. J. Read, C. A. Campbell, M. J. Robbins, and M. N. Pangalos. 2000. The use of TaqMan RT-PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J. Neurosci. Methods 98:9-20. [DOI] [PubMed] [Google Scholar]
- 34.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 35.Nolte, K. B., R. M. Feddersen, K. Foucar, S. R. Zaki, F. T. Koster, D. Madar, T. L. Merlin, P. J. McFeeley, E. T. Umland, and R. E. Zumwalt. 1995. Hantavirus pulmonary syndrome in the United States: a pathological description of a disease caused by a new agent. Hum. Pathol. 26:110-120. [DOI] [PubMed] [Google Scholar]
- 36.Pensiero, M. N., and J. Hay. 1992. The Hantaan virus M-segment glycoproteins G1 and G2 can be expressed independently. J. Virol. 66:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pensiero, M. N., G. B. Jennings, C. S. Schmaljohn, and J. Hay. 1988. Expression of the Hantaan virus M genome segment by using a vaccinia virus recombinant. J. Virol. 62:696-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pensiero, M. N., J. B. Sharefkin, C. W. Dieffenbach, and J. Hay. 1992. Hantaan virus infection of human endothelial cells. J. Virol. 66:5929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plyusnin, A., O. Vapalahti, and A. Vaheri. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677-2687. [DOI] [PubMed] [Google Scholar]
- 40.Pomerantz, J. L., and D. Baltimore. 1999. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18:6694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prescott, J., C. Ye, G. Sen, and B. Hjelle. 2005. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J. Virol. 79:15007-15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raftery, M. J., A. A. Kraus, R. Ulrich, D. H. Kruger, and G. Schonrich. 2002. Hantavirus infection of dendritic cells. J. Virol. 76:10724-10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raymond, T., E. Gorbunova, I. N. Gavrilovskaya, and E. R. Mackow. 2005. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc. Natl. Acad. Sci. USA 102:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reich, N. C. 2002. Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J. Interferon Cytokine Res. 22:103-109. [DOI] [PubMed] [Google Scholar]
- 45.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304-4310. [DOI] [PubMed] [Google Scholar]
- 47.Schmaljohn, C. 1996. Bunyaviridae and their replication, p. 1447-1471. In B. N. Fields et al. (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven Press, Philadelphia, Pa. [Google Scholar]
- 48.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmaljohn, C. S., G. B. Jennings, J. Hay, and J. M. Dalrymple. 1986. Coding strategy of the S genome segment of Hantaan virus. Virology 155:633-643. [DOI] [PubMed] [Google Scholar]
- 50.Schmaljohn, C. S., A. L. Schmaljohn, and J. M. Dalrymple. 1987. Hantaan virus M RNA: coding strategy, nucleotide sequence, and gene order. Virology 157:31-39. [DOI] [PubMed] [Google Scholar]
- 51.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 52.Song, J. W., L. J. Baek, I. N. Gavrilovskaya, E. R. Mackow, B. Hjelle, and R. Yanigahara. 1996. Sequence analysis of the complete S genomic segment of a newly identified hantavirus isolated from the white-footed mouse (Peromyscus leucopus): phylogenetic relationship with other sigmadontine rodent borne hantaviruses. Virus Genes 12:249-256. [DOI] [PubMed] [Google Scholar]
- 53.Spiropoulou, C. F., S. Morzunov, H. Feldmann, A. Sanchez, C. J. Peters, and S. T. Nichol. 1994. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology 200:715-723. [DOI] [PubMed] [Google Scholar]
- 54.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 55.Sumpter, R., Jr., Y.-M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sundstrom, J. B., L. K. McMullan, C. F. Spiropoulou, W. C. Hooper, A. A. Ansari, C. J. Peters, and P. E. Rollin. 2001. Hantavirus infection induces the expression of RANTES and IP-10 without causing increased permeability in human lung microvascular endothelial cells. J. Virol. 75:6070-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vapalahti, O., A. Lundkvist, S. K. Kukkonen, Y. Cheng, M. Gilljam, M. Kanerva, T. Manni, M. Pejcoch, J. Niemimaa, A. Kaikusalo, H. Henttonen, A. Vaheri, and A. Plyusnin. 1996. Isolation and characterization of Tula virus, a distinct serotype in the genus Hantavirus, family Bunyaviridae. J. Gen. Virol. 77:3063-3067. [DOI] [PubMed] [Google Scholar]
- 58.Vapalahti, O., A. Lundkvist, and A. Vaheri. 2001. Human immune response, host genetics, and severity of disease. Curr. Top. Microbiol. Immunol. 256:153-169. [DOI] [PubMed] [Google Scholar]
- 59.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 60.Weaver, B. K., O. Ando, K. P. Kumar, and N. C. Reich. 2001. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. 15:501-515. [DOI] [PubMed] [Google Scholar]
- 61.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 63.Yanagihara, R., C. A. Daum, P. W. Lee, L. J. Baek, H. L. Amyx, D. C. Gajdusek, and C. J. Gibbs. 1987. Serological survey of Prospect Hill virus infection in indigenous wild rodents in the USA. Trans. R. Soc. Trop. Med. Hyg. 81:42-45. [DOI] [PubMed] [Google Scholar]
- 64.Yanagihara, R., and D. J. Silverman. 1990. Experimental infection of human vascular endothelial cells by pathogenic and nonpathogenic hantaviruses. Arch. Virol. 111:281-286. [DOI] [PubMed] [Google Scholar]
- 65.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 66.Zaki, S. R., P. W. Greer, L. M. Coffield, C. S. Goldsmith, K. B. Nolte, K. Foucar, R. M. Feddersen, R. E. Zumwalt, G. L. Miller, A. S. Khan, et al. 1995. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am. J. Pathol. 146:552-579. [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, P. 1989. Infection of human endothelial cells by the virus causing hemorrhagic fever with renal syndrome. Zhonghua Yi Xue Za Zhi 69:588-590. (In Chinese.) [PubMed] [Google Scholar]