Abstract
Conciliating biosafety with efficient gene transfer remains a constant concern in the development of retroviral vectors. Semliki Forest virus (SFV) replicons allow important retroviral vector production with interesting features. It is noteworthy that retroviruses have the ability to package Ψ+ and, to some extent, Ψ− cellular RNAs. Therefore, it was important to study the retroviral transfer of highly abundant SFV genomes expressing retroviral proteins. Here, we show that full-length SFV-vector replicons, with or without Ψ, are efficiently packaged into retrovirus particles. Mechanistically, our data suggest that SFV packaging is the sum of its retroviral nucleocapsid-dependent recruitment together with a passive hijacking of membrane-anchored SFV replicon. A direct consequence of this phenomenon is the formation of particles harboring autonomous replicative abilities and contaminating vector preparations. Importantly, we confirm that retroviral SFV mobilization is not an exclusive feature of murine gamma retroviruses, since it is also observed using lentivectors.
It is well established that retroviral vectors derived from simple oncoretroviruses, murine leukemia virus (MLV), or lentiviruses, such as human immunodeficiency virus type 1 (HIV-1), are effective and versatile gene delivery systems (31). Recombinant particle production involves the expression of two components, either continuously in stable packaging cell lines or transiently by cotransfection of naïve cells (31). Vector production by stable cell lines can be maximized using bioreactors or various chemical boosts (4, 14, 23, 26, 32). Most transient production protocols rely on DNA transfection, with the rest involving delivery of retroviral components using heterologous viral vectors (20, 31, 38, 40, 43, 44). Among these is the Semliki Forest virus (SFV) vector system (20, 43). It was initially developed for robust protein production in mammalian cells (1). SFV-positive RNA is capable of autoamplification through self-primed replication (42). In addition, the SFV genome contains an internal promoter, which prompts a further amplification via transcription of a subgenomic RNA (42). Within vectors, the subgenomic RNA encodes the transgene (25). Li and Garoff and others have developed an SFV system allowing retroviral vector production (20, 43). Interestingly, Li and collaborators have shown that the Semliki-derived system, which displays an exclusive cytoplasmic replication, allows the transfer of intron-containing retroviral vectors (19).
An important consideration when using retroviral vectors as therapeutic tools is biosafety (31). Treatment of severe combined immunodeficiency (SCID) patients has emphasized the risk of insertional mutagenesis with retroviral vector based on MLV (12). As a consequence, targeted integration is now a major goal (2). Replication-competent retrovirus (RCR) in vector preparations is also to be considered. In animal models, RCRs have been shown to trigger cancer (3, 13, 24). Using classical retroviral production systems, the emergence of replication-competent virus comes essentially from recombination events (10, 35). Therefore, regulatory agencies demand a thorough evaluation of RCR contaminants throughout the process of vector production. Accordingly, the biosafety of retroviral vector produced using the SFV system must be exhaustively evaluated. Muriaux and collaborators have shown that the retroviral packaging of subgenomic Semliki genome is possible (28). Moreover, one of the recombinant SFVs involved in retroviral vector production contains a complete retroviral packaging signal. Thus, the packaging of this full-length replicon should be measured. Furthermore, Rolls et al. and Lebedeva et al. have shown that envelope-encoding SFV vectors can autonomously replicate and spread (17, 37). It was therefore important to evaluate the presence of all types of SFV contaminants in the supernatant of retrovirus-producing cells. Here, we show that full-length SFV vectors are efficiently packaged into retrovirus particles. Importantly, we demonstrate that this phenomenon is packaging sequence independent. Moreover, using the SFV-based system for retroviral production, we show that transduced cells acquire the ability to produce new retrovirus particles. Lastly, we also establish that lentiviruses are fully competent for SFV vector packaging.
SFV mobilization during SFV-driven retrovirus production.
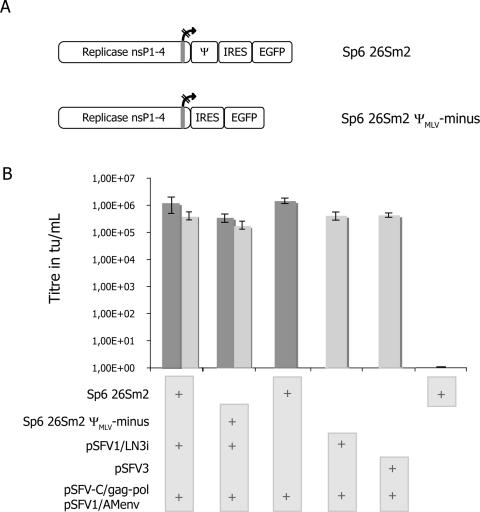
In this study, our aim was to evaluate the biosafety of an SFV-based retrovirus system described by Li and Garoff (20). Since Muriaux and collaborators have shown the presence of SFV genomic RNA within retrovirus particles (27, 28), we first tested the efficiency of SFV retroviral packaging. For this, we added to the standard mixture an in vitro-transcribed Sp6 26Sm2 RNA expressing green fluorescent protein (GFP) as a marker. This SFV replicon contained a retroviral packaging signal and a nonfunctional internal 26S promoter, leading to the exclusive production of full-length SFV genomic RNAs (6) (Fig. 1A). The transfer of 26Sm2 during retrovirus production was measured by counting GFP-expressing cells (Fig. 1B). Parallel samples allowed the enumeration of LacZ-expressing cells (Fig. 1B). Surprisingly, we noticed that the GFP and the LacZ titers were comparable (Fig. 1B). As the pSFV1/LN3i LacZ contained a retroviral packaging sequence, this result therefore suggested that the LacZ titer was a mix of retroviral vector and SFV β-galactosidase mobilization events.
FIG. 1.
(A) Schematic representation of the two full-length GFP vectors. For details, see the text and reference 6. IRES, internal ribosome entry site. (B) Transmobilization of full-length SFV replicon by retrovirus particles. BHK-21 cells were cotransfected by a mix of RNAs. Supernatants from transfected cells were harvested to transduce 293T cells. Titers are expressed in transducing units per milliliter. Each column in the graph is associated with a specific mix of plasmids identified by plus symbols in light gray boxes. Within the graph, titers in dark gray represent GFP mobilization, while light gray columns reflect LacZ titers. GFP titers obtained with the 26Sm2 construct, in the presence or absence of a competitive LacZ SFV vector, were in the same order of magnitude. 26Sm2 ΨMLV-minus vector was almost as efficiently packaged as 26Sm2, its Ψ-positive counterpart. pSFV3, a LacZ-expressing, ΨMLV-minus SFV vector harboring a functional 26S internal promoter, was also efficiently mobilized. Cell debris-releasing RNA or GFP do not support transfer, as proved by the absence of GFP-positive cells when using 26Sm2 alone. Altogether, these data indicated that retrovirus particles are a vehicle for SFV vectors. Titers are means of results from at least five independent experiments.
To examine the role of the MLV packaging sequence for SFV mobilization, we next used an Sp6 26Sm2 construct in which we had deleted the packaging sequence (26Sm2 ΨMLV-minus) (Fig. 1A). Compared to the Ψ-containing vector, the 26Sm2 ΨMLV-minus vector was only slightly less efficiently mobilized (Fig. 1B). This indicated that the Ψ sequence did not play a significant role in the retroviral SFV vector mobilization.
In the previous assays, there was packaging competition between the two reporter constructs SFV-GFP (26Sm2) and SFV-LacZ (pSFV1/LN3i LacZ). It was also impossible to distinguish LacZ expression resulting from the integrated retroviral vector from that resulting from the mobilized full-length SFV replicon. To more accurately evaluate the retroviral packaging of ΨMLV-minus SFV vector, we used a standard replicon expressing β-galactosidase, pSFV3. Using pSFV3, we obtained LacZ titers similar to those generated in the previous assays or to those obtained with 26Sm2 and pSFV1/LN3i LacZ alone (Fig. 1B). This confirmed the efficiency of retroviral SFV vector packaging and the fact that a retroviral packaging signal was unnecessary for SFV vector retroviral mobilization. Of note is that within producing cells, the subgenomic RNA is susceptible to competition with the full-length RNA for retroviral packaging (29). While we cannot exclude a moderate participation of subgenomic RNA expression, as is the case for retroviral RNAs (9), only full-length SFV could drive the high transgene expression we observed upon transduction.
SFV vectors are released from primary target cells via secondary mobilization.
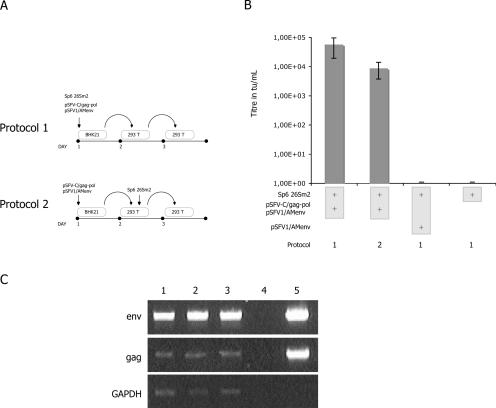
The above data suggested that any SFV replicon had the ability to be packaged into a retrovirus particle. Thus, in the triple transfection system, pSFV-C/gag-pol or pSFV1/AMenv SFV vectors may also be packaged. To address this question, we designed a series of secondary transduction experiments (Fig. 2A). The simplest assay was designed to confirm that transduced cells were able to produce recombinant SFV-containing retrovirus particles. We collected supernatant from transduced 293T cells and used it to transduce naïve 293T cells (Fig. 2A, protocol 1). We detected GFP-positive cells after the second transduction (0.5 × 104 transducing units [tu]/ml [Fig. 2B]). However, we could not rule out the possibility that initial particles adsorbed on the primarily transduced cells did not contribute to the secondary titer. To further confirm mobilization of pSFV-C/gag-pol and pSFV1/AMenv, we developed a second assay (Fig. 2A, protocol 2). Only RNAs from pSFV-C/gag-pol and pSFV1/AMenv were electroporated into BHK-21 cells. Fifteen hours after transfection, supernatant was harvested and delivered to target cells. Five hours following transduction, the cells were electroporated with 26Sm2 RNAs. Fifteen hours after that, supernatant was collected to transduce naïve 293T cells. GFP expression was detectable 24 h after this second transduction (104 tu/ml [Fig. 2B]). None of the cells transduced using supernatants from cells solely electroporated by Sp6 26Sm2 RNAs, alone or in combination with pSFV1/AMenv, were GFP positive (Fig. 2B). The amphotropic envelope needed a specific proteolytic cleavage to promote receptor-mediated cell fusion (17, 36). We cannot exclude the possibility that spontaneous mutations of the envelope intracytoplasmic domain could generate autonomous particles. However, this is a rare event (33) which could not explain the titers we observed (Fig. 2B). Moreover, using RNAs from secondary transduced cells in a reverse transcriptase PCR (RT-PCR) assay, we confirmed the mobilization of pSFV-C/gag-pol and pSFV1/AMenv (Fig. 2C).
FIG. 2.
(A) Schematic representation of the transduction protocols. Two protocols were designed to challenge secondary mobilization. Protocol 1 was based on the harvesting of supernatant from transduced cells. After filtration, this supernatant served for the transduction of secondary cells. Constructs are indicated over the vertical upper arrow. In protocol 2, BHK-21 cells were transfected using pSFV-C/gag-pol and pSFV1/AMenv. One day later, supernatant was collected to transduce 293T cells. Five hours later, these cells were electroporated with 26Sm2 RNA. At day 3, the supernatant was harvested to transduce new 293T cells. (B) Efficiency of the secondary mobilization. Each column in the graph is associated with a specific mix of plasmids identified by plus symbols in light gray boxes. For protocol 1, we observed titers only 1 log lower compared to those of a simple transduction (B). For protocol 2, we still observed a GFP titer, confirming the secondary mobilization, as a result of the transfer of pSFV-C/gag-pol and pSFV1/AMenv replicons into target cells. A control panel was obtained using protocol 1: we observed no cell transduction with supernatant from 293T cells transfected with 26Sm2 alone or together with pSFV1/AMenv. This indicates that the SFV transfer needs retroviral functions, such as Env cytoplasmic cleavage. Results are means from three independent experiments. (C) RT-PCR analysis on total cellular RNA from secondary transduction. Primer localizations on vectors were chosen to detect the transcomplementing transgenes (Env and Gag). To confirm mobilization of transcomplementing sequences, we performed an RT-PCR on total RNAs extracted from secondary transduced cells (protocol 1). Lanes 1 to 3, RNAs from secondary cells from three independent experiments; lane 4, H2O-negative control; lane 5, amplification on a mix of 10 ng/ml of pSFV1/AMenv or pSFV-C/gag-pol plasmids. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Molecular determinants for SFV retroviral mobilization.
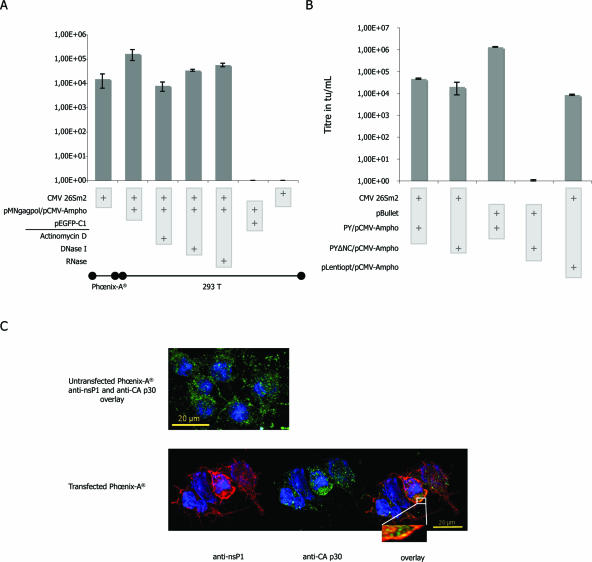
We next confirmed SFV mobilization by classical retroviral production systems. When supernatant from Phœnix-A cells, transfected with cytomegalovirus (CMV) 26Sm2 (6), was added to naïve 293T cells, we observed a transduction (Fig. 3A). Interestingly, titers could be improved by transiently expressing the MLV packaging system (Fig. 3A).
FIG. 3.
(A and B) Titers, means of results from five independent experiments, are expressed in transducing units per milliliter. Each column in the graphs is associated with a specific mix of plasmids identified by plus symbols in light gray boxes. For these experiments, we used the CMV-driven 26Sm2 SFV-derived vector. (A) Titers obtained using an RNA Pol II system for retroviral transcomplementation. Phœnix-A packaging cells were transfected using CMV 26Sm2. Tritransfection experiments were performed as described in the text. A series of controls was performed to eliminate passive transfer of plasmid or RNA: Pol II-driven transcription in transduced cells was inhibited through addition of actinomycin D; GFP detection titer at 104 tu/ml confirmed SFV-driven expression; supernatants were also treated with DNase I (125 IU) and RNase A (10 μg) before transduction with no effect on titers (columns 4 and 5, respectively); finally, the absence of titer obtained with pEGFP-C1 or 26Sm2 alone confirmed SFV retroviral mobilization. (B) Exploring the molecular requirement for SFV retroviral mobilization. The 26Sm2 replicon was mobilized using a retroviral transcomplementation system harboring a wild-type NC or a deleted NC (ΔNC). As opposed to what is observed using a classical GFP-expressing retroviral vector, pBullet (columns 3 and 4 of the graph), deletion of the NC had no effect on SFV mobilization (first two columns). In the last column, the lentiviral mobilization was tested with a Rev-independent Gag-PolHIV expression system (pLentiopt). The Gag-PolHIV titers were comparable to those obtained with the MLV system. Results are means from five independent experiments. (C) Cellular localization of MLV Gag and nsP1. Phœnix-A cells were transfected with the nsP1-expressing CMV 26Sm2 plasmid. Specific antibodies for MLV CA p30 and nsP1 were used to determine the cellular localization of the two proteins. Pictures are superpositions of six 0.25-μm-wide slices. (Upper panel) Overlay for control Phœnix-A cells not transfected but incubated with the two antibodies: only the green CA p30 signal is observed. (Lower panel) Pictures of transfected Phœnix-A cells: detection of nsP1 (in red), CA p30 (in green). The presence of several yellow dots on the overlay suggested the vicinity of the two proteins.
As the SFV system can give rise to huge amounts of transgene proteins, we considered pseudotransduction, the transfer of GFP rather than the transfer of the GFP-encoding SFV replicon (22). Substitution of CMV 26Sm2 by pEGFP-C1, which gave equivalent GFP signal in producing cells, did not allow GFP detection within target cells (Fig. 3A). To further investigate pseudotransduction, target 293T cells were incubated with the translation inhibitor puromycin before transduction (41). Under these conditions, GFP was not detected in cells exposed to supernatant from tritransfected cells (data not shown). To eliminate expression from passive transfer of GFP-expressing plasmid DNA, target cells were preincubated with actinomycin D, an RNA Pol II transcription inhibitor with no effect on SFV-driven replication (15). GFP expression was only slightly affected by actinomycin D (Fig. 3A).
The above results invited a more precise evaluation of the need for an interaction between the retroviral Gag protein and the SFV RNA to promote packaging. We first used a Gag retroviral transcomplementation vector bearing a nucleocapsid deletion (ΔNC). NC is considered to be the master of retroviral RNA recruitment (7, 8, 27). As expected, the ΔNC construct was unable to package retroviral vector, leading to undetectable GFP expression (Fig. 3B). Conversely, the use of the ΔNC construct had little effect on the retroviral mobilization of the 26Sm2 replicon (Fig. 3B). This confirmed that the replicon was mostly recruited by a Gag-independent process.
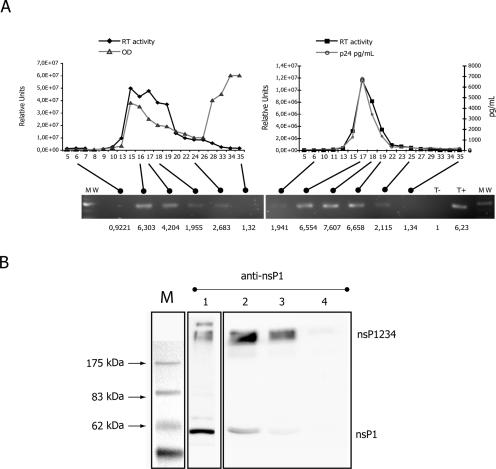
It is noteworthy that nsP1, Gag, and the retroviral envelope are acylated (16, 21, 30). By sharing such membrane-targeting domains, the proteins could have a common localization within the cell membrane. Confocal examination of CMV 26Sm2 expressing Phœnix-A cells revealed several domains where Gag and nsP1 proteins colocalized (Fig. 3C). To confirm that this could result in the packaging of an SFV replicon, we purified chimerical vectors using a sucrose gradient. RT-PCR analysis revealed the presence of 26Sm2 RNA in the retroviral RT-positive fractions (Fig. 4A). We also detected nsP precursor by Western blotting on particles purified on a sucrose cushion. As shown in Fig. 4B, we could detect the cleaved nsP1 and the uncleaved nsP precursor within the retrovirus particles. Of note, cells simply transfected by 26Sm2 and pSFV1/AMenv RNAs also led to particles containing the nsP precursor (Fig. 4B, lane 1). This result was in accordance with the observation made by Lebedeva and collaborators when using SFV replicon expressing a retroviral envelope (17).
FIG. 4.
(A) Purification of chimerical particles. Particles were purified using a sucrose gradient. Supernatant from 293T cells transfected with pMN gag-pol or pLentiopt, pCMV-Ampho, and CMV 26Sm2 was deposited on a continuous sucrose gradient, 20% to 60%, in 35-ml tubes. Tubes were centrifuged at 100,000 × g for 2 h using an SW 28 swinging rotor. Fractions were collected using a Pharmacia collector equipped with a 260-nm UV detector for protein detection (optical density [OD]). Each fraction was checked for RT activity using rA/dT oligonucleotides in the presence of αdATP32. Black lozenges and squares represent RT activity for MLV and lentivirus vectors, respectively. For HIV-based vectors, we also measured the CA p24 concentration in each fraction (gray dots on the right panel). In the indicated fractions, we detected the presence of SFV genome by RT-PCR. Primer localization on vectors was chosen to detect full-length RNA by targeting the nsP1. MW, molecular weight marker. The lower panel gives a densitometric measurement of PCR fragment signal in arbitrary units. (B) Western blot detection of nsP1 in vectors. Vector proteins were extracted after ultracentrifugation on a sucrose cushion of supernatant harvested from producing tritransfected BHK-21 cells. Proteins were separated on sodium dodecyl sulfate-polyacrylamide gels. nsP1-specific antibody recognized the two forms of the protein, the uncleaved precursor and the free cleaved protein. Lane 1, supernatant from pSFV1/AMenv-transfected BHK-21 cells; lane 2, supernatant from pSFV1/AMenv- and pSFV-C/gag-pol-transfected BHK-21 cells; lane 3, supernatant from pSFV1/AMenv-, pSFV-C/gag-pol-, and 26Sm2-transfected BHK-21 cells; lane 4, negative control, supernatant from 293T cells transfected with pMN gag-pol, pCMV-Ampho, and pBullet. Only vectors produced using the SFV system contained both the uncleaved and cleaved forms of nsP1, as shown by the detection in lanes 1 to 3 of 250-kDa and 61-kDa bands, respectively.
From a clinical perspective, the interest in retroviral vectors has recently shifted from MLV vectors to lentivectors. Furthermore, former data suggested a main contribution of nonspecific mechanisms in SFV retroviral mobilization. It was therefore important to check the ability of lentivectors to mobilize an SFV replicon. For that purpose, we performed a tritransfection assay using an optimized Rev-independent Gag-PolHIV expression system (pLentiopt), pCMV-Ampho, and CMV 26Sm2. Supernatants from transfected 293T cells were collected to transduce naïve 293T cells. Remarkably, we obtained GFP-expressing cells with a titer equivalent to those obtained with MLV vectors (Fig. 3A and B). Sucrose gradient purification of chimerical particles indicated the presence of 26Sm2 RNA in the p24- and RT-positive fractions (Fig. 4A). This confirmed that lentivectors were also fully competent to transfer HIV Ψ-minus SFV vectors.
In conclusion, our data establish that SFV-based systems must be prohibited for the production of clinical-grade high-titer retroviral vectors for MLV as well as for HIV. One unanticipated aspect of the present study was that retroviral vectors could be an attractive vehicle for SFV vectors. An important restriction to that is the need for an approach leading to efficiency and safety at the same time. Using constitutive packaging cells, such as Phœnix-A, we have a safe but inefficient system (Fig. 3A). Conversely, the Semliki system offers high titers, albeit with equivalent amounts of Gag-PolMLV and Env contaminating SFV replicons (Fig. 2). We therefore have to envision alternative solutions or modifications of the different partners. Noncytotoxic SFV replicon might be efficient for packaging in Phœnix-A cells (25, 33). However, all the constructs we have been able to test to date were not satisfactory to provide efficient replication and thus delivery (data not shown). Moreover, one main interest of SFV vectors comes from the transient aspect of their huge expression. This is useful in eliminating a target cell or in triggering an immune response (11, 18). The use of noncytotoxic SFV vectors would lead to long-lasting but weak expression (39). Alternatively, cells that do not support SFV replication exist, and provided that they allow a high retroviral budding, the use of a Pol II-dependent expression system for a ΨMLV SFV vector (5, 18, 34) should promote the efficient formation of retrovirus particles containing the full-length vector.
Acknowledgments
We thank the AFM for its financial support.
We thank H. Garoff and A. Merits for SFV reagents and J. L. Darlix for the gift of NC variants and CA p30 antibodies. Greg Towers remains a patient and helpful reader.
REFERENCES
- 1.Berglund, P., M. Sjoberg, H. Garoff, G. J. Atkins, B. J. Sheahan, and P. Liljestrom. 1993. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Biotechnology (NY) 11:916-920. [DOI] [PubMed] [Google Scholar]
- 2.Bushman, F. 2002. Targeting retroviral integration? Mol. Ther. 6:570-571. [PubMed] [Google Scholar]
- 3.Cornetta, K., R. A. Morgan, and W. F. Anderson. 1991. Safety issues related to retroviral mediated gene transfer in humans. Hum. Gene Ther. 2:5-14. [DOI] [PubMed] [Google Scholar]
- 4.Cosset, F.-L., Y. Takeuchi, J. L. Battini, R. A. Weiss, and M. K. L. Collins. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diatta, A., E. Piver, C. Collin, P. Vaudin, and J. C. Pages. 2005. Semliki Forest virus-derived virus-like particles: characterization of their production and transduction pathways. J. Gen. Virol. 86:3129-3136. [DOI] [PubMed] [Google Scholar]
- 6.Dorange, F., E. Piver, T. Bru, C. Collin, P. Roingeard, and J. C. Pages. 2004. Vesicular stomatitis virus glycoprotein: a transducing coat for SFV-based RNA vectors. J. Gene Med. 6:1014-1022. [DOI] [PubMed] [Google Scholar]
- 7.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643-655. [DOI] [PubMed] [Google Scholar]
- 8.D'Souza, V., and M. F. Summers. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586-590. [DOI] [PubMed] [Google Scholar]
- 9.Galla, M., E. Will, J. Kraunus, L. Chen, and C. Baum. 2004. Retroviral pseudotransduction for targeted cell manipulation. Mol. Cell 16:309-315. [DOI] [PubMed] [Google Scholar]
- 10.Garrett, E., A. R. Miller, J. M. Goldman, J. F. Apperley, and J. V. Melo. 2000. Characterization of recombination events leading to the production of an ecotropic replication-competent retrovirus in a GP+envAM12-derived producer cell line. Virology 266:170-179. [DOI] [PubMed] [Google Scholar]
- 11.Guan, M., J. R. Rodriguez-Madoz, P. Alzuguren, C. Gomar, M. G. Kramer, S. Kochanek, J. Prieto, C. Smerdou, and C. Qian. 2006. Increased efficacy and safety in the treatment of experimental liver cancer with a novel adenovirus-alphavirus hybrid vector. Cancer Res. 66:1620-1629. [DOI] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basileqq, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302:415-419. [DOI] [PubMed] [Google Scholar]
- 13.Kim, R., A. Trubetskoy, T. Suzuki, N. A. Jenkins, N. G. Copeland, and J. Lenz. 2003. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J. Virol. 77:2056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotani, H., P. B. Newton III, S. Zhang, Y. L. Chiang, E. Otto, L. Weaver, R. M. Blaese, W. F. Anderson, and G. J. McGarrity. 1994. Improved methods of retroviral vector transduction and production for gene therapy. Hum. Gene Ther. 5:19-28. [DOI] [PubMed] [Google Scholar]
- 15.Kujala, P., A. Ikaheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kaariainen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakkonen, P., T. Ahola, and L. Kaariainen. 1996. The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J. Biol. Chem. 271:28567-28571. [DOI] [PubMed] [Google Scholar]
- 17.Lebedeva, I., K. Fujita, A. Nihrane, and J. Silver. 1997. Infectious particles derived from Semliki Forest virus vectors encoding murine leukemia virus envelopes. J. Virol. 71:7061-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leitner, W. W., L. N. Hwang, M. J. DeVeer, A. Zhou, R. H. Silverman, B. R. Williams, T. W. Dubensky, H. Ying, and N. P. Restifo. 2003. Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways. Nat. Med. 9:33-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, K. J., and H. Garoff. 1998. Packaging of intron-containing genes into retrovirus vectors by alphavirus vectors. Proc. Natl. Acad. Sci. USA 95:3650-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, K. J., and H. Garoff. 1996. Production of infectious recombinant Moloney murine leukemia virus particles in BHK cells using Semliki Forest virus-derived RNA expression vectors. Proc. Natl. Acad. Sci. USA 93:11658-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M., C. Yang, S. Tong, A. Weidmann, and R. W. Compans. 2002. Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J. Virol. 76:11845-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, M. L., B. L. Winther, and M. A. Kay. 1996. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)-Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J. Virol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loew, R., N. Selevsek, B. Fehse, D. von Laer, C. Baum, A. Fauser, and K. Kuehlcke. 2004. Simplified generation of high-titer retrovirus producer cells for clinically relevant retroviral vectors by reversible inclusion of a lox-P-flanked marker gene. Mol. Ther. 9:738-746. [DOI] [PubMed] [Google Scholar]
- 24.Lund, A. H., G. Turner, A. Trubetskoy, E. Verhoeven, E. Wientjens, D. Hulsman, R. Russell, R. A. DePinho, J. Lenz, and M. van Lohuizen. 2002. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 32:160-165. [DOI] [PubMed] [Google Scholar]
- 25.Lundstrom, K. 2003. Semliki Forest virus vectors for gene therapy. Expert Opin. Biol. Ther. 3:771-777. [DOI] [PubMed] [Google Scholar]
- 26.Merten, O. W., P. E. Cruz, C. Rochette, C. Geny-Fiamma, C. Bouquet, D. Goncalves, O. Danos, and M. J. Carrondo. 2001. Comparison of different bioreactor systems for the production of high titer retroviral vectors. Biotechnol. Prog. 17:326-335. [DOI] [PubMed] [Google Scholar]
- 27.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 78:12378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76:11405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen, K. E., and K. B. Andersen. 1999. Palmitoylation of the intracytoplasmic R peptide of the transmembrane envelope protein in Moloney murine leukemia virus. J. Virol. 73:8975-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pages, J. C., and T. Bru. 2004. Toolbox for retrovectorologists. J. Gene Med. 6(Suppl. 1):S67-S82. [DOI] [PubMed] [Google Scholar]
- 32.Pagès, J. C., N. Loux, D. Farge, P. Briand, and A. Weber. 1995. Activation of Moloney murine leukemia virus LTR enhances the titer of recombinant retrovirus in psi CRIP packaging cells. Gene Ther. 2:547-551. [PubMed] [Google Scholar]
- 33.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piver, E., C. Collin, A. Diatta, P. Vaudin, and J. C. Pages. 2005. Cellular factors influencing Semliki Forest virus vector biology. Gene Ther. 12(Suppl. 1):S111-S117. [DOI] [PubMed] [Google Scholar]
- 35.Reeves, L., L. Duffy, S. Koop, J. Fyffe, and K. Cornetta. 2002. Detection of ecotropic replication-competent retroviruses: comparison of s(+)/l(-) and marker rescue assays. Hum. Gene Ther. 13:1783-1790. [DOI] [PubMed] [Google Scholar]
- 36.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rolls, M. M., P. Webster, N. H. Balba, and J. K. Rose. 1994. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell 79:497-506. [DOI] [PubMed] [Google Scholar]
- 38.Russell, R. A., G. Vassaux, P. Martin-Duque, and M. O. McClure. 2004. Transient foamy virus vector production by adenovirus vectors. Gene Ther. 11:310-316. [DOI] [PubMed] [Google Scholar]
- 39.Sawicki, D. L., S. Perri, J. M. Polo, and S. G. Sawicki. 2006. Role for nsP2 proteins in the cessation of alphavirus minus-strand synthesis by host cells. J. Virol. 80:360-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sena-Esteves, M., J. C. Tebbets, S. Steffens, T. Crombleholme, and A. W. Flake. 2004. Optimized large-scale production of high titer lentivirus vector pseudotypes. J. Virol. Methods 122:131-139. [DOI] [PubMed] [Google Scholar]
- 41.Starck, S. R., and R. W. Roberts. 2002. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. RNA 8:890-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahlfors, J. J., K. G. Xanthopoulos, and R. A. Morgan. 1997. Semliki Forest virus-mediated production of retroviral vector RNA in retroviral packaging cells. Hum. Gene Ther. 8:2031-2041. [DOI] [PubMed] [Google Scholar]
- 44.Yee, J. K., A. Miyanohara, P. P. La, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]