Abstract
Marburg virus (MARV) has been associated with sporadic episodes of hemorrhagic fever, including a recent highly publicized outbreak in Angola that produced severe disease and significant mortality in infected patients. MARV is also considered to have potential as a biological weapon. Recently, we reported the development of a promising attenuated, replication-competent vaccine against MARV based on recombinant vesicular stomatitis virus (VSV) expressing the glycoprotein of the Musoke strain of MARV (VSVΔG/MARVGP-Musoke). We used this vaccine to demonstrate complete protection of cynomolgus monkeys against a homologous MARV challenge. While these results are highly encouraging, an effective vaccine would need to confer protection against all relevant strains of MARV. Here, we evaluated the protective efficacy of the VSVΔG/MARVGP-Musoke vaccine against two heterologous MARV strains, the seemingly more pathogenic Angola strain and the more distantly related Ravn strain. In this study, seven cynomolgus monkeys were vaccinated with the VSVΔG/MARVGP-Musoke vector. Three of these animals were challenged with the Angola strain, three with the Ravn strain, and a single animal with the Musoke strain of MARV. Two animals served as controls and were each injected with a nonspecific VSV vector; these controls were challenged with the Angola and Ravn strains, respectively. Both controls succumbed to challenge by day 8. However, none of the specifically vaccinated animals showed any evidence of illness either from the vaccination or from the MARV challenges and all of these animals survived. These data suggest that the VSVΔG/MARVGP-Musoke vaccine should be sufficient to protect against all known MARV strains.
Marburg virus (MARV) causes severe and often fatal infections in humans and nonhuman primates. Historically, several strains of MARV have produced confirmed case fatality rates ranging from 23% to slightly greater than 50% (4, 16). However, in 2004 and 2005 a new strain of MARV caused even more significant mortality during a large outbreak in Angola. Case fatality rates during this episode fluctuated around 90%. An initial report by the World Health Organization noted that the epidemic killed 329 people of the 374 that were infected; the outbreak was recently declared over, and these numbers were revised, with the updated report noting that there were 227 deaths among the 252 reported cases (12). The reasons for the increased lethality of this new strain of MARV are presently unknown but are of significant concern.
While there are currently no licensed vaccines or antivirals to prevent or treat MARV infections, we recently described the development of a promising new replication-competent vaccine against MARV based on recombinant vesicular stomatitis virus (VSV) (7, 14). In one study, we demonstrated complete protection of cynomolgus macaques against a high-dose (1,000 PFU) lethal MARV challenge by use of a single injection of recombinant VSV vectors expressing the glycoprotein (GP) of the homologous Musoke strain of MARV (MARV-Musoke) (14). More recently, we demonstrated that the same vaccine vector used as postexposure treatment for rhesus monkeys (Macaca mulatta) infected with 1,000 PFU of strain Musoke was also able to protect all animals from clinical disease and death (5).
MARV and Ebola virus (EBOV) comprise the two genera that make up the family Filoviridae (6). There is a single species, Lake Victoria marburgvirus, within the MARV genus. There are a number of different strains of MARV. Comparative analyses of the GP and viral protein (VP) 35 genes of MARV strains showed that there are two distinct lineages within the Lake Victoria marburgvirus species of MARV. The original MARV isolates from the 1967 episodes in Marburg, Germany (Popp and Ratayczak strains), from a case in 1975 in South Africa (Ozolin strain), and from 1980 in Kenya (Musoke strain) comprise one lineage. An isolate from Kenya in 1987 (Ravn strain) represents a second genetic lineage within the Lake Victoria marburgvirus species (21 to 23% amino acid difference) (21).
MARV is composed of seven structural proteins and the nonsegmented negative-sense viral RNA genome. Four proteins (NP, VP35, VP30, and L) make up the helical nucleocapsid, which is surrounded by a matrix that is composed of the viral proteins VP40 and VP24. The surface of MARV virions is coated with spikes that consist of the structural GP. The GP plays a role in virus entry and pathogenesis and serves as a major and logical target for vaccine strategies, including our recombinant VSV-based system (14).
While most strains of MARV produce lethal infections in nonhuman primates, the disease courses vary among the strains and are in general more protracted than what is seen with EBOV. Recent studies have suggested that the new Angola isolate of MARV appears to produce a disease in nonhuman primates that is more rapid and severe than that produced by other MARV strains and, in fact, appears to be as virulent as that produced by Zaire ebolavirus (ZEBOV) in rhesus macaques (T. W. Geisbert, unpublished observation).
In developing vaccines against any virus, there are always concerns about broad protection and the ability to protect against different strains or isolates of the same virus. Of particular concern regarding cross-protection among the MARV strains are results reported using a different vaccine vector system. Notably, a study using a platform based on Venezuelan equine encephalitis virus (VEEV) replicons showed that cynomolgus monkeys vaccinated with VEEV (MARV GP) or VEEV (MARV NP) replicons based on strain Musoke were protected against lethal homologous challenge (9) but not against a challenge with the Ravn strain of MARV (MARV-Ravn) (10). This result raises the concern that the 21 to 23% difference in amino acids between the two MARV lineages may be significant enough to affect the ability of a candidate vaccine to confer cross-protection against the different MARV strains. Here, we tested the ability of our recombinant VSV vaccine expressing the MARV-Musoke strain GP to protect nonhuman primates against a lethal challenge with either the more genetically diverse Ravn strain or the seemingly more pathogenic Angola strain (MARV-Angola).
MATERIALS AND METHODS
Vaccine vectors and viruses.
The recombinant VSVs expressing the GPs of ZEBOV (VSVΔG/ZEBOVGP) and MARV-Musoke (VSVΔG/MARVGP-Musoke) were generated by use of the infectious clone for the VSV Indiana serotype as described recently (7, 14). Briefly, the appropriate open reading frames (ORF) for the GPs were generated by PCR, cloned into the VSV genomic vectors lacking the VSV GP gene, sequence confirmed, and originally rescued using the method described earlier. MARV-Musoke was isolated from a human case in 1980 in Kenya (22), MARV-Ravn was isolated from a human case in 1987 in Kenya (13), and MARV-Angola was isolated from a patient of the recent outbreak in Angola in 2005 (26).
Sequencing and phylogenetic analysis.
MARV-Angola was isolated from clinical specimens (whole blood) from several patients. The sequence of the GP gene was determined using primers based on the GP sequences of the Musoke strain. The deduced amino acid sequence of the Angola GP ORF was compared with the amino acid sequences of different filovirus GPs which were retrieved from GenBank. Phylogenetic analysis was performed with MEGA version 3.1 (www.megasoftware.net) using a neighbor-joining tree and 1,000 bootstrap replicates. The GenBank accession numbers for protein sequences included in the analysis are NP_042028 (MARV, Popp strain), ABA87127 (MARV, Musoke strain), ABE27085 (MARV, Durba strain), AAQ55258 (MARV, Ozolin strain), ABE27071 (MARV, Ravn strain), ABE27092 (MARV, Durba strain), AAB37093 (Ivory Coast ebolavirus, Ivory Coast strain), BAB69006 (Reston ebolavirus, Reston strain), AAU43887 (Sudan ebolavirus [SEBOV], Gulu strain), AAC54889 (Reston ebolavirus, Philippine strain), AAB37096 (SEBOV, Boniface strain), AAC54882 (SEBOV, Maleo strain), AAC57992 (ZEBOV, Eckron strain), AAQ55048 (ZEBOV, Zaire strain), AAN37507 (ZEBOV, Mayinga strain), and AAL25818 (ZEBOV, Gabon strain).
Animal studies.
Nine healthy adult cynomolgus macaques (Macaca fascicularis) (5 to 9 kg) were used for these studies. Seven of these animals were vaccinated intramuscularly with ∼2 × 107 PFU of VSVΔG/MARVGP-Musoke, while two animals served as experimental controls and received ∼2 × 107 PFU of VSVΔG/ZEBOVGP. All animals were challenged 28 days after the single-dose immunization with 1,000 PFU of MARV as follows. One group of three macaques that were vaccinated with VSVΔG/MARVGP-Musoke was challenged with MARV-Ravn, a second group of three animals vaccinated with VSVΔG/MARVGP-Musoke was challenged with MARV-Angola, and the remaining macaque that was vaccinated with VSVΔG/MARVGP-Musoke was challenged with MARV-Musoke. One of the control animals receiving VSVΔG/ZEBOVGP was challenged with MARV-Ravn, while the second control animal was challenged with MARV-Angola.
Swab samples (oral, nasal, and rectal) and/or blood were taken before vaccination (day −28); at days 3 (day −25), 6 (day −22), 14 (day −14), and 28 (day 0) after vaccination; and at days 3, 6, 10, 14, and 28 after the MARV challenges. Animal studies were performed in biosafety level 4 biocontainment facilities at USAMRIID and were approved by the USAMRIID Laboratory Animal Care and Use Committee. Animal research was conducted in compliance with the Animal Welfare Act and other federal statues and regulations relating to animals and experiments involving animals and adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals (17). The facility used is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Hematology and serum biochemistry.
Total white blood cell counts, red blood cell counts, platelet counts, hematocrit values, total hemoglobin, mean cell volume, mean corpuscular volume, and mean corpuscular hemoglobin concentration were determined from blood samples collected in tubes containing EDTA by using a laser-based hematologic analyzer (Coulter Electronics, Hialeah, FL). The white blood cell differentials were performed manually on Wright-stained blood smears. Serum samples were tested for concentrations of albumin, amylase, alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, gamma-glutamyltransferase, glucose, cholesterol, total protein, total bilirubin, blood urea nitrogen, and creatinine by using a Piccolo point-of-care blood analyzer (Abaxis, Sunnyvale, CA).
Virus detection.
RNA was isolated from blood and swabs by use of appropriate RNA isolation kits (QIAGEN, Mississauga, Ontario, Canada). For the detection of VSV, we used a reverse transcriptase PCR (RT-PCR) assay targeting the matrix gene (nucleotide [nt] positions 2355 to 2661, GenBank accession number NC_001560). MARV RNA was detected using primer pairs targeting the L gene (GenBank accession number X 68494) (for RT-PCR, nt positions 1966 to 2243, and for nested PCR, nt positions 2017 to 2213). The low detection limit for this MARV assay is 0.1 PFU/ml of plasma. Virus titration was performed by plaque assay on Vero E6 cells from all blood and selected organ (adrenal, ovary, lymph nodes, liver, spleen, pancreas, lung, heart, and brain) and swab samples. Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero E6 monolayers in duplicate wells (0.2 ml per well); thus, the limit for detection was 25 PFU/ml.
Humoral immune responses.
Immunoglobulin G (IgG) antibodies against MARV were detected with an enzyme-linked immunosorbent assay (ELISA) by using purified virus particles as an antigen source (14). Neutralization assays were performed by measuring plaque reduction in a constant virus:serum dilution format as previously described (14). Briefly, a standard amount of MARV-Musoke (∼100 PFU) was incubated with serial twofold dilutions of the serum sample for 60 min. The mixture was used to inoculate Vero E6 cells for 60 min. Cells were overlaid with an agar medium and incubated for 8 days, and plaques were counted 48 h after neutral red staining. Endpoint titers were determined by the dilution of serum which neutralized 50% of the plaques (50% plaque reduction neutralization titer [PRNT50]).
Cellular immune responses.
The method for assessment of T-cell responses to MARV was published previously (14). Briefly, peripheral blood mononuclear cells were isolated from cynomolgus macaque whole-blood samples by Histopaque gradient (Sigma, St Louis, MO). Approximately 1 × 106 cells were stimulated in 200 μl RPMI medium (Gibco/Invitrogen, Carlsbad, CA) for 6 h at 37°C with anti-CD28 and anti-CD49d antibodies and either dimethyl sulfoxide or a pool of MARV-Musoke GP-specific peptides in the presence of brefeldin A. The peptides were 15 amino acids in length, overlapping by 11 and spanning the entire MARV GP at a final concentration of 2 μg/ml. Cells were fixed and permeabilized with FACSlyse (Becton Dickinson, San Jose, CA) supplemented with Tween 20 and stained with a mixture of antibodies against lineage markers (phycoerythrin-conjugated CD3, peridinin chlorophyll protein-conjugated CD4, or fluorescein isothiocyanate-conjugated CD8) and either allophycocyanin-conjugated tumor necrosis factor alpha or allophycocyanin-conjugated gamma interferon. Samples were run on a FACSCalibur instrument and analyzed using FlowJo software. Positive gating for lymphocytes by using forward versus side scatter was followed by CD3+/CD8− and CD3+/CD4− gating, and specific populations were further defined by anti-CD4 and anti-CD8 positivity, respectively. Cytokine-positive cells were defined as percentages within these individual lymphocyte subsets, and at least 200,000 events were analyzed for each sample.
RESULTS
Sequence analysis.
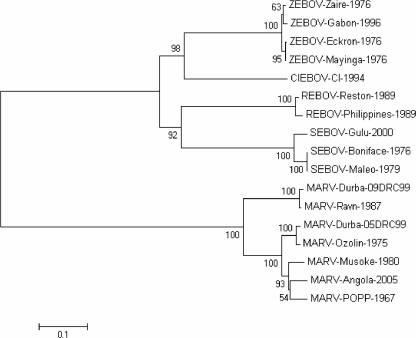
At the beginning of this study, the GP sequence of MARV-Angola was not determined. Therefore, we sequenced the GP ORF of several isolates which were obtained in Winnipeg, Manitoba, Canada, by inoculation of Vero E6 cells with whole blood collected from patients during the MARV-Angola outbreak. Comparative analysis showed that all GP gene sequences were identical. Thus, the deduced amino acid sequence of the GP ORF of one of the isolates was used for phylogenetic analysis with the deduced amino acid sequences of several EBOV and MARV GPs, including two recently deposited sequences from the MARV outbreak in Durba/Watsa, Democratic Republic of the Congo (Fig. 1). The phylogenetic analysis demonstrated that MARV strains separated into two major branches. One branch included strains from 1987 (Ravn) and 1998 to 2000 (Durba/Watsa). The second major branch again separated into two further branches, with strains from 1975 (Ozolin) and 1998 to 2000 (Durba/Watsa) separated from strains originating in 1967 (Popp and Ratayczak), 1980 (Musoke), and 2004 to 2005 (Angola). The close relationship between the Angola and Musoke strains indicated a likelihood of cross-protection by the VSVΔG/MARVGP-Musoke vaccine against a heterologous challenge with the Angola strain. However, cross-protection against a challenge with the more distantly related Ravn strain seemed less likely. We recently demonstrated that the VSVΔG/MARVGP-Musoke vaccine does not provide any cross-protection against ZEBOV strains (14).
FIG. 1.
Phylogenetic tree analysis for the GPs of filoviruses. The amino acid sequences of filovirus GPs present in the protein database of GenBank were analyzed using MEGA version 3.1 (www.megasoftware.net). A neighbor-joining tree and 1,000 bootstrap replicates for branch points were prepared. The analysis shows that the GP of MARV-Angola is more closely related to that of MARV-Musoke than to that of MARV-Ravn and has substantial differences with the GPs of ZEBOV species. The numbers to the left of the strains show percentages of bootstrap values. With the exception of the two Durba strains, the assigned designations are from reference 6. CIEBOV, Ivory Coast ebolavirus; REBOV, Reston ebolavirus.
Clinical observations.
A total of nine cynomolgus monkeys were used to evaluate whether a preventative vaccination strategy employing a single injection of VSVΔG/MARVGP-Musoke could cross-protect against MARV hemorrhagic fever caused by the Ravn and Angola strains (Fig. 2A). We vaccinated seven animals by intramuscular injection of VSVΔG/MARVGP-Musoke vectors (subjects no. 1 to 7) and two control animals with nonspecific VSVΔG/ZEBOVGP (controls no. 1 and 2). Animals were challenged 28 days after the single-dose vaccine with either heterologous MARV-Ravn (subjects no. 1 to 3 and control no. 1) or heterologous MARV-Angola (subjects no. 4 to 6 and control no. 2). One animal (subject no. 7) was challenged with homologous MARV-Musoke and served as an internal vaccine control, as we have shown previously that the VSVΔG/MARVGP-Musoke vaccine can provide complete protection against a homologous MARV challenge (14).
FIG. 2.
Vaccination and challenge of nonhuman primates. (A) Flow chart of the experimental design. Arrows indicate the days of sampling (blood and swabs). Days preceded by a minus indicate days prior to challenge. (B) Kaplan-Meier mortality chart of the MARV vaccine study. Open triangles, animal vaccinated with VSVΔG/EBOVGP and challenged with MARV-Ravn (control no. 1); X's, animal vaccinated with VSVΔG/EBOVGP and challenged with MARV-Angola (control no. 2); filled diamonds, animals vaccinated with VSVΔG/MARVGP and challenged with MARV-Ravn (subjects no. 1 to 3); open squares, animals vaccinated with VSVΔG/MARVGP-Musoke and challenged with MARV-Angola (subjects no. 4 to 6); open circles, animal vaccinated with VSVΔG/MARVGP-Musoke and challenged with MARV-Musoke (subject no. 7).
Animals were monitored closely after both vaccination and MARV challenge for clinical symptoms of illness, viremia from either the vaccine or MARV, and shedding of the recombinant VSVs (Fig. 2A). None of the animals vaccinated with VSVΔG/MARVGP-Musoke showed any evidence of clinical illness either after vaccination or after the MARV challenge. Importantly, all of the animals vaccinated with VSVΔG/MARVGP-Musoke survived against a heterologous challenge with either MARV-Ravn or MARV-Angola with no observable clinical changes. In contrast, both the MARV-Ravn-challenged (control no. 1) and the MARV-Angola-challenged (control no. 2) control animals followed a typical disease course, developing fevers, macular rashes, and signs of depression by day 6 postinfection, and succumbed to MARV hemorrhagic fever on day 8 postinfection (Fig. 2B).
Viremia and blood chemistry.
To determine whether viremia or shedding of the recombinant VSVs occurred after immunization, whole-blood and swab samples from all nine of the vaccinated animals were analyzed by RT-PCR and virus isolation. A transient and low-level (≤1.7 log10 PFU/ml) recombinant VSV viremia was detected by virus isolation at day 3 after vaccination in plasma from four of the VSVΔG/MARVGP-Musoke vaccinated animals (subjects no. 1, 4, 5, and 7) (Fig. 3A). Also, a low level of VSVΔG/ZEBOVGP was detected by virus isolation from a nasal swab of one of the control animals at day 3. However, this animal had evidence of self-inflicted bleeding around the nares so we are uncertain as to the exact significance of this particular finding. In addition, we did not detect VSVΔG/ZEBOVGP in plasma of this control animal by virus isolation, further questioning the importance of the low level of recombinant virus detected from the nasal swab.
FIG. 3.
Viremia levels in nonhuman primates after vaccination and MARV challenge. (A) VSV viremia levels were determined after vaccination with VSVΔG/MARVGP-Musoke (subjects no. 1 to 7) or VSVΔG/ZEBOVGP (controls no. 1 and 2). (B) MARV viremia levels from plasma, determined at the indicated time points after challenge with MARV-Ravn, MARV-Angola, or MARV-Musoke. Viremias were determined by plaque assay.
Blood samples were also analyzed after MARV challenge for evidence of MARV replication and shedding by plaque assay (Fig. 3B) and by RT-PCR (data not shown). By day 6, both the MARV-Ravn-challenged control animal (control no. 1) and the MARV-Angola-challenged control animal (control no. 2) developed high MARV titers in the blood as detected by plaque assay (>107 log PFU/ml). RT-PCR was more sensitive and showed evidence of MARV in plasma of these two control animals by day 3 postinfection. In contrast, no MARV was detected in the plasma by virus isolation or RT-PCR in any of the animals vaccinated with VSVΔG/MARVGP-Musoke.
Analysis of blood chemistry and hematology was performed before and after the MARV challenges. Again, in accordance with no clinical symptoms, none of the animals vaccinated with VSVΔG/MARVGP-Musoke showed any evidence of changes in blood chemistry and hematology either after vaccination or after the MARV challenges. In contrast, while neither of the control animals showed any evidence of changes in blood chemistry or hematology after vaccination with VSVΔG/ZEBOVGP, both controls developed lymphopenia and thrombocytopenia after the MARV challenges. In addition, both of these controls showed significant elevations in circulating levels of enzymes (alkaline phosphatase, aspartate aminotransferase, and alanine aminotransferase) associated with impairment of the liver by day 6 postinfection.
Evaluation of antibody and cellular immune responses.
The antibody responses of the cynomolgus macaques immunized with VSVΔG/MARVGP-Musoke were evaluated after vaccination (day −14 and day 0) and after MARV challenge (day 14 and day 28) by IgG ELISA and by plaque neutralization tests (PRNT50). All of the animals developed high anti-MARV IgG antibody levels (≥1:1,000) by the day of the MARV challenges (day 0) (Fig. 4A). Low levels of anti-MARV neutralizing antibodies (1:10 to 1:20) were observed for four of the seven animals vaccinated with VSVΔG/MARVGP-Musoke at the day of MARV challenge (Fig. 4B). All seven of these animals developed low levels of neutralizing antibodies (1:10 to 1:80) by day 28 after the MARV challenges (Fig. 4B).
FIG. 4.
Humoral immune responses to MARV before and after challenge in nonhuman primates. (A) IgG responses to MARV measured using an established ELISA (see Materials and Methods). Titers are presented as endpoint dilutions. (B) MARV neutralizing antibodies detected using a plaque reduction neutralization assay (PRNT50) as described in Materials and Methods. Titers are presented as endpoint dilutions. Days preceded by a minus indicate days prior to challenge, and day 0 is the day of challenge.
To better understand the cellular responses of T-cell populations found in peripheral blood mononuclear cell fractions of specifically and nonspecifically vaccinated animals in mediating protection against MARV challenge, flow cytometry was employed during the course of study. There was no evidence of either gamma interferon or tumor necrosis factor alpha production in CD4 or CD8 T-cell populations either before or after the MARV challenges in any of the animals used in this study (data not shown).
DISCUSSION
The recent outbreak of MARV in Angola generated significant interest from the popular press and reinforces the danger of emerging viruses, such as MARV, as significant public health threats. However, MARV also poses a threat as a potential biological weapon (1). It is primarily for this reason that there has been an increased investment in developing countermeasures against this highly lethal pathogen.
Significant advances in developing countermeasures against the filoviruses, particularly regarding the creation of promising vaccines, have been made over the last decade (14, 23, 24). Recently, we showed that a replication-competent vaccine based on recombinant VSV expressing either the GP of EBOV or the GP of MARV could completely protect nonhuman primates against a homologous filovirus challenge (14). While the EBOV vaccine based on the Zaire species GP completely protected macaques against a lethal ZEBOV challenge, it was unable to protect macaques against challenge with another EBOV species, SEBOV. This was not an unexpected finding since macaques that survive experimental challenge with wild-type SEBOV are not protected against a subsequent back-challenge with ZEBOV (2). Indeed, there is a 37 to 44% difference between SEBOV and ZEBOV at the nucleotide and amino acid levels (20), further supporting the view that there would be little if any cross-protection among these species of EBOV.
From the perspective of vaccine development, it is apparent that a vaccine that would protect against both SEBOV and ZEBOV would likely need to include SEBOV-specific antigens as well as ZEBOV-specific antigens. While there is only a single species of MARV, as noted previously, there are two genetically disparate lineages of MARV with 21 to 23% difference in amino acids (21). This variation raised concern that the recombinant VSV vaccine based on MARV-Musoke may not protect against more-divergent strains of MARV. Importantly, we show in the current study that the VSVΔG/MARVGP-Musoke vector does in fact protect nonhuman primates against a lethal challenge with one of the most divergent MARV strains, Ravn, and also against challenge with the more closely related but ostensibly most pathogenic strain of MARV, Angola. Thus, the VSV-based approach seems to be superior to the only other successful approach, using VEEV MARV GP and/or VEEV MARV NP, which protected nonhuman primates against a lethal homologous challenge (9) but failed to protect against a lethal heterologous challenge using the Ravn strain (10).
Regarding the mechanism by which the VSVΔG/MARVGP-Musoke vaccine protects, results of the current effort are consistent with our original vaccination study (14) as well as a recent postexposure treatment study (5), as protection of monkeys by the VSVΔG/MARVGP-Musoke vaccine in both cases appeared to be associated primarily with the humoral but not the cellular immune response. Notably, neutralizing antibodies were poorly induced in the animals in this study as well as in both previous studies (5, 14), suggesting that protection may in part be due to nonneutralizing antibodies which were present at high levels in these animals.
The replication-competent VSV-based vaccine platform has a number of advantages over successful but replication-defective systems, such as the adenovirus system using human adenovirus 5 vectors (23, 24). First, a significant concern regarding many vector-based vaccine systems, in particular, adenovirus 5, is antivector immunity (3). Importantly, there is a very low percentage of VSV seropositivity in the general population (25). In addition, any antivector immunity that may be present in a very small number of individuals may not be important at all, because in VSV infections, neutralizing antibodies are directed against the VSV glycoprotein, which is not expressed using the recombinant vector employed here (7). Second, durability is a major concern of any vaccine platform, in particular, for vaccines designed to be used against exotic pathogens, such as viral hemorrhagic fevers, which are found primarily in remote geographic locations where boosting is often difficult and not practicable. In general, live, attenuated vaccines give long-lasting immunity after a single administration. For example, the highly attenuated yellow fever vaccine confers near-complete protection that persists for 30 or more years (15). While replication-defective viruses have a number of advantages over classical inactivated virion vaccine approaches, the durability of such replication-defective vaccines compared to that of live, attenuated vaccines is largely unknown. Safety is of course a serious concern that is associated with the use of any live vaccine. Currently, replication-defective VSV vectors capable of only a single cycle of replication are being developed as an alternative to replication-competent VSV vectors (18). Whether these vectors can confer protective immunity against highly lethal pathogens, such as the hemorrhagic fever viruses, and whether there will be any trade-off regarding durability for potential safety remain to be determined.
Not only have live, attenuated VSV-based vaccines shown promise in nonhuman primate models of EBOV and MARV infections, we also recently demonstrated the success of this platform in protecting monkeys against a lethal Lassa virus challenge (8). Others are using replication-competent VSV-based systems as candidate vaccines for a variety of viruses, including human immunodeficiency virus (HIV). In fact, a candidate recombinant VSV-based HIV vaccine was shown previously to prevent AIDS-like disease in monkeys (19). Moreover, in an effort to address safety concerns with live vaccines, Wyeth is currently engineering VSV vectors that are highly attenuated in animals. The company plans to begin human trials with these vectors expressing HIV antigens within a year (11).
The use of attenuated recombinant VSV-based vectors has proven to be an effective and promising platform for the development of preventive vaccines against a number of pathogenic viruses. Findings from the current study taken together with observations from our previous work (5, 14) suggest that a vaccine that would confer protection against all relevant species or strains of filoviruses would likely require three antigens, including those specific for the ZEBOV GP, the SEBOV GP, and the MARV-Musoke GP.
Acknowledgments
We thank Denise Braun, Daryl Dick, Lisa Fernando, Andrea Paille, and Carlton Rice for technical assistance and assistance with animal care. We also thank Gabriela Dveksler, Robert Friedman, Peter Jahrling, Elliott Kagan, and Martin Ottolini for helpful suggestions. We are grateful to Pierre Rollin and Tom Ksiazek for providing the Angola isolate of Marburg virus.
The study was supported in part by a grant from the Canadian Institute of Health Research (CIHR MOP-43921) awarded to H.F. and by the Medical Chemical/Biological Defense Research Program and Military Infectious Diseases Research Program, U.S. Army Medical Research and Material Command (project number 04-4-7J-012).
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
REFERENCES
- 1.Borio, L., T. Inglesby, C. J. Peters, A. L. Schmaljohn, J. M. Hughes, P. B. Jahrling, T. Ksiazek, K. M. Johnson, A. Meyerhoff, T. O'Toole, M. S. Ascher, J. Bartlett, J. G. Breman, E. M. Eitzen, Jr., M. Hamburg, J. Hauer, D. A. Henderson, R. T. Johnson, G. Kwik, M. Layton, S. Lillibridge, G. J. Nabel, M. T. Osterholm, T. M. Perl, P. Russell, and K. Tonat. 2002. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287:2391-2405. [DOI] [PubMed] [Google Scholar]
- 2.Bowen, E. T., G. S. Platt, G. Lloyd, R. T. Raymond, and D. I. Simpson. 1980. A comparative study of strains of Ebola virus isolated from southern Sudan and northern Zaire in 1976. J. Med. Virol. 6:129-138. [DOI] [PubMed] [Google Scholar]
- 3.Chirmule, N., K. Propert, S. Magosin, Y. Qian, R. Qian, and J. Wilson. 1999. Immune responses to adenovirus and adeno-associated virus in humans. Gene Ther. 6:1574-1583. [DOI] [PubMed] [Google Scholar]
- 4.Colebunders, R., H. Sleurs, P. Pirard, M. Borchert, M. Libande, J. P. Mustin, A. Tshomba, L. Kinuani, L. A. Olinda, F. Tshioko, and J. J. Muyembe-Tamfum. 2004. Organisation of health care during an outbreak of Marburg haemorrhagic fever in the Democratic Republic of Congo, 1999. J. Infect. 48:347-353. [DOI] [PubMed] [Google Scholar]
- 5.Daddario-DiCaprio, K. M., T. W. Geisbert, U. Stroher, J. B. Geisbert, A. Grolla, E. A. Fritz, L. Fernando, E. Kagan, P. B. Jahrling, L. E. Hensley, S. M. Jones, and H. Feldmann. 2006. Postexposure protection against Marburg haemorrhagic fever with recombinant vesicular stomatitis virus vectors in non-human primates: an efficacy assessment. Lancet 367:1399-1404. [DOI] [PubMed] [Google Scholar]
- 6.Feldmann, H., T. W. Geisbert, P. B. Jahrling, H. D. Klenk, S. V. Netesov, C. J. Peters, A. Sanchez, R. Swanepoel, and V. E. Volchkov. 2004. Filoviridae, p. 645-653. In C. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier, London, United Kingdom.
- 7.Garbutt, M., R. Liebscher, V. Wahl-Jensen, S. Jones, P. Moller, R. Wagner, V. Volchkov, H. D. Klenk, H. Feldmann, and U. Stroher. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 78:5458-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geisbert, T. W., S. Jones, E. A. Fritz, A. C. Shurtleff, J. B. Geisbert, R. Liebscher, A. Grolla, U. Stroher, L. Fernando, K. M. Daddario, M. C. Guttieri, B. R. Mothe, T. Larsen, L. E. Hensley, P. B. Jahrling, and H. Feldmann. 2005. Development of a new vaccine for the prevention of Lassa fever. PLoS Med. 2:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hevey, M., D. Negley, P. Pushko, J. Smith, and A. Schmaljohn. 1998. Marburg virus vaccines based upon alphavirus replicons protect guinea pigs and nonhuman primates. Virology 251:28-37. [DOI] [PubMed] [Google Scholar]
- 10.Hevey, M., D. Negley, A. Staley, and A. Schmaljohn. 2001. Determination of vaccine components required for protecting cynomolgus macaques against genotypically divergent isolates of Marburg virus, abstr. W36-4. Abstr. 20th Annu. Meet. Am. Soc. Virol., Madison, Wis.
- 11.International AIDS Vaccine Initiative. 2005. Annual AIDS vaccine meeting highlights recent data from clinical trials and lessons on recruitment and retention of volunteers. International AIDS Vaccine Initiative, New York, N.Y. [Online.] http://www.iavireport.org/Issues/Issue9-4/promise.asp. [PubMed]
- 12.International Society for Infectious Diseases. 2005. Marburg hemorrhagic fever—Angola, archive no. 20051108.3269. International Society for Infectious Diseases, Brookline, Mass. [Online.] http://www.promedmail.org.
- 13.Johnson, E. D., B. K. Johnson, D. Silverstein, P. Tukei, T. W. Geisbert, A. N. Sanchez, and P. B. Jahrling. 1996. Characterization of a new Marburg virus isolated from a 1987 fatal case in Kenya. Arch. Virol. Suppl. 11:101-114. [DOI] [PubMed] [Google Scholar]
- 14.Jones, S. M., H. Feldmann, U. Stroher, J. B. Geisbert, L. Fernando, A. Grolla, H. D. Klenk, N. J. Sullivan, V. E. Volchkov, E. A. Fritz, K. M. Daddario, L. E. Hensley, P. B. Jahrling, and T. W. Geisbert. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11:786-790. [DOI] [PubMed] [Google Scholar]
- 15.Lefeuvre, A., P. Marianneau, and V. Deubel. 2004. Current assessment of yellow fever and yellow fever vaccine. Curr. Infect. Dis. Rep. 6:96-104. [DOI] [PubMed] [Google Scholar]
- 16.Martini, G. 1971. Marburg virus disease: clinical syndrome, p. 1-9. In G. A. Martini and R. Siegert (ed.), Marburg virus disease. Springer-Verlag, New York, N.Y.
- 17.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 18.Publicover, J., E. Ramsburg, and J. K. Rose. 2005. A single-cycle vaccine vector based on vesicular stomatitis virus can induce immune responses comparable to those generated by a replication-competent vector. J. Virol. 79:13231-13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez, A., S. G. Trappier, B. W. Mahy, C. J. Peters, and S. T. Nichol. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA 93:3602-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez, A., S. G. Trappier, U. Stroher, S. T. Nichol, M. D. Bowen, and H. Feldmann. 1998. Variation in the glycoprotein and VP35 genes of Marburg virus strains. Virology 240:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, D. H., B. K. Johnson, M. Isaacson, R. Swanapoel, K. M. Johnson, M. Killey, A. Bagshawe, T. Siongok, and W. K. Keruga. 1982. Marburg-virus disease in Kenya. Lancet i:816-820. [DOI] [PubMed] [Google Scholar]
- 23.Sullivan, N. J., T. W. Geisbert, J. B. Geisbert, L. Xu, Z. Y. Yang, M. Roederer, R. A. Koup, P. B. Jahrling, and G. J. Nabel. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 25.Wagner, R. R., and J. K. Rose. 1996. Rhabdoviridae: the viruses and their replication, p. 1121-1135. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.World Health Organization. 2005. Marburg haemorrhagic fever, Angola. Wkly. Epidemiol. Rec. 80:158-159. [PubMed] [Google Scholar]