Abstract
The role of NF-κB in regulating human cytomegalovirus (HCMV) replication and gene transcription remains controversial. Multiple, functional NF-κB response elements exist in the major immediate-early promoter (MIEP) enhancer of HCMV, suggesting a possible requirement for this transcription factor in lytic viral replication. Here we demonstrate by generating and analyzing HCMVs with alterations in the MIEP-enhancer that, although this region is essential for HCMV growth, none of the four NF-κB response elements contained within the enhancer are required for MIE gene expression or HCMV replication in multiple cell types. These data reveal the robustness of the regulatory network controlling the MIEP enhancer.
The major immediate-early promoter (MIEP) of human cytomegalovirus (HCMV) is responsive to a multitude of transcription factors and plays a pivotal role in initiating the viral transcription/replication cycle (7, 16; reviewed in references 22 and 23). Regulation of the MIEP has been postulated to be critical in determining HCMV permissiveness and the transition between latent and lytic infection. Thus, deciphering the molecular mechanisms of the MIEP regulation may reveal key control points contributing to HCMV pathogenesis.
The MIEP enhancer includes four cognate NF-κB recognition sites, and NF-κB activates MIEP transcription in transient-transfection assays (20, 25-27). HCMV infection results in rapid induction of cellular NF-κB (19, 27, 30), and several groups have reported a potential contribution of NF-κB to the replication strategy of HCMV through regulation of the MIEP (8, 13). In contrast, we and others have reported a neutral or even a negative role of NF-κB activation on HCMV transcription/replication cycle in different cell types (3, 4, 11, 14, 15). However, the basis for these experimental discrepancies is currently unclear. Importantly, a direct test of the requirement for the MIEP NF-κB binding sites in HCMV transcription/replication has still not been performed. Here we report on formally assessing the direct requirement of the cognate binding sites for NF-κB in contributing to major immediate-early (MIE) transcription and viral growth.
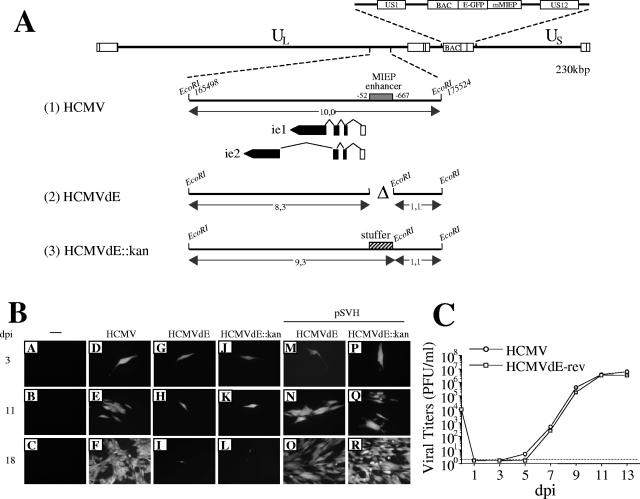
As a first step toward understanding NF-κB regulation of the HCMV MIEP, we deleted enhancer sequences from −52 to −667 (including all NF-κB response elements), in HCMV AD169. A parental HCMV bacterial artificial chromosome (BAC) (5, 6) containing the E-GFP open reading frame (ORF) under control of the murine cytomegalovirus (MCMV) MIEP (Fig. 1A, line 1) was used to construct two enhancerless HCMV recombinant mutants. In HCMVdE, MIEP sequences from −52 to −667 were removed (Fig. 1A, line 2), and in HCMVdE::Kan, enhancer sequences were replaced with a 1-kbp stuffer region to maintain the genomic spatial integrity of the ie1/ie2 and UL127 promoters (Fig. 1A, line 3). Once the integrity of constructed HCMV genomes was confirmed by restriction analysis (data not shown), they were transfected in MRC-5 fibroblasts. Three days posttransfection, ∼100 single cells expressing green fluorescent protein (GFP) could be detected in all cultures (see Fig. 1B, panels D, G, and J). Cells transfected with the parental HCMV BAC yielded viral plaques (Fig. 1B, panel E) that progressed to complete cytopathicity (panel F), whereas cultures transfected with the enhancerless HCMV BACs did not result in viral spread (panels H, I, K, and L). These data indicate that deletion of the entire MIEP enhancer region of HCMV genome is lethal.
FIG. 1.
Effect of a deletion of the entire MIEP enhancer on HCMV. (A) Schematic representation of the enhancerless HCMV BAC genomes constructed. The top line represents the map of the parental HCMV BAC genome with the US1-US12 region expanded above, indicating the BAC sequences and the E-GFP gene under control of the MCMV MIEP (mMIEP) that replace the US2-US11 region. Below, the EcoRI J fragment (nucleotide sequences from 165498 to 175524 of the HCMV genome [28]) encompassing the HCMV MIE region is enlarged (line 1), with the structures of the ie1 and ie2 transcripts indicated. The first noncoding exon of the ie1/ie2 transcription unit is depicted as an open rectangle, and coding exons are shown solid. The shaded box marks the HCMV MIEP enhancer (extending from nucleotides −52 to −667 relative to the ie1/ie2 HCMV transcription start site). The two enhancerless HCMV BAC genomes, HCMVdE and HCMVdE::Kan, were derived from the parental HCMV BAC by the ET BAC mutagenesis method (5, 6, 24). The deletion of the entire HCMV enhancer (nucleotide sequences from 174713 to 175328 of the HCMV genome) in HCMVdE (line 2) is indicated by a “Δ” symbol. In HCMVdE::Kan (line 3), the 1-kbp fragment from the kanamycin resistance gene that replaces the HCMV MIEP enhancer is represented by a cross-hatched box. A new EcoRI restriction site was introduced at the location of the enhancer deletion (in HCMVdE) or the stuffer insertion (in HCMVdE::Kan) to facilitate the characterization of the mutant genomes by an EcoRI restriction digestion (data not shown). Sizes of the natural and new EcoRI J DNA fragments for each recombinant BAC are indicated. The illustration is not drawn to scale. (B) Transfection of HCMV enhancerless genomes in cultured fibroblasts. MRC-5 cells were cotransfected by the calcium phosphate precipitation technique with 2 μg of either HCMV (D to F), HCMVdE (G to I and M to O), or HCMVdE::Kan (J to L and P to R) BAC DNAs together with 1 μg of a vector expressing the tegument protein pp71 (2), and when indicated with 1 μg of the HCMV IE1 and IE2 expression vector pSVH (29). GFP expression was detected by fluorescence microscopy at the indicated days after infection (dpi). Magnification, ×20. (C) Growth kinetics of HCMVdE-rev. MRC-5 cells were infected at a multiplicity of infection (MOI) of 0.025 with HCMV or HCMVdE-rev, a revertant of HCMVdE in which sequences from −52 to −667 of the MIEP were reintroduced in the HCMVdE genome by the ET BAC mutagenesis method. At the indicated days postinfection supernatants from the infected cultures were harvested, and titers were determined by standard plaque assays on MRC-5 cells. Each datum point represents the average and standard deviation from three separate cultures. The dashed line represents the limit of detection.
To verify that the generated HCMV mutants were defective due to deletion of the enhancer, the ability of an IE1/IE2 expression plasmid (pSVH) (29) to rescue replication was tested. Cotransfection of enhancerless BACs with pSVH resulted in spread of GFP-expressing virus to adjacent cells (Fig. 1B, panels M to O and P to R), ultimately leading to a complete cytopathic effect. In addition, a revertant HCMVdE virus was generated by the ET BAC mutagenesis method (5, 24) and shown to replicate with identical kinetics to the parental HCMV in MRC-5 cells (Fig. 1C). Thus, these results indicate that deleting the entire MIEP enhancer in HCMV AD169 abolishes lytic viral replication in cultured fibroblasts and are consistent with previous results resecting MIEP enhancer sequences in the Towne strain of HCMV (18, 21). Here, through the rescue experiments, we have eliminated the possibility that the replication defects seen in the enhancerless HCMV recombinants were due at least in part to alterations in other regions of their genome. These observations are also in line with the absolute requirement of the enhancer during the acute MCMV infection (17).
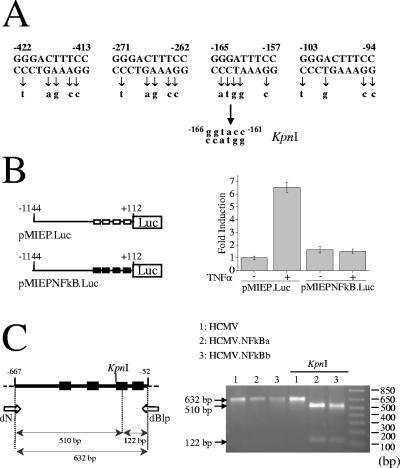
To directly analyze the role of NF-κB in regulating the MIEP, we specifically disrupted the four enhancer NF-κB binding sites. The point mutations introduced in the MIEP (3) are illustrated in Fig. 2A. It should be noted that these four sites are the only NF-κB binding sites present in the MIE enhancer and that, in the context of the whole genome, NF-κB recognition sites have only been described to date in another location, the US3 immediately-early (IE) enhancer (9). Transfection of human U937 cells with reporter plasmids containing either the wild-type or the NF-κB mutant MIEP revealed that tumor necrosis factor alpha (TNF-α)-induced MIEP activity was abolished in the mutant, whereas basal transcription was not affected (Fig. 2B). These results are consistent with previous studies indicating a role for NF-κB in regulating MIEP activity in transfection-based assays. To then test whether the four cognate NF-κB response elements in the MIEP are required for HCMV replication, two independent HCMV recombinants mutated in the NF-κB binding sites were generated (HCMV.NFkBa and HCMV.NFkBb). HCMV.NFkBa was constructed by using the ET mutagenesis method and subsequent transfection of the mutant BAC in MRC-5 cells, whereas HCMV.NFkBb was generated by cotransfecting MRC-5 cells with HCMVdE, pSVH, and pUChEnh.NFkB, a plasmid that carries HCMV sequences from nucleotide 171443 to 176844 (10) in which the four NF-κB elements of the enhancer were mutated. Infectious recombinant viruses were recovered from the transfections, used to infect new cell monolayers, and plaque purified three times. Viral stocks of both HCMV.NFkBa and HCMV.NFkBb were prepared, their genomic integrity was verified by restriction digestion (data not shown), and the successful disruption of the enhancer NF-κB binding sites was confirmed by PCR analysis (Fig. 2C) and the nucleotide sequence of the MIEP region (data not shown).
FIG. 2.
Construction of recombinant HCMVs with point mutations in the NF-κB binding sites of the enhancer region. (A) The sequence and location of the four NF-κB binding sites within the HCMV MIEP are shown, and the point mutations introduced in each specific element are indicated below the wild-type sequence. Mutation of the NF-κB recognition site at position −157 to −165 generates a KpnI restriction site at position −161 to −166, which is shown. Coordinates refer to the HCMV ie1/ie2 transcription start site. (B) The structures of the luciferase (Luc) reporter constructs pMIEP.Luc and pMIEPNFkB.Luc containing MIEP sequences from −1144 to +112 (relative to the HCMV ie1/ie2 transcription start site) without (□) and with (▪) the NF-κB binding sites disrupted, respectively, are shown. 5 × 105 U937 cells were electroporated with 2 μg of pMIEP.Luc or pMIEPNFkB.Luc, along with 0.6 μg of the internal control plasmid pRL-TK, and cultured for 46 h before the luciferase activity was assayed. Six hours before harvesting, cultures were treated with TNF-α (10 ng/ml; +) or vehicle (phosphate-buffered saline; −). The results are presented as the fold induction, taking “1” as the activity presented by the pMIEP.Luc in the absence of TNF-α. The values shown represent the average ± the standard deviation (bars) of four determinations. (C) Schematic diagram of the HCMV MIEP enhancer region, with the location of the four NF-κB binding sites marked by solid boxes. The KpnI restriction site at position −161 to −166 introduced when the NF-κB binding site located between positions −157 and −165 is mutated is indicated, as well as the sizes of the expected KpnI fragments derived from the PCR-amplified enhancer fragment with primers dN and dBlp (3) (flanking the enhancer and indicated with white arrows). Coordinates refer to the HCMV ie1/ie2 transcription start site. To confirm the correct mutagenesis of the NF-κB binding sites, enhancer sequences were amplified from stocks of HCMV (lanes 1 and 4), HCMV.NFkBa (lanes 2 and 5), and HCMV.NFkBb (lanes 3 and 6) by PCR. Marked by arrows are the amplified products before (lanes 1 to 3) or after (lanes 4 to 6) digestion with KpnI resolved by gel electrophoresis. Size markers are shown at the right margin.
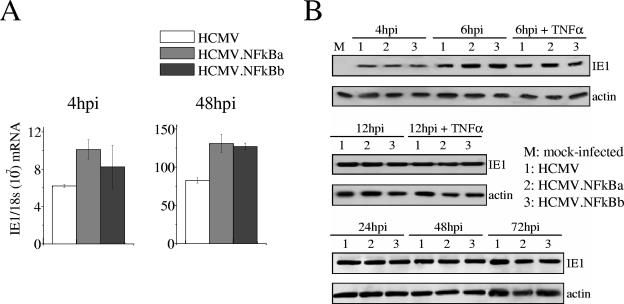
We next assessed the effect of abrogating the enhancer NF-κB recognition sites on expression emanating from the MIEP in the context of the infection of human embryonic lung (HEL) fibroblasts with either wild-type or mutant viruses. As shown by real-time PCR, IE1 mRNA levels were comparable or even slightly higher (48 h postinfection [hpi]) in cells infected with HCMV.NFkBa and HCMV.NFkBb than in parental HCMV-infected cells (Fig. 3A). In addition, HEL cells were infected throughout a 72-h period with the three viruses and subjected to Western blots by using a monoclonal antibody specific for the IE1 protein. We could not detect significant differences in the expression of the IE1 protein between HCMV and the HCMV.NFkB mutants at any of the time points analyzed (Fig. 3B). Furthermore, treatment of cells with the NF-κB inducer TNF-α (Fig. 3B) did not result in differential expression of IE1 in cells infected with wild-type or mutant viruses. Consequently, we conclude that the NF-κB binding sites in the MIEP do not significantly influence ie1 gene transcription or expression in lytically infected fibroblasts.
FIG. 3.
Analysis of ie1 gene expression in HCMV.NFkB-infected cells. (A) Real-time PCR analysis of ie1 RNA expression in cells infected with HCMV.NFkB. HEL fibroblasts were infected at an MOI of 0.01 with HCMV, HCMV.NFkBa, or HCMV.NFkBb and harvested at the time points after infection indicated for isolation of RNA and subsequent analysis by real-time PCR using primers within exon 4 of the HCMV ie1 gene as previously described (3). The results are presented as the relative amount of ie1 mRNA normalized to 18S rRNA, and error bars represent the standard errors of the means. (B) Expression kinetics of the IE1 protein by HCMV.NFkB mutants. HEL fibroblasts were mock infected or infected at an MOI of 0.6 (for the 4-, 6-, and 12-h time points) or 0.1 (for the 24, 48, and 72 h time points). Where indicated, cells were treated with 10 ng of TNF-α/ml 2 h before infection, during, and immediately after the adsorption period. At the indicated time (in hours) postinfection (hpi), samples were lysed, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 7% gels, transferred to nitrocellulose as previously described (1), and probed with an HCMV IE1 specific monoclonal antibody (MAB810; Chemicon, Temecula, CA). As an internal control, actin immunodetection was performed with a monoclonal antibody (A2066; Sigma, St. Louis, MO).
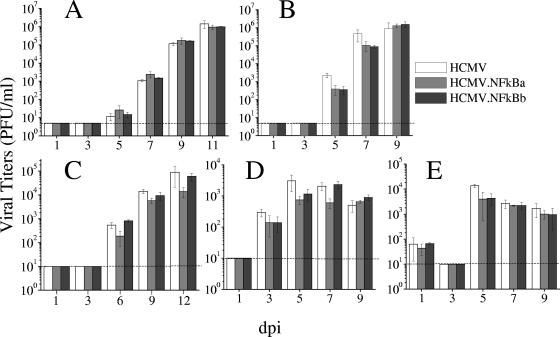
To examine the requirement for the enhancer NF-κB binding elements on HCMV growth, kinetic studies with HCMV, HCMV.NFkBa, or HCMV.NFkBb were performed on HEL fibroblasts. No difference in viral production at any time point was observed between the mutants and the parental virus (Fig. 4A). Treatment of cultures with TNF-α drastically inhibited HCMV growth, likely due to the induction of beta interferon (4; data not shown). In order to examine the replication capacity of HCMV.NFkB mutants in other cell types, we tested lung fibroblasts (MRC-5), U373 MG cells derived from glioblastoma, retinal pigment epithelium (RPE) cells, and differentiated embryonal carcinoma cells NTERA2 (NT2/D1). Importantly, parental and mutant viruses replicated in a comparable manner (Fig. 4B to E), strongly suggesting a neutral role of NF-κB for HCMV acute transcription/replication cycle in different cell types in culture.
FIG. 4.
Growth kinetics of HCMV.NFkB mutants. HEL (A), MRC-5 (B), RPE (C), U373 (D), and NT2/D1 cells (E; differentiated for 5 days with 10−5 M retinoic acid) were infected at an MOI of 0.025 (HEL and MRC-5) or 1 (RPE, U373, and NT2/D1, resulting in ca. 10 to 25% of GFP-positive cells in the culture at 48 hpi) with HCMV, HCMV.NFkBa, or HCMV.NFkBb. At the indicated time in days postinfection, the amount of extracellular (HEL, MRC-5, and RPE) or cell associated (U373 and NT2/D1) infectious virus present in the cultures was determined by plaque titration assays on MRC-5 cells. Each datum point represents the average and standard deviation from three separate cultures. Dashed lines represent the limits of detection.
The molecular details on the activation of the MIEP during HCMV infection are still poorly understood. In the present study, we demonstrate by generating and characterizing HCMV recombinants with alterations in the MIEP enhancer that (i) the enhancer region is necessary for HCMV growth and (ii) the MIEP NF-κB response elements do not contribute to lytic replication of HCMV in multiple cell types.
The involvement of NF-κB activity on MIE expression and HCMV replication has been a controversial issue (3, 4, 8, 11-15). The discrepancy pivots around whether NF-κB directly or indirectly influences HCMV transcription and/or replication. It is generally assumed in the field that the activation of NF-κB results in the direct stimulation of the MIEP and, as a consequence, in increased, i.e., gene expression and viral replication. Our observations are in agreement with the fact that NF-κB contributes to HCMV MIEP-enhancer activation in transient-transfection assays, as has been documented in a number of reports. However, we clearly show here that the cognate NF-κB binding sites within the enhancer do not play a major independent role in the transcription/replication strategies of HCMV in a variety of cell types. We demonstrate in a direct manner that, in the context of the infection, the enhancer is insensitive to mutations in the NF-κB binding sites, underlining a high level of robustness of the associated regulatory network controlling this region. While these results are in line with previous observations (3, 11, 14, 15), they are in contrast with others (8, 12, 13). In these apparently contradictory studies, a positive involvement of NF-κB in MIEP transcription and HCMV replication has been observed by blocking NF-κB activity using a variety of pharmacologic agents exhibiting a range of specificity and selectivity. These studies provide an indirect test and, moreover, the selectivity of the agents (e.g., aspirin or MG-132) used to inhibit NF-κB signaling pathways should be taken in consideration. It must be also noted that although to date only NF-κB sites have been found in the MIEP and in the US3 IE enhancer (9), the presence of additional NF-κB responsive genes in the HCMV genome could account in part for some of the discrepancies found in different studies.
Our data do not exclude the possibility that NF-κB may regulate HCMV MIE gene expression or growth in cell types not examined here or are required for in vivo replication and/or reactivation from latency. The fact that the viruses used in the present study derive from the HCMV laboratory strain AD169 prevented their analysis in other cell types more relevant for HCMV infection, including macrophages, endothelial, or dendritic cells. Mouse CMV recombinants containing the HCMV MIEP (with or without mutations in the NF-κB response elements) replicate and establish latency in the mouse (1, 3; A. Angulo unpublished results), permitting us to explore these aspects in the context of an in vivo infection.
Acknowledgments
We thank Anett Rassmann for technical assistance.
This study was supported by grants from the Ministerio de Ciencia y Tecnología (MCYT; SAF2002-00270 and SAF2005-05633 to A.A.), the Wellcome Trust (to P.G.), and the American Heart Association (0330064N to C.A.B.) and by the Deutsche Forschungsgemeinschaft (SFB 587, project A13 to M.M.). A.A. is a fellow from the Ramón y Cajal program (MCYT).
REFERENCES
- 1.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedict, C. A., A. Angulo, G. Patterson, S. Ha, H. Huang, M. Messerle, C. F. Ware, and P. Ghazal. 2004. Neutrality of the NF-κB-dependent pathway for human and murine cytomegalovirus transcription and replication in vitro. J. Virol. 78:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict, C. A., T. A. Banks, L. Senderowicz, M. Ko, W. J. Britt, A. Angulo, P. Ghazal, and C. F. Ware. 2001. Lymphotoxins and cytomegalovirus cooperatively induce interferon-beta, establishing host-virus detente. Immunity 15:617-626. [DOI] [PubMed] [Google Scholar]
- 5.Borst, E. M., and M. Messerle. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 79:3615-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, E., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate-early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 8.Caposio, P., M. Dreano, G. Garotta, G. Gribaudo, and S. Landolfo. 2004. Human cytomegalovirus stimulates cellular IKK2 activity and requires the enzyme for productive replication. J. Virol. 78:3190-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, Y.-J., W.-P. Tseng, and G. S. Hayward. 1996. Two distinct upstream regulatory domains containing multicopy cellular transcription factor binding sites provide basal repression and inducible enhancer characteristics to the immediate-early IES (US3) promoter from human cytomegalovirus. J. Virol. 70:5312-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M., S. A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barel. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microb. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 11.Cinatl, J., S. Margraf, J.-U. Vogel, M. Scholz, J. Cinatl, and H. W. Doerr. 2001. Human cytomegalovirus circumvents NF-κB dependence in retinal pigment epithelial cells. J. Immunol. 167:1900-1908. [DOI] [PubMed] [Google Scholar]
- 12.DeMeritt, I. B., J. P. Podduturi, A. M. Tilley, M. T. Nogalski, and A. D. Yurochko. 2006. Prolonged activation of NF-κB by human cytomegalovirus promotes efficient viral replication and late gene expression. Virology 346:15-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMeritt, I. B., L. E. Milford, and A. D. Yurochko. 2004. Activation of the NFκB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eickhoff, J. E., and M. Cotten. 2005. NF-κB activation can mediate inhibition of human cytomegalovirus replication. J. Gen. Virol. 86:285-295. [DOI] [PubMed] [Google Scholar]
- 15.Eickhoff, J., M. Hanke, M. Stein-Gerlach, T. P. Kiang, K. Herzberger, P. Habenberger, S. Muller, B. Klebl, M. Marschall, T. Stamminger, and M. Cotton. 2004. RICK activates a NF-κB-dependent anti-human cytomegalovirus response. J. Biol. Chem. 279:9642-9652. [DOI] [PubMed] [Google Scholar]
- 16.Ghazal, P., and J. A. Nelson. 1993. Transcription factors and viral regulatory proteins as potential mediators of human cytomegalovirus pathogenesis, p. 360-383. In Y. Becker, G. Darai, and E.-S. Huang (ed.), Molecular aspects of human cytomegalovirus diseases. Springer-Verlag Publishers, Heidelberg, Germany.
- 17.Ghazal, P., M. Messerle, K. Osborn, and A. Angulo. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isomura, H., T. Tsurumi, and M. F. Stinski. 2004. Role of the proximal enhancer of the major immediate-early promoter in human cytomegalovirus replication. J. Virol. 78:12788-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalik, T. F., B. Wing, J. S. Haskill, J. C. Azizkhan, A. S. Baldwin, and E. S. Huang. 1993. Multiple mechanisms are implicated in the regulation of NF-κB activity during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 90:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, Y., W.-J. Sohn, D.-S. Kim, and H.-J. Kwon. 2004. NF-κB- and c-Jun-dependent regulation of human cytomegalovirus immediate-early gene enhancer/promoter in response to lipopolysaccharide and bacterial CpG-oligodeoxynucleotides in macrophage cell line RAW 264.7. Eur. J. Biochem. 271:1094-1105. [DOI] [PubMed] [Google Scholar]
- 21.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier, J. L., and M. F. Stinski. 1996. Regulation of cytomegalovirus immediate-early genes. Intervirology 39:331-342. [DOI] [PubMed] [Google Scholar]
- 23.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 24.Muyrers, J. P., Y. Zhang, V. Benes, G. Testa, W. Ansorge, and A. F. Stewart. 2000. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 3:239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosch, S., K. Staak, J. Stein, C. Liebenthal, T. Stamminger, H.-D. Volk, and D. H. Kruger. 1995. Stimulation of HCMV cytomegalovirus IE enhancer/promoter in HL-60 cells by TNFα is mediated via induction of NF-κB. Virology 208:197-206. [DOI] [PubMed] [Google Scholar]
- 26.Prosch, S., R. Wuttke, D. H. Kruger, and H.-D. Volk. 2002. NF-κB-A potential therapeutic target for inhibition of human cytomegalovirus (re)activation? Biol. Chem. 383:1601-1609. [DOI] [PubMed] [Google Scholar]
- 27.Sambucetti, L. C., J. M. Cherrington, G. W. G. Wilkinson, and E. S. Mocarski. 1989. NF-κB activation of the human cytomegalovirus enhancer is mediated by a viral transactivator and by T-cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spector, D. H., L. Hock, and J. C. Tamashiro. 1982. Cleavage maps for human cytomegalovirus DNA strain AD169 for restriction endonucleases EcoRI, BglII, and HindIII. J. Virol. 42:558-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yurochko, A. D., E. S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E. S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]