Abstract
The Rous sarcoma virus gag gene contains a cis-acting negative regulator of splicing (NRS) element that is implicated in viral polyadenylation regulation. To study the mechanism of polyadenylation promotion at the viral poly(A) site located over 8 kb downstream, we performed in vitro polyadenylation analysis. RNA containing only the poly(A) site and flanking sequences in the 3′ long terminal repeat (LTR) was not polyadenylated detectably in vitro; however, if the transcript contained the NRS upstream of the LTR, polyadenylation was observed. Insertion of the viral env 3′ splice site sequence between the NRS and the LTR did not alter the level of polyadenylation appreciably. We conclude that the NRS promotes polyadenylation in vitro and can do so without formation of a splicing complex with a 3′ splice site. We then explored the roles of several cellular factors in NRS-mediated polyadenylation. Mutation of the binding sites of U1 and U11 snRNPs to the NRS did not affect polyadenylation, whereas hnRNP H strongly inhibited polyadenylation. We propose a model in which hnRNP H and SR proteins compete for binding to the NRS. Bound SR proteins may bridge between the NRS and the 3′ LTR and aid in the recruitment of the 3′-end processing machinery.
The replication of Rous sarcoma virus (RSV), like that of all retroviruses, requires unspliced RNA in the cytoplasm, where it functions in a dual role as mRNA for gag-pol translation and as packaged genomic RNA (reviewed in reference 8). Because retroviruses depend on the cellular machinery for RNA transcription, processing, export, and translation, they have evolved special mechanisms to aid in the accumulation of unspliced RNA. Some complex retroviruses, such as human immunodeficiency virus type 1, encode accessory proteins to promote export of incompletely spliced RNAs; however, simple retroviruses like RSV have not been found to encode accessory proteins (6, 8) and instead use cis-acting RNA sequences to interact with cellular factors. Avian retroviruses have been found to contain weak 3′ splice sites and a cis-acting negative regulator of splicing (NRS) element within the gag gene, which contribute to the maintenance of unspliced retroviral RNA (2, 13, 33). In addition, two direct repeat sequences, which flank the src gene of RSV, are involved in unspliced RNA export (37). The RSV polyadenylation signal (AAUAAA) is in the U3 region of the 3′ long terminal repeat (LTR) and is associated with a downstream GU-rich sequence (18). However, polyadenylation of avian retroviruses is relatively inefficient, and the viral poly(A) site is read through about 15% of the time (19).
The NRS is a 230-nucleotide (nt) RNA sequence which has two functional domains: (i) SR and hnRNP H protein-binding domains in its purine-rich 5′ portion (12, 30) and (ii) overlapping U1 and U11 small nuclear ribonucleoprotein (snRNP) binding sites at its 3′ end (5, 15, 20, 32, 39). The NRS element has been proposed to act as a decoy 5′ splice site, which generates a nonproductively spliced complex with a downstream 3′ splice site (9, 13, 14). Previous evidence suggests that, in addition to its role in splicing inhibition, the NRS may also stimulate RSV polyadenylation. Miller and Stoltzfus (34) showed that large deletions, which overlapped in a 606-nt region including the NRS, led to a large increase in readthrough transcription of the viral RNA. In addition, deletion of a region containing the env 3′ splice site sequence also led to increased readthrough (34).
Recently, more-limited mutations in the NRS have also been shown to increase the level of readthrough transcription. O'Sullivan et al. (38) showed that point mutations in the NRS pseudo-5′ splice site sequence lead to increased readthrough of the viral poly(A) site. A 42-nt deletion in the purine-rich 5′ portion of the NRS also causes increased readthrough (40). Avian leukosis viruses (ALVs) bearing NRS mutations induce short-latency B-cell lymphomas at a high frequency after infection of chicken embryos (23, 40, 43). In these clonal tumors, readthrough transcripts from ALV into the downstream cellular myb gene have been observed; the presence of these transcripts results in overexpression of an oncogenic, truncated Myb protein (23, 43). Finally, Fogel et al. (11) carried out extensive mutagenesis of the 5′ portion of the NRS and found that the polyadenylation-stimulatory activity of the NRS in vivo does not require hnRNP H but is dependent upon the binding sites for SR proteins and snRNPs. Thus, there is evidence to show that RSV RNA splicing and polyadenylation regulation are coupled by the NRS, but it is not known how this occurs mechanistically or what NRS-bound factors are functionally important.
mRNA 3′-end processing has been shown many times to be closely coupled with other gene expression events in the nucleus. Recognition of the cleavage and polyadenylation signal by cleavage factors is important for transcript termination, and several general polyadenylation factors can be found in association with the C-terminal domain of RNA polymerase II (21, 28). In addition, the efficiency of removal of upstream introns is promoted by polyadenylation (35, 36), just as splicing factors that interact with the terminal 3′ intron reciprocally promote 3′-end processing (28, 29, 44). Polyadenylation itself can be a regulated event; for example, several cellular and viral genes contain alternative polyadenylation signals that are used differentially during development or in different tissues (10). It is of interest to study how polyadenylation signals are regulated and gain further insight into the tightly coupled interactions in gene expression. The mechanism by which the NRS promotes 3′-end processing is novel because most auxiliary elements previously shown to regulate polyadenylation are located within close proximity to the core polyadenylation elements (1, 17, 25), whereas the NRS is located over 8 kilobases upstream of the polyadenylation site of the primary RSV RNA transcript.
To investigate the mechanism by which the NRS promotes polyadenylation at the 3′ LTR from a distance, we used in vitro analysis. We found that the NRS is able to promote polyadenylation directly without the need for an intervening, downstream 3′ splice site. Additionally, we found that hnRNP H strongly inhibits polyadenylation. We suggest that there is a competition between hnRNP H and a polyadenylation-stimulatory factor(s), possibly SR proteins, for binding to the NRS. Further, we propose that SR proteins are able to form a bridging interaction between the NRS and the polyadenylation site and use protein-protein interactions to recruit the 3′-end processing machinery.
MATERIALS AND METHODS
Plasmid constructions.
The 3′ LTR sequence of the PrC strain of RSV from nt 9196 to 9392 was synthesized by PCR using forward (5′-GATTCTAGGATCCGAAGGTATCAGACGGGTCTAACATG) and reverse (5′-CTTAGGAATTCAAATGAAGCCTTCTG) oligonucleotide primers (Operon). This PCR product was inserted into the BamHI and EcoRI sites of pBluescript II KS(+) (Stratagene) to generate the LTR construct. The NRS sequence from nt 706 to 1007 was synthesized by PCR using forward (5′-CATGCTTCTAGACCGGAGTGCATCGAG) and reverse (5′-CTAGCAGGATCCCTGGCTCCGCCC) oligonucleotide primers and inserted into the XbaI and BamHI sites of the LTR construct to produce the NRS LTR construct. The triple point mutant NRS.1 was described previously (15, 20, 38). The env sequence from nt 4995 to 5130, including the 3′ splice site at nt 5078, was synthesized by PCR using forward (5′-GATCGTGGATCCGGTACCCTCTCGAAAAG) and reverse (5′-GCTACAGGGATCCTTTCTCCTTGGAGTCC) oligonucleotide primers and inserted into the BamHI restriction site of the LTR construct to yield the ENV LTR construct. The env 3′ splice site fragment was similarly amplified and inserted into the NRS LTR construct to make the NRS ENV LTR construct.
In vitro polyadenylation assays and poly(A) selections.
Capped, 32P-labeled runoff transcripts were synthesized by in vitro transcription using T7 RNA polymerase as previously described (13). Plasmid templates were linearized with HindIII, and transcripts were purified from 6% polyacrylamide-8 M urea gels prior to use. HeLa nuclear extracts were prepared as previously described (27) from HeLa cells obtained from the National Cell Culture Center. A typical 13-μl in vitro polyadenylation reaction mixture contained a final concentration of 2.3% polyvinyl alcohol, 15 mM creatine phosphate, 1 mM ATP, 1 mM magnesium chloride, and 56% (vol/vol) HeLa nuclear extract. We used adeno-associated virus (AAV) RNA made from a template obtained from Jeff Wilusz (7) as a positive control in the polyadenylation assays. Inactivation of U1 and U2 snRNAs using complementary oligonucleotides was carried out as described by Bruzik and Steitz (4). In reactions where hnRNP H protein in the extract was sequestered by GRS competitor RNA, a synthetic RNA oligomer (5′-GGGGGAGGUGUGGG) was added directly to reaction mixtures, as described previously (1).
Before addition of RNA, the reaction mixtures were preincubated for 20 min at 30 degrees. After the RNA was added, polyadenylation reaction mixtures were incubated for 60 min at 30°C. RNAs recovered from the reactions were poly(A) selected using an oligo(dT)-cellulose column to isolate poly(A)+ RNA (42), separated on a 6% polyacrylamide-8 M urea gel, and quantified on an InstantImager (Packard).
Cross-linking assays.
32P-labeled NRS RNAs were UV cross-linked to proteins in HeLa nuclear extract, digested with RNase A, and electrophoresed on sodium dodecyl sulfate-polyacrylamide gels as described by Wilusz and Shenk (45).
RESULTS
The NRS promotes and is required for polyadenylation at the 3′ LTR in vitro.
In previous studies, it has been shown that mutations in the NRS element of RSV affect splicing regulation and cause readthrough of the cleavage/polyadenylation site in the 3′ LTR (11, 34, 38, 40). We wanted to use in vitro analysis to determine the mechanism by which the NRS promotes viral RNA polyadenylation at a site more than 8 kb downstream. We first wished to determine if a transcript containing the poly(A) signal in the RSV LTR, together with flanking sequences, can be polyadenylated in vitro.
For this purpose, capped, 32P-labeled runoff transcripts were synthesized; these transcripts contained a portion of the LTR (nucleotides 9196 to 9392) that flanks the major poly(A) site at nt 9312 (Fig. 1A). In vitro polyadenylation assays were then performed using HeLa nuclear extract. After incubation with the extract, RNAs were recovered and analyzed directly by polyacrylamide gel electrophoresis. In some cases, the recovered RNAs were also fractionated on an oligo(dT)-cellulose column to select for poly(A)+ RNA. As a positive control, we analyzed the polyadenylation of an AAV polyadenylation substrate (7).
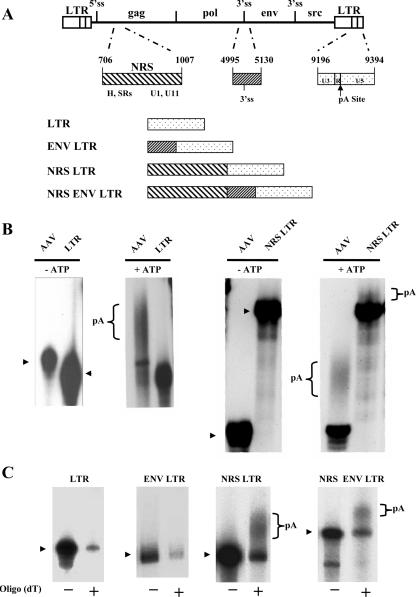
FIG. 1.
The NRS promotes polyadenylation in vitro. (A) Schematic representation of the RSV proviral genome with an expanded view of the NRS, the region flanking the env 3′ splice site, and the portion of the LTR used in this study. The U3 region of the LTR begins at nucleotide 9058, and the major cleavage/polyadenylation (pA) site is at nt 9312 (arrow). hnRNP H and SR proteins have been shown to bind the 5′ end of the NRS, whereas U1 and U11 snRNPs bind at the 3′ end. Polyadenylation substrates are schematically represented. (B) In vitro polyadenylation assays. The indicated radiolabeled, capped RNAs were incubated in in vitro polyadenylation reaction mixtures containing HeLa cell nuclear extract in the presence or absence of ATP. The arrowheads indicate the precursor RNA. (C) After in vitro polyadenylation reactions were performed with the indicated transcripts, poly(A)+ RNA was isolated using an oligo(dT) column. −, unpolyadenylated RNA that flowed through the column; +, poly(A)+ RNA eluted. The arrowhead at the left of each autoradiogram indicates the precursor RNA. Polyadenylated transcripts in the NRS LTR and NRS ENV LTR are denoted reactions.
While the control AAV RNA was efficiently polyadenylated in our in vitro reactions, the RSV LTR substrate was not detectably polyadenylated (Fig. 1B). To further analyze the LTR RNA after incubation in the polyadenylation reaction, it was run through an oligo(dT)-cellulose column to select for poly(A)+ RNA. While a small amount of LTR RNA was eluted from the oligo(dT) column, it was the same size as the input RNA (Fig. 1C). Thus, the 3′ LTR alone showed no detectable polyadenylation in this assay, suggesting that other viral elements may be necessary.
Miller and Stoltzfus (34) previously showed that the efficiency of RSV 3′-end processing was decreased by deleting either of two upstream RSV RNA sequences: (i) a 520-nucleotide sequence (nt 630 to 1149) in the gag gene containing the NRS as well as flanking sequences and (ii) a 136-nucleotide sequence (nt 4995 to 5130) spanning the env 3′ splice site (nt 5078), which included exonic splicing enhancer sequences (24). Consequently, we wanted to determine whether either of these sequences promotes polyadenylation in vitro.
First, the region spanning the env 3′ splice site from nucleotides 4995 to 5130 was cloned in front of the 3′ LTR to generate the ENV LTR construct (Fig. 1A). Like the 3′ LTR alone, this construct was not polyadenylated (Fig. 1C), suggesting that this env fragment is not sufficient to promote polyadenylation in vitro. It is possible that a larger region, encompassing this env gene sequence, may be necessary to play a role in polyadenylation. Alternatively, additional viral elements not included in our ENV LTR construct may be required for such an effect.
Next, to test whether the NRS promotes polyadenylation in our in vitro system, a 301-nucleotide fragment containing the NRS was cloned in front of the 3′ LTR to generate the NRS LTR construct (Fig. 1A). Because the NRS has been proposed to act as a decoy 5′ splice site, which generates a nonproductively spliced complex with a downstream 3′ splice site (9, 13, 14), we also made a construct, named NRS ENV LTR. This construct contained the NRS, the env 3′ splice site fragment described above, and the 3′ LTR, to test whether formation of a splicing complex is necessary to promote polyadenylation. As shown in Fig. 1B and C, the NRS LTR and NRS ENV LTR transcripts were both polyadenylated, showing that the NRS promotes polyadenylation in vitro (greater-than-10-fold activation compared to the LTR alone) and that it does so directly without the need to form a complex with a 3′ splice site.
It should be noted that the polyadenylation efficiency of the NRS LTR transcript was appreciably less than that of the AAV RNA analyzed in parallel as a positive control (Fig. 1B). Averaging four experiments, the NRS ENV LTR transcript was polyadenylated at a level similar to the NRS LTR (6.1% ± 4.5% more efficiently than NRS LTR transcript, which is not statistically significant). The nuclear extract and conditions used for the in vitro polyadenylation assays are nearly identical to those used in our laboratory for in vitro splicing (13, 15). Because the addition of the env fragment between the NRS and LTR had a minimal effect on polyadenylation efficiency, we focused our next studies on the NRS LTR construct to determine how the NRS promotes polyadenylation.
A splicing complex containing U1 or U11 snRNP is not necessary for NRS-mediated polyadenylation.
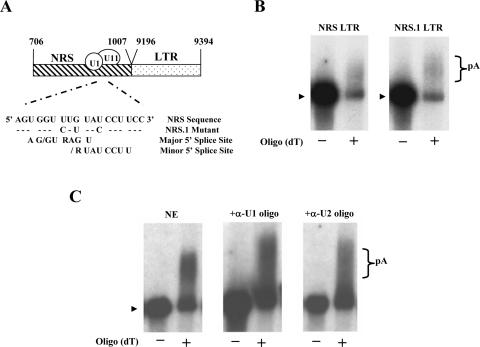
After showing that the NRS promotes polyadenylation in vitro in the absence of the env splice site sequence, we investigated the role that known NRS-binding factors may play in polyadenylation regulation in vitro. At the 3′ end of the NRS, there are overlapping U1 and U11 snRNP binding sites (Fig. 2A) (20). U1 snRNP binding has been previously shown to both promote and inhibit polyadenylation depending on the substrate (17, 25). Our laboratory has shown that point mutations in the U1 binding site of the NRS in full-length viral constructs cause increased readthrough transcription in vivo, thus implicating U1 binding in polyadenylation regulation in RSV (38). In particular, the NRS.1 triple point mutation (diagrammed in Fig. 2A), which disrupts both the U1 and U11 binding sites, caused the greatest effect in that study by increasing readthrough transcription from the wild-type level of 15% to 23%. We thus chose the NRS.1 mutation to use in this study to investigate what effect U1/U11 binding may have on polyadenylation in vitro.
FIG. 2.
Effect of U1/U11 snRNP binding on polyadenylation. (A) Schematic of the NRS LTR construct used in this study and the mutation introduced to disrupt U1/U11 snRNP binding. The consensus major and minor 5′ splice site sequences are indicated. (B) In vitro polyadenylation assays. The indicated capped, radiolabeled RNAs were incubated in in vitro polyadenylation reaction mixtures containing HeLa cell nuclear extract, and then poly(A)+ RNA was isolated using an oligo(dT) column. −, unpolyadenylated RNA that flowed through the column; +, poly(A)+ RNA eluted after washes. (C) U1 and U2 snRNAs in the nuclear extract were inactivated using complementary oligonucleotides. In vitro polyadenylation reactions using the NRS LTR substrate were then performed, followed by isolation of poly(A)+ RNA using an oligo(dT) column.
The NRS.1 mutant NRS sequence was cloned next to the 3′ LTR, and we performed an in vitro polyadenylation assay followed by a selection for poly(A)+ RNA. We found that NRS.1 LTR was able to be polyadenylated only 2% ± 10% more efficiently than NRS LTR, which is a statistically negligible difference (Fig. 2B). This result, therefore, suggests that the U1/U11 binding sites of the NRS do not play an important role in the polyadenylation promotion activity of the NRS in vitro. It also supports the finding above that formation of a splicing complex is not necessary for the NRS to promote polyadenylation in vitro.
A possible role for U1 snRNP in polyadenylation regulation was further investigated using an oligonucleotide directed against the 5′ end of U1 snRNA to inactivate the U1 snRNP (4). As a control, we used a similar oligonucleotide directed against U2 snRNP. Polyadenylation of the NRS LTR transcript in vitro was not detectably affected by preincubation of the nuclear extract with either oligonucleotide (Fig. 2C).
hnRNP H binding to the NRS inhibits polyadenylation.
Because intact U1 and U11 snRNP binding sites were not necessary for the polyadenylation-stimulatory activity of the NRS, we next investigated the role that factors that bind the 5′ purine-rich region of the NRS may play. It has previously been shown that hnRNP H interacts with the NRS (12, 40). Additionally, hnRNP H is known to promote polyadenylation by binding downstream of the core polyadenylation signal of some cellular mRNAs (1, 3). hnRNP H-specific mutations in the RSV NRS do not affect the polyadenylation-stimulatory activity of the NRS in vivo; nevertheless, Fogel et al. suggested that hnRNP H may inhibit polyadenylation (11). We were thus interested in investigating the role of hnRNP H binding to the NRS in 3′-end processing in vitro.
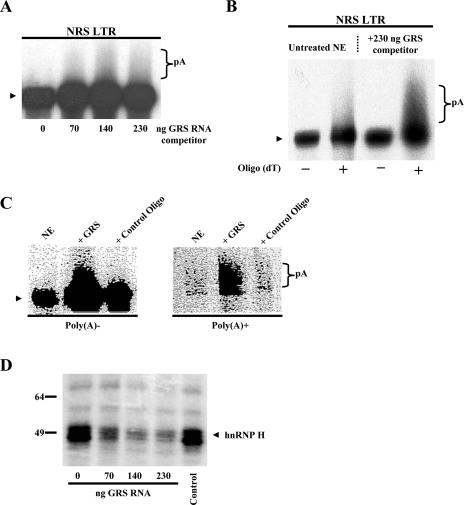
Rather than performing extensive mutagenesis of the NRS sequence, we chose to alter the level of available hnRNP H proteins present in the HeLa nuclear extract. This was carried out by adding a synthetic RNA oligonucleotide, GRS (5′-GGGGGAGGUGUGGG), which contains a G-rich binding site for hnRNP H protein, as described previously by Arhin et al. (1). To determine the effect of sequestration of hnRNP H protein on the processing efficiency of the NRS LTR construct, increasing amounts of GRS RNA were added to the in vitro polyadenylation reaction mixtures (Fig. 3A). When 230 ng of competitor RNA was added to the nuclear extract, NRS LTR polyadenylation was 1.9-fold ± 0.1-fold greater than that observed in an untreated extract (Fig. 3B). As a control, an oligomer containing an unrelated sequence was also incubated with nuclear extract and found to have no effect on polyadenylation efficiency (Fig. 3C). We also observed dose-dependent inhibition of UV cross-linking of hnRNP H to the NRS RNA after incubation of the extract with the GRS competitor RNA but not with the control oligonucleotide (Fig. 3D). Thus, we propose that the increase in polyadenylation in the presence of the GRS RNA was likely due to the prevention of hnRNP H binding to the NRS.
FIG. 3.
hnRNP H inhibits polyadenylation. (A) To sequester endogenous hnRNP H, nuclear extract was supplemented with increasing amounts of GRS competitor RNA. In vitro polyadenylation reactions using the NRS LTR substrate were performed as described in Materials and Methods. (B) Poly(A)+ RNA was subsequently isolated using an oligo(dT) column. −, unpolyadenylated RNA that flowed through the column; +, poly(A)+ RNA eluted. (C) A control oligonucleotide containing an unrelated sequence had no effect on the polyadenylation efficiency of the NRS LTR substrate. In vitro polyadenylation reactions were performed followed by fractionation on an oligo(dT) column to separate poly(A)− and poly(A)+ RNA. Two hundred thirty nanograms of oligonucleotide was used. (D) UV cross-linking of hnRNP H (∼53 kDa) to the NRS RNA is inhibited after incubation of the extract with increasing amounts of GRS competitor RNA but not with 230 ng of the control oligonucleotide. Protein molecular weight markers are indicated.
To verify that the increase in polyadenylation efficiency was due to decreased hnRNP H binding to the NRS rather than to the LTR RNA itself, we measured the processing efficiency of the LTR construct when GRS RNA was added to the polyadenylation reaction mixtures. Sequestering hnRNP H did not activate polyadenylation of the LTR alone (data not shown), suggesting that it is hnRNP H binding to the NRS that is regulating polyadenylation. We thus can conclude that hnRNP H binding to the NRS greatly inhibits polyadenylation and define a novel role for hnRNP H as an inhibitor of polyadenylation.
DISCUSSION
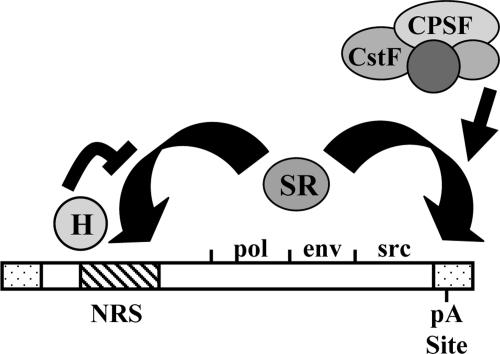
In this study, we investigated the mechanism by which the avian retroviral NRS element promotes polyadenylation at a site over 8 kb downstream in the 3′ LTR. While no detectable polyadenylation was observed in vitro with the LTR RNA substrate alone, addition of the NRS sequence upstream of the LTR enhanced polyadenylation. Surprisingly, the viral env 3′ splice site failed to enhance polyadenylation in vitro. Further, hnRNP H strongly inhibited the polyadenylation-stimulatory activity of the NRS. Since SR proteins have been shown previously to compete with hnRNP H for binding to the NRS (11, 12, 30), we suggest that SR proteins are responsible for the polyadenylation promotion activity of the NRS. We propose a model in which the NRS directly promotes polyadenylation by binding SR proteins, which form a bridge between the NRS and the 3′ LTR and help to recruit the polyadenylation machinery (Fig. 4).
FIG. 4.
Model for direct polyadenylation stimulation by the NRS. hnRNP H and SR proteins both competitively bind to the NRS. hnRNP H inhibits SR protein binding and thus inhibits the polyadenylation-stimulatory effect. We propose that SR proteins are able to bridge between the NRS and the polyadenylation site in the 3′ LTR and aid in the recruitment of the 3′-end processing machinery through protein-protein interactions.
Fogel et al. (11) previously suggested an alternative model in which the NRS promotes polyadenylation by forming a complex with downstream 3′ splice sites, thus stabilizing the binding of splicing factors at the weak viral 3′ splice sites. In contrast, our data show that an interaction with a 3′ splice site is not necessary for the NRS to promote polyadenylation; insertion of the env 3′ splice site between the NRS and the poly(A) signal did not increase levels of polyadenylation in vitro. Furthermore, neither U1 or U11 snRNP binding sites on the NRS nor an active U1 snRNP in the nuclear extract is required to promote polyadenylation of the NRS LTR transcript. It is possible that the env fragment we used was too small to have an effect or that a full-length viral RNA transcript is needed. Thus, we cannot rule out the possibility that additional polyadenylation enhancement is provided in vivo by the interaction of the NRS with the 3′ splice site fragment, which contains ESE sequences (24) that also bind SR proteins.
Sequestration of hnRNP H with an oligonucleotide prevented its cross-linking to the NRS RNA and dramatically increased the polyadenylation efficiency of the NRS LTR transcript. This suggests that binding of hnRNP H to the NRS interferes with the NRS polyadenylation-stimulatory activity. The purine-rich 5′ end of the NRS contains overlapping binding sites for hnRNP H and SR proteins (12, 30, 31, 40). SR proteins are essential, multifunctional splicing factors required for different steps of spliceosome assembly and are thought to mediate cross-exon and cross-intron interactions between splicing factors bound at the 5′ and 3′ splice sites (16). In addition, SR proteins have been demonstrated to be involved in RNA nuclear export, RNA stability, mRNA quality control, and translation (reviewed in reference 22). However, SR proteins have not been shown previously to be involved in promoting 3′-end processing.
We have previously shown that a 42-nt deletion in the purine-rich 5′ portion of the NRS causes reduced hnRNP H and SR protein binding as well as an increase in viral readthrough transcripts in vivo (40). This deletion in ALVs is associated with an increased incidence of rapid-onset lymphomas associated with readthrough into downstream cellular oncogenes (43). Integrating previous work with our current data, we propose the following model of how the NRS regulates polyadenylation at the 3′ LTR. hnRNP H and SR proteins compete for binding to the 5′ half of the NRS. When SR proteins, including SF2/ASF, bind to the NRS, they form a bridge between the NRS and the 3′ LTR. The arginine/serine-rich (RS) domains of the SR proteins then make protein-protein interactions with members of the cleavage and polyadenylation machinery, recruiting it to the cleavage/polyadenylation site in the 3′ LTR (Fig. 4).
We were surprised to observe that U1 snRNA inactivation had no effect in our study, despite a U1 binding site being only 170 nt upstream of the polyadenylation site in the NRS LTR construct. Our laboratory has previously shown that U1 snRNP binding site mutations in full-length viral constructs cause increased readthrough, suggesting that U1 promotes polyadenylation in vivo (38). It is also known that U1 binding sites near a polyadenylation site can strongly inhibit 3′-end processing, even at distances much longer than 170 nt (17, 41). However, in our in vitro study, mutating the U1 binding site had no effect on polyadenylation efficiency. We thus can only suggest that U1 may play a minor role in stimulating polyadenylation in vitro, but the stronger effects of SR proteins, hnRNP H, and other effectors are masking any observable effects. In addition, a spliceosome-like complex, involving U1 snRNP and 3′ splice sites (13), may aid in bringing the NRS and 3′ LTR spatially close together, but we propose that the SR proteins themselves play the major role in helping to recruit the 3′-end processing machinery.
It is now becoming clear that polyadenylation signals are very diverse and that AAUAAA is not ubiquitous at the 3′ ends of all eukaryotic mRNAs as once thought. Data from expressed sequence tag databases have suggested that the actual incidence of AAUAAA is much lower, perhaps as low as 50 to 60% (26). For proper gene expression, the correct cleavage and polyadenylation site must be identified, just as the proper 5′ and 3′ splice sites must be recognized within vast stretches of intronic RNA. SR proteins are known to bind to exonic splicing enhancers to recruit the splicing machinery to the proper splice sites. Similarly, SR proteins may bind to specific polyadenylation enhancer sequences (like the NRS) and help to define the proper cleavage/polyadenylation site and recruit the 3′-end processing machinery. In summary, we propose that SR proteins may be major regulators of all steps of mRNA processing, recognizing the proper RNA sequences and recruiting the proper machinery to those sites.
Acknowledgments
We thank Jeff Wilusz for reagents and much helpful advice, Yingying Li for technical assistance, and Yun-Xing Wang for T7 polymerase.
This work was supported by NIH research grant RO1 CA238796 to K.L.B. and a Pfizer Summer Undergraduate Research fellowship to J.E.W.
REFERENCES
- 1.Arhin, G., M. Boots, P. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrigo, S., and K. Beemon. 1988. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 8:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagga, P., G. Arhin, and J. Wilusz. 1998. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 26:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruzik, J. P., and J. A. Steitz. 1990. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell. 62:889-999. [DOI] [PubMed] [Google Scholar]
- 5.Cabello-Villegas, J., K. Giles, A. M. Soto, P. Yu, A. Mougin, K. L. Beemon, and Y. X. Wang. 2004. Solution structure of the pseudo-5′ splice site of a retroviral splicing suppressor. RNA 10:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlberg, K., and K. Beemon. 1988. Proposed gag-encoded transcriptional activator is not necessary for Rous sarcoma virus replication or transformation. J. Virol. 62:4003-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., and J. Wilusz. 1998. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 26:2891-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J., S. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 9.Cook, C., and M. McNally. 1999. Interaction between the negative regulator of splicing element and a 3′ splice site: requirement for U1 small nuclear ribonucleoprotein and the 3′ splice site branch point/pyrimidine tract. J. Virol. 73:2394-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards-Gilbert, G., K. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25:2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogel, B., L. McNally, and M. McNally. 2002. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 30:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogel, B., and M. McNally. 2000. A cellular protein, hnRNP H, binds to the negative regulator of splicing element for Rous sarcoma virus. J. Biol. Chem. 275:32371-32378. [DOI] [PubMed] [Google Scholar]
- 13.Giles, K. E., and K. L. Beemon. 2005. Retroviral splicing suppressor sequesters a 3′ splice site in a 50S aberrant splicing complex. Mol. Cell. Biol. 25:4397-4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gontarek, R. 1994. A Rous sarcoma virus intronic element negatively regulates splicing by binding snRNPs. Ph.D. thesis. Johns Hopkins University, Baltimore, Md.
- 15.Gontarek, R. R., M. T. McNally, and K. Beemon. 1993. Mutation of an RSV intronic element abolishes both U11/U12 binding and negative regulation of splicing. Genes Dev. 7:1926-1936. [DOI] [PubMed] [Google Scholar]
- 16.Graveley, B. 2000. Sorting out the complexity of SR protein functions. RNA. 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunderson, S., M. Polucarpou-Schwarz, and I. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70k and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 18.Guntaka, R. V. 1993. Transcription termination and polyadenylation in retroviruses. Microbiol. Rev. 57:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman, S. A., and J. M. Coffin. 1986. Differential transcription from the long terminal repeats of avian leukosis virus DNA. J. Virol. 60:497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibbert, C., R. Gontarek, and K. Beemon. 1999. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA 5:333-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose, Y., and J. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., and J. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, W., M. R. Kanter, I. Dunkel, R. G. Ramsay, K. L. Beemon, and W. S. Hayward. 1997. Minimal truncation of the c-myb gene product in rapid-onset B-cell lymphoma. J. Virol. 71:6526-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz, R. A., and A. M. Skalka. 1990. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz, C., K. Murthy, N. Schek, J. O'Conner, J. Manley, and J. Alwine. 1996. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 10:325-337. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald, C., and J. Redondo. 2002. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol. 190:1-8. [DOI] [PubMed] [Google Scholar]
- 27.Mayeda, A., and A. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 28.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. Patterson, M. Wickens, and D. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 29.McCracken, S., D. Longman, I. Johnstone, J. Caceres, and B. Blencowe. 2003. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 278:44153-44160. [DOI] [PubMed] [Google Scholar]
- 30.McNally, L., and M. McNally. 1996. SR protein factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 70:1163-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNally, L., and M. McNally. 1998. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 18:3103-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNally, L., and M. McNally. 1999. U1 small nuclear ribonucleoprotein and splicing inhibition by the Rous sarcoma virus negative regulator of splicing element. J. Virol. 73:2385-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNally, M., and K. Beemon. 1992. Intronic sequences and 3′ splice sites control Rous sarcoma virus splicing. J. Virol. 66:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J., and C. M. Stoltzfus. 1992. Two distant upstream regions containing a cis-acting signals regulating splicing facilitate 3′-end processing of avian sarcoma virus RNA. J. Virol. 66:4242-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesic, D., and L. Maquat. 1994. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 8:363-375. [DOI] [PubMed] [Google Scholar]
- 36.Niwa, M., S. Rose, and S. Berget. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4:1552-1559. [DOI] [PubMed] [Google Scholar]
- 37.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm and gag gene expression. J. Virol. 70:3834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Sullivan, C., T. Polony, R. Paca, and K. Beemon. 2002. Rous sarcoma virus negative regulator of splicing selectively suppresses src mRNA splicing and promotes polyadenylation. Virology 302:405-412. [DOI] [PubMed] [Google Scholar]
- 39.Paca, R. E., C. S. Hibbert, C. T. O'Sullivan, and K. L. Beemon. 2001. Retroviral splicing suppressor requires three nonconsensus uridines in a 5′ splice site-like sequence. J. Virol. 75:7763-7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polony, T., S. Bowers, P. Neiman, and K. Beemon. 2003. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J. Virol. 77:9378-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu, J., and D. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by the distance between the promoter and the intron and is inhibited by U1 small nuclear RNP binding to the intervening donor. J. Biol. Chem. 279:14889-14898. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Smith, M. R., R. E. Smith, I. Dunkel, V. Hou, K. L. Beemon, and W. S. Hayward. 1997. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J. Virol. 71:6534-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagner, S., C. Vagner, and I. Mattaj. 2000. The carboxy terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 14:403-413. [PMC free article] [PubMed] [Google Scholar]
- 45.Wilusz, J., and T. Shenk. 1988. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell 52:221-228. [DOI] [PubMed] [Google Scholar]