Abstract
The phosphoprotein (P protein) of vesicular stomatitis virus (VSV) is an essential subunit of the viral RNA-dependent RNA polymerase complex and plays a central role in viral transcription and replication. Using both the yeast two-hybrid system and coimmunoprecipitation assays, we confirmed the self-association of the P protein of Indiana serotype (Pind) and heterotypic interaction between Pind and the P protein of New Jersey serotype (Pnj). Furthermore, by using various truncation and deletion mutants of Pind, the self-association domain of the Pind protein was mapped to amino acids 161 to 210 within the hinge region. The self-association domain of Pind protein is not required for its binding to nucleocapsid and large proteins. We further demonstrated that the self-association domain of Pind protein is essential for VSV transcription in a minireplicon system and that a synthetic peptide spanning amino acids 191 to 210 in the self-association domain of Pind protein strongly inhibited the transcription of the VSV genome in vitro in a dose-dependent manner. These results indicated that the self-association domain of Pind protein plays a critical role in VSV transcription.
Vesicular stomatitis virus (VSV) is a negative-stranded RNA virus belonging to family Rhabdoviridae. It packages within the mature virion an RNA-dependent RNA polymerase that transcribes the negative-sense genome RNA into mRNAs (2) in the infected cells that are subsequently translated into five proteins: the glycoprotein (G protein), the matrix protein (M protein), the nucleoprotein (N protein), the RNA-dependent RNA polymerase large protein (L protein), and the phosphoprotein (P protein) (42). The N-RNA complex and the P and L proteins, which form the ribonucleoprotein (RNP) core complex, can be separately isolated and purified from the virions and can effectively reconstitute mRNA synthesis in vitro when these components are mixed (17, 20, 21). L protein by itself fails to transcribe the N-RNA template; P protein is absolutely required to elicit its RNA polymerase activity (17, 21, 36). P protein thus seems to act as a transcription factor for L protein. In addition, it may also help to stabilize the L protein from proteolytic degradation (7) and may be required for productive virion assembly (13). P protein also associates with newly synthesized N protein during infection to keep the latter in a soluble, replication-competent form (16, 34, 39) to enwrap the nascent RNA during replication (34). P protein also forms a tripartite replicase complex with L and N proteins, as L-N-P, which has been shown to convert N-RNA template to positive-sense genome RNP in vivo.
Pind protein has been arbitrarily divided into three domains, domains I, II, and III. Domain I (residues 1 to 137) is highly acidic in nature, and binds to the L protein (22). Domain I contains three phosphorylation sites (Ser-60, Thr-62, and Ser-64) that are phosphorylated by cellular casein kinase II and are indispensable for the transcriptional role of the P protein (5, 6, 9, 14, 28, 38, 44, 46). Domain I also contains the N protein binding site, which is important for viral replication (47). Domain II (residues 211 to 244) also binds to the L protein and contains two phosphorylation sites (Ser-226 and Ser-227), which have been shown to be required for optimal replication (9, 31). The protein kinase that carries out phosphorylation at these sites still remains unknown. The extreme C terminus (residues 245 to 265), which is designated domain III, is basic in nature and is essential for binding of the N-RNA template for genome RNA transcription (15). The fourth region of Pind protein is an undefined hypervariable hinge region that spans amino acid residues 138 to 210. This region is not required for binding to N or L protein (22, 47, 13) but seems to play a critical role in the assembly of infectious VSV (13). By X-ray crystal structure analysis, a proteinase K-resistant region spanning part of domain I and the hinge region was defined as the central domain (residues 107 to 177). This region was crystallized and designated the oligomerization domain (18, 19), and its structure was established.
We have recently undertaken a biochemical approach to precisely locate and map the self-association domain of the Pind protein. By a combination of a yeast two-hybrid system and an immunoprecipitation assay using a battery of mutants of Pind proteins, we have been able to precisely map the self-association domain, which spans residues 161 to 210 within the hinge domain. We have further shown that this domain is functionally essential for transcription in vitro and in vivo.
MATERIALS AND METHODS
Cells and viruses.
HeLa and BHK-21 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. VSV (Indiana serotype) Mudd Summers strains were propagated as described previously (3, 4) from BHK-21 cells by inoculation at a multiplicity of infection of 0.05. Recombinant vaccinia virus (vTF7-3) carrying the bacteriophage T7 RNA polymerase gene (23) was propagated as described by Li and Pattnaik (32).
Plasmid constructs.
pET3a-Pind (4), containing Pind cDNA, was used as a template in all PCR amplifications for construction of plasmids encoding wild-type Pind and mutant proteins (Fig. 1), and pET3a-Pnj (4), carrying Pnj cDNA, was used as a template in PCR amplification for construction of plasmids encoding wild-type Pnj protein. PCR products were subcloned in frame with the open reading frame (ORF) encoding the GAL4 DNA binding domain (BD) into pGBKT7 (Clontech) and in frame with the ORF encoding the GAL4 activating domain (AD) into pGADT7 (Clontech). The plasmid-encoded recombinant proteins were named BD-baits (proteins which contain a Myc tag between the N-terminal BD and C-terminal bait proteins) and AD-preys (proteins which contain a hemagglutinin [HA] tag between the N-terminal AD and C-terminal prey proteins). Detailed cloning strategies are available upon request. All constructs were verified by sequencing. pVSV-CAT2 was a kind gift from Sue A. Moyer (29). pBS-N, pBS-P, and pBS-L were kindly provided by John K. Rose (45).
FIG. 1.

Schematic representation of Pind protein and its mutants used in this study. The Pind protein is divided into four regions, here designated I, Hinge, II, and III. 60S, 62T, 64S, 226S, and 227S represent the phosphorylation sites in Pind. The numbers represent amino acid positions.
Yeast mating.
Plasmids encoding BD-baits and AD-preys were transformed into Saccharomyces cerevisiae AH109 (type a) and Y187 (type α) (Clontech), respectively, by using the lithium acetate method (26). All transformant clones were confirmed for correct protein expression before yeast mating assay. A mixture of a selected clone from AH109 and a selected clone from Y187 in 0.5 ml of YPDA medium (Clontech) was incubated at 30°C for 24 h with shaking at 200 rpm, and then 100-μl aliquots of the mating culture were spread onto an SD/−Trp/−Leu plate for diploid cell growth and onto an SD/−Trp/−Leu/−His/−Ade plate for selecting protein interaction, respectively. Clones growing on the SD/−Trp/−Leu or SD/−Trp/−Leu/−His/−Ade plates were further replicated on an SD/−Trp/−Leu/−His/−Ade plate containing 5-bromo-4-chloro-3-indolyl-α-galactopyranoside (X-α-Gal) for detecting blue colony growth. Empty AD and BD were used as negative controls. AD-T (simian virus 40 T antigen) and BD-p53 were used as positive controls.
In vivo coimmunoprecipitation.
Recombinant proteins were expressed by transient transfection. HeLa cells in six-well plates were infected with vTF7-3 for 1 h at a multiplicity of infection of 10, washed, and then transfected with the appropriate plasmids in the presence of Lipofectamine 2000 (Invitrogen). After 24 h of transfection, cell lysates were prepared as described elsewhere (10). In experiments to map the self-association domain of Pind protein, Myc-tagged Pind was coexpressed with HA-tagged Pind or mutants in various combinations. To detect interactions between mutants with the self-association domain deleted and N protein, Myc-tagged N protein of Indiana serotype (Nind) was coexpressed with HA-tagged Pind or mutants. Precleared supernatants of lysates were incubated with polyclonal anti-Myc antibody sc-789 (Santa Cruz) for 2 h at 4°C with rotation. After centrifugation, supernatants were mixed with protein G Sepharose 4 Fast Flow medium (Amersham Pharmacia Biotech) and incubated overnight. To detect interactions between mutants with the self-association domain deleted and L protein, Flag-tagged L protein (a kind gift from Daniel Shaji) was coexpressed with HA-tagged Pind or mutants. Precleared supernatants of lysates were incubated with polyclonal anti-L antibody (antiserum produced from immunized rabbits by using synthesized peptides) and mixed with protein G Sepharose 4 Fast Flow medium as described above. Beads were collected and washed five times with washing buffer as described elsewhere (10). The beads were boiled in Laemmli buffer and bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting using anti-Myc monoclonal antibody sc-40 (Santa Cruz), anti-HA antibody (Sigma), or anti-Flag antibody (Santa Cruz).
VSV CAT minigenome assay.
BHK-21 cells in six-well plates were infected with vTF7-3 at an multiplicity of infection of 4 for 1 h at 37°C and transfected with 1 μg pBS-N, 0.5 μg pBS-P or various quantities of the plasmids encoding HA-Pind or mutant proteins, 0.3 μg pBS-L, and 0.5 μg pVSV-CAT2 in Opti-MEM medium with Lipofectamine 2000. The cells were incubated for 40 h at 37°C, and then the transfection medium was aspirated. The cells were washed with cold phosphate-buffered saline twice and then lysed. After centrifugation of the cell lysates, the resulting supernatants were subjected to a chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA) for the detection of CAT expression according to the manufacturer's protocol (Roche) to monitor transcription of the minigenome mediated by L, P, and N proteins. All assays were tested at least twice in two separate experiments. Results were calculated as relative CAT expression levels.
Effects of peptides derived from the self-association domain in vitro transcription of VSV genome.
The N-RNA and L-P complexes were purified from VSV (Indiana serotype, Mudd Summers strain) particles as described elsewhere (T. Ogino and A. K. Banerjee, unpublished data). An in vitro transcription reaction was carried out under standard conditions with the N-RNA complex (75 ng of protein), the L-P complex (25 ng), and 100 μM [α-32P]GTP (1,400 cpm/pmol) as the labeled substrate in the presence or absence of various concentrations of synthetic peptides (peptide of residues 161 to 190 [P161-190], P191-210, and scrambled P191-210). 32P-labeled transcripts, treated with RNase H (Roche Molecular Biochemicals) in the presence of oligo(dT), were analyzed by electrophoresis in a 5% polyacrylamide gel containing 7 M urea, followed by autoradiography.
RESULTS
Pind self-association in a yeast two-hybrid system.
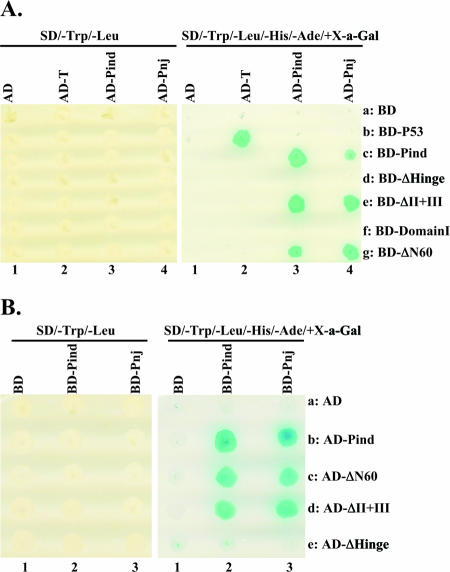
Fig. 1 depicts the boundaries of the proposed domains of Pind protein (265 amino acids) described above with three and two phosphorylation sites within domain I and domain II, respectively. Various deletion mutants were generated as described in Materials and Methods and were used to locate and map the self-association domain within the Pind protein. First, a yeast two-hybrid system was used to test protein-protein interaction. The cDNAs encoding the full length of Pind, Pnj, and some selected mutants of Pind proteins were fused in frame with the cDNA encoding GAL4-BD individually. The cDNA encoding the full length of Pind was also fused in frame with the cDNA encoding GAL4-AD. The recombinant plasmids encoding the BD- and AD-fused proteins were transformed into Saccharomyces cerevisiae AH109 and Y187, respectively. The expected chimeric proteins were expressed adequately in transformed yeasts (data not shown). Yeast mating was then performed, and the results are shown in Fig. 2A. Blue yeast growth on SD/−Trp/−Leu/−His/−Ade/+X-α-Gal plates was observed in the presence of BD-Pind plus AD-Pind or BD-p53 plus AD-T (positive control) (Fig. 2A, right panel, lanes 3c and 2b). No yeast growth was observed in SD/−Trp/−Leu/−His/−Ade-X-α-Gal plates in the presence of BD-Pind plus AD or AD-Pind plus BD (Fig. 2A, right panel, lanes 1c and 3a). Furthermore, heterotypic interaction was also observed between BD-Pind and AD-Pnj (Fig. 2A, right panel, lane 4c). Diploid cells were all able to grow on SD/−Trp/−Leu plates, indicating that yeasts were correctly mated in all mating combinations (Fig. 2A, left panel). These data clearly show that Pind protein could interact with itself and with Pnj protein and self-associate in the yeast two-hybrid system. Next, several deletion mutants of Pind protein (ΔII+III [with domains II and III deleted], ΔHinge [with the hinge region deleted], ΔN60 [with the N-terminal 60 amino acids deleted], and Domain I [consisting of residues 1 to 137]) (Fig. 1) fused with BD were used to study whether they also interact with AD-Pind. No yeast growth in SD/−Trp/−Leu/−His/−Ade/+X-α-Gal was observed when BD-Domain I and BD-ΔHinge were used as baits in yeast mating with AD-Pind (Fig. 2A, right panel, lanes 3d and 3f). However, BD-ΔII+III and BD-ΔN60 interacted efficiently with AD-Pind (Fig. 2A, right panel, lanes 3e and 3g), indicating that the N-terminal 60 amino acids and the C-terminal 55 amino acids (domains II and III) are not required for the self-association of Pind protein in the yeast two-hybrid system. Similar results were obtained when we switched Pind mutants from BD to AD (Fig. 2B), and a yeast two-hybrid assay was carried out as described above. Since BD-Domain I and BD-ΔHinge were unable to interact with Pind protein, we wanted to test whether only the hinge region was sufficient to mediate self-interaction of Pind protein. A plasmid encoding the ΔDomain I mutant (with residues 1 to 137 deleted) fused to BD was transformed into yeast strain AH109. However, we were consistently unable to detect any protein expression from this plasmid (data not shown) strongly implicating that domain I must contain a region(s) which is essential for Pind mRNA or protein stability.
FIG. 2.
Homotypic and heterotypic interactions of VSV P protein in the yeast two-hybrid assay. (A) The yeast AH109 strains (type a) expressing BD-baits were mated with the yeast Y187 strains (type α) expressing AD-preys in the indicated combinations. The yeast diploid cells (AH109/Y187) were plated onto the selective plates with SD/−Trp/−Leu, to confirm the presence of plasmids for BD-baits and AD-preys (left panel), and SD/−Trp/−Leu/−His/−Ade/+X-α-Gal (X-a-Gal), to detect interactions between BD-baits and AD-preys (right panel). Yeast coexpressing interacting proteins which activate the expression of reporter genes (HIS, ADE2, and MEL1) grows as blue colonies on the SD/−Trp/−Leu/−His/−Ade/+X-α-Gal plates. The diploid cells coexpressing AD-T and BD-p53 were used as positive controls. Experiments were repeated at least three times for consistency. (B) Plasmids encoding Pind and mutants were fused to AD (AD-preys). Plasmids encoding Pind and Pnj were fused to BD (BD-baits). The yeast two-hybrid assay was performed as described for panel A.
Mapping of the self-association domain of Pind protein by coimmunoprecipitation in vivo.
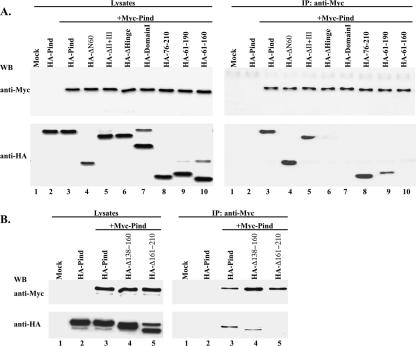
To further confirm Pind-Pind interaction and narrow down the self-association site of Pind protein, coimmunoprecipitation analyses in vivo were carried out using additional deletion and truncation mutants of Pind protein (Fig. 1). First, we tested Pind-Pind interaction in vivo by coexpressing Myc-Pind and HA-Pind in HeLa cells. Since the plasmids encoding BD-Pind and AD-Pind all use T7 promoter, which is located after the ORF of BD and AD, Pind protein was efficiently expressed in Myc- and HA-tagged forms, respectively, in mammalian cells expressing T7 polymerase, whereas the fusion proteins described above containing BD and AD were not expressed. As shown in Fig. 3A, left panel, using vTF7-3 expressing T7 polymerase, only Myc- and HA-tagged Pind were expressed in HeLa cells. When whole cell lysates were subjected to immunoprecipitation by anti-Myc polyclonal antibody, HA-Pind could be coimmunoprecipitated with Myc-Pind (Fig. 3A, right panel, lane 3), but HA-Pind alone could not be coimmunoprecipitated in the absence of Myc-Pind (Fig. 3A, right panel, lane 2). These results confirm that Pind protein self-association also occurs in vivo. Next, several Pind mutants that were used in the yeast two-hybrid system were coexpressed with Myc-Pind in vivo and tested for interactions by a coimmunoprecipitation assay. HA-ΔN60 and HA-ΔII+III, but not HA-Domain I and HA-ΔHinge, could be coimmunoprecipitated by Myc-Pind (Fig. 3A, right panel, lanes 4, 5, 6, and 7), confirming the results obtained from the yeast two-hybrid assay. Consistent with our findings described above, we failed to detect any protein expression of Myc-ΔDomain I or HA-ΔDomain I in HeLa cells lysates in the presence of T7 polymerase, supporting our contention that part of domain I contains elements that stabilize the Pind protein or its mRNA. Therefore, we concluded that amino acids 61 to 210, which span part of domain I, and the entire hinge region mediate self-association of Pind protein. To precisely map the self-association domain of Pind protein, three truncation mutants of Pind protein, HA with Pind residues 76 to 210 (HA-76-210), HA-61-190, and HA-61-160, were coexpressed with Myc-Pind, and the interaction was analyzed by a coimmunoprecipitation assay. As shown in Fig. 3A (right panel, lanes 8, 9, and 10), HA-76-210 was efficiently coimmunoprecipitated with Myc-Pind, but mutant HA-61-190 was coimmunoprecipitated only very little with Myc-Pind. Furthermore, HA-61-160 failed to interact with Myc-Pind in spite of a level of protein expression similar to that of HA-76-210 (Fig. 3A, left panel, lanes 8 and 10). These results establish that amino acid residues 161 to 210 are necessary for Pind protein self-association. To further confirm this result, two additional deletion mutants, HA plus Pind with residues 138 to 160 deleted (HA-Δ138-160) and HA-Δ161-210, were coexpressed with Myc-Pind (Fig. 3B, left panel, lanes 4 and 5). Only HA-Δ138-160, not HA-Δ161-210, could be coimmunoprecipitated with Myc-Pind (Fig. 3B, right panel, lanes 4 and 5), indicating that amino acid residues 161 to 210 (50 amino acid residues) indeed mediate self-association of Pind protein in vivo.
FIG. 3.
Mapping of the self-association domain of Pind protein. Myc-Pind and Pind or its mutants tagged with HA were coexpressed in HeLa cells as indicated. Cell lysates were assayed by Western blotting (WB) using anti-Myc and anti-HA monoclonal antibodies to confirm protein expression (A and B, left panel). Immunoprecipitation (IP) was performed using anti-Myc polyclonal antibodies, and the immune complexes were detected by anti-Myc and anti-HA monoclonal antibodies, respectively (right panels of panels A and B).
Monomeric Pind protein interacts with the N and L proteins.
Next, we were interested in determining whether Pind mutants that were not able to self-associate could still interact with N and L proteins. It has been previously shown that the N-terminal region of P protein binds to free N protein and that the C-terminal of P protein binds to N-RNA template (47). Our coimmunoprecipitation experiments clearly show that both HA-Δ161-210 and HA-ΔII+III could be coimmunoprecipitated with Myc-Nind when coexpressed with Myc-Nind (Fig. 4A, right panel, lanes 4 and 5). These results strongly suggest that the self-association domain of Pind protein is not required for its binding to Nind. Similarly, following coexpression of HA-Δ161-210 with Flag-L and HA-ΔII+III with Flag-L, both Pind mutant proteins were also coimmunoprecipated with anti-L antibody (Fig. 4B, right panel, lanes 4 and 5), indicating that the self-association domain of Pind protein is also not required for its interaction with L protein.
FIG. 4.

Self-association of Pind protein is not required for its binding to N and L proteins. HA-Pind, HA-Δ161-210, or HA-ΔII+III was coexpressed with Myc-Nind (A) or Flag-L (B) in HeLa cells as indicated. Cell lysates were assayed for protein expression by Western blotting (WB) using anti-Myc and anti-HA monoclonal antibodies (A) or anti-Flag and anti-HA monoclonal antibodies (B). Immunoprecipitation (IP) was performed using anti-Myc (A) or anti-L (B) polyclonal antibody, and proteins in immune complexes were detected using anti-Myc and anti-HA (A) or anti-Flag and anti-HA (B) monoclonal antibodies.
Role of Pind protein self-association in VSV minigenome transcription.
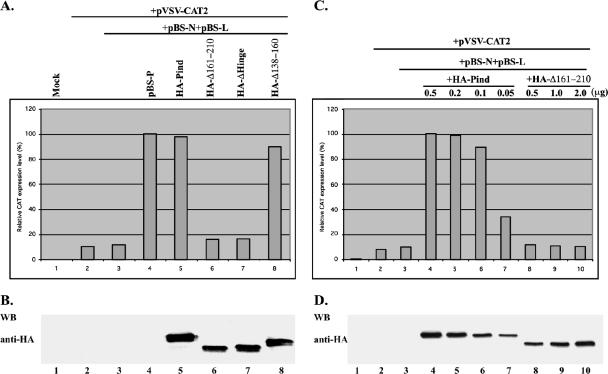
To study the role of self-association of Pind protein in transcription of viral RNA, an in vivo VSV minigenome transcription assay in which a VSV minigenome containing the 5′ and 3′ cis elements of VSV genome and a reporter gene (the CAT gene) flanked by these elements expresses the CAT protein when cotransfected with support plasmids (pBS-N, pBS-P, and pBS-L) encoding VSV N, P, and L proteins, respectively, was used (29). First, to test whether the HA-tagged Pind was functional, vTF7-3-infected BHK-21 cells were transfected with the plasmid pVSV-CAT, pBS-P or plasmid encoding HA-Pind, and other support plasmids. Following incubation at 37°C for 40 h, genomic-sense CAT mRNA was synthesized by the T7 RNA polymerase, and subsequently, the N protein encapsidated the genomic RNA which in turn was used as the template for transcription by expressed L and P proteins, which resulted in CAT reporter gene expression. The CAT expression was then analyzed by ELISA. As shown in Fig. 5A, the relative CAT expression observed using support plasmid pBS-P (defined as 100%) (top panel, column 4) was about the same as with the support plasmid encoding HA-Pind (98%) (top panel, column 5), indicating that the latter clone is as functionally active as the former. When HA-Δ138-160 replaced HA-Pind, it was still capable of supporting minigenome transcription, although less efficiently than HA-Pind (85%) (Fig. 5A, top panel, column 8). In contrast, when the mutant HA-Δ161-210, which failed to oligomerize, was used, the relative CAT expression level was significantly decreased to 16.1%, close to the background level (Fig. 5A, top panel, column 6). The protein expression levels of HA-Pind and all mutants were also assayed by Western blotting with anti-HA antibody. As shown in Fig. 5B, the expression level of HA-Δ161-210 was slightly lower than that of HA-Pind (lanes 5 and 6). To exclude the possibility that the lower CAT expression level observed for the support plasmid encoding HA-Δ161-210 was not due to lower protein expression, we decreased the amounts of transfected plasmids encoding HA-Pind and increased the amounts of plasmid encoding HA-Δ161-210. As shown in Fig. 5C and D, with the decrease in HA-Pind expression level (plasmid from 0.5 to 0.1 μg), the relative CAT expression level was only slightly decreased (Fig. 5C, column 4, 5, and 6, and D, lanes 4, 5, and 6). However, with the increasing expression level of HA-Δ161-210 (Fig. 5D, lanes 8, 9, and 10), CAT expression level still remained significantly low, virtually equal to the background expression level of CAT (Fig. 5C, columns 3, 8, 9, and 10). Taken together, the data strongly suggest that the self-association of Pind protein is a prerequisite for the function of the Pind protein in viral RNA transcription.
FIG. 5.
Role of self-association of Pind protein in VSV transcription. (A) BHK cells expressing T7 polymerase were transfected with pBS-N, pBS-L, pVSV-CAT2, and pBS-P or the plasmids encoding HA-tagged Pind or mutants in the indicated combinations. A CAT ELISA was performed using lysates of the transfected cells to measure relative CAT enzyme expression levels as viral minigenome transcription activities. The CAT expression level in the cells transfected with pBS-P, a positive control vector, was defined as 100%. (B) Pind and mutants expressed in the transfected cells were detected by Western blotting (WB) with anti-HA antibody. (C) BHK cells expressing T7 polymerase were transfected with decreasing quantities of Pind plasmids or increasing quantities of Δ161-210-coding plasmids together with the indicated plasmids in a minigenome assay. Cell lysates were assayed for relative CAT expression and HA-Pind and mutant expression by Western blotting (WB) with anti-HA antibody.
Effects of peptides derived from Pind protein self-association domain in VSV mRNA synthesis in vitro.
Having confirmed that amino acid residues 161 to 210 are necessary for Pind protein self-association and VSV transcription, it was of interest to determine whether synthetic peptides spanning the self-association domain were able to inhibit VSV mRNA synthesis in vitro. To address this question, we performed an in vitro VSV transcription reconstitution assay which is based on the transcription of purified N-RNA template and purified L-P polymerase complexes from VSV virions. Peptides P161-190 and P191-210, along with a scrambled peptide derived from P191-210, were synthesized and purified; the scrambled peptide served as the negative control (Fig. 6A). Various concentrations (8 nM, 80 nM, and 800 nM) of the peptides were incubated with the N-RNA template and L-P complex in the presence of [α-32P]GTP at 30°C, and the resulting mRNA products were analyzed by urea-polyacrylamide gel electrophoresis. As shown in Fig. 6B, the added N-RNA template alone had no VSV polymerase activity (lane 1), and the accumulated mRNAs (G, N, and P/M) were clearly discerned in the presence of the L-P complex (lane 2). The mRNA synthesis was slightly inhibited (by 29%) in the presence of 800 nM P161-190, which is a concentration 100-fold higher than the concentration of Pind (lane 5); [α-32P]GMP incorporation into mRNAs was reduced from 2.5 pmol to 1.7 pmol (Fig. 6C). However, mRNA synthesis was virtually inhibited (from 18 to 97%) when the concentration of P191-210 was increased from 8 nM to 800 nM (lanes 7 and 8), and [α-32P]GMP incorporation into mRNA was reduced from 2.5 pmol to nearly 0 pmol. As expected, the synthesis of mRNA remained essentially unchanged with increasing concentrations of the scrambled P191-210 peptide (Fig. 6B, lanes 8, 9, and 10, and C). Thus, peptide P191-210 derived from the self-association domain of Pind inhibited VSV mRNA synthesis in vitro in a dose-dependent manner. Particularly important is that only a 100-fold-higher concentration of P191-210 compared to the Pind concentration resulted in the almost complete cessation of VSV genome transcription in vitro. These results indicate that amino acid residues 191 to 210 play a vital role in the transcription of VSV genome RNA.
FIG. 6.
A peptide derived from the oligomerization domain of Pind protein inhibits transcription of the VSV genome in vitro. (A) The location of the self-association domain in Pind protein is shown. The amino acid sequences of the P161-190, P191-210, and scrambled P191-210 synthetic peptides are shown. (B) In vitro transcription was performed with purified L-P and N-RNA complexes in the presence or absence of indicated concentrations of P161-190, P191-210, and scrambled P191-210 peptides. 32P-labeled transcripts were analyzed by urea-polyacrylamide gel electrophoresis followed by autoradiography. The positions of viral mRNAs are shown on the left. (C) Transcription activities (shown in panel B) are expressed as pmol of [α-32P]GMP incorporated into mRNAs. The x axis indicates concentrations of the peptides in log scale.
DISCUSSION
It has been well established that the oligomeric P protein binds to the L protein to impart the transcriptase function to the latter and that phosphorylation within the highly acidic domain I is required for the self-association process (25). It is generally believed that a trimer form of P protein is bound to the L protein to form the L-P3 holoenzyme complex (24). Although the apparent self-association of P protein can be observed by biochemical and biophysical techniques, the precise domain within the P protein responsible for this process remained unknown.
In the present study, we have shown Pind-Pind interaction using a yeast two-hybrid system, as well as a coimmunoprecipitation assay. Using a series of Pind mutants, we have been able to precisely map the self-association domain of Pind protein to amino acids 161 to 210 within the hinge region. Furthermore, we have demonstrated that the self-association domain of Pind protein is essential for VSV transcription by a minigenome transcription system. Interestingly, a peptide containing residues 191 to 210 located within the self-association domain was able to inhibit VSV transcription in vitro in a dose-dependent manner. Although the precise reason(s) underlying this inhibitory effect of the peptide in transcription remains unclear, it is reasonable to speculate that the peptide somehow mimics a part of the self-association domain of Pind and disturbs functions of the wild-type Pind either by its direct incorporation into the oligomer or by inhibiting the oligomer formation. Thus, we conclude that self-association of P protein is essential for its functional ability, although our data do not provide a direct measurement of the number of P monomers present in the oligomer that is required for it to be functionally active.
It has been previously shown that unphosphorylated VSV P protein, which is in a monomeric form, was unable to support transcription in the presence of RNA polymerase L (25), suggesting that monomeric P protein may not bind to L protein. However, from the present study, it is quite evident that monomeric Pind protein lacking the self-association domain would still bind to the L protein, but the transcription function of L protein is lost. Consistent with this conclusion, it was reported that the inactive monomeric P protein of human parainfluenza virus type 3 maintains its ability to bind to its L protein (11). Similar data have been published for rinderpest virus belonging to the family Paramyxoviridae, although the oligomerization-incompetent P protein did not support transcription, but it still bound to its L protein (41). However, a different situation was reported for Marburg virus, which belongs to the family Filoviridae, in which monomeric VP35 (a counterpart of P protein) fails to interact with its RNA polymerase L (35). Thus, the self-association state of P protein seems to be a necessary condition but not sufficient to keep the RNA polymerase complex in the functional state and perform the function of transcriptase while bound to the L protein.
Most of the P proteins in the order Mononegavirales oligomerize via predicted coiled-coil motifs. These motifs are located in different positions in various P proteins, e.g., in the P protein of the family Paramyxoviridae, the coiled-coil motifs are mostly located in the C terminus (1, 10, 11, 12, 30, 37, 43, 48, 49), whereas the coiled-coil motif in respiratory syncytial virus is situated in the central part of the P protein (8) and the putative coiled-coil motif of Marburg virus and Ebola virus VP35 (family Filoviridae) is located at the N terminus which was shown to be essential for self-association (35). It is interesting that the VSV Pind also contains a predominant coiled-coil motif (12, 27), which is located at the amino-terminal region as predicted by COILS prediction software (33); the region spans residues 16 to 36 (data not shown). However, the mutational analyses with the Pind protein clearly show that this region is not involved in self-association of Pind since mutant ΔN60 still interacts efficiently with wild-type Pind protein (Fig. 3). Similar data were also shown for rabies virus P protein, in which the N-terminal part containing the coiled-coil motif was not required for self-association (27). In contrast, the P protein of Chandipura virus, a member of the Rhabdoviridae family, isolated from a human patient was shown to oligomerize via the N-terminal 46 amino acids which cover a coiled-coil region (40). Thus, the coiled-coil motifs in the P proteins may be necessary for their functions but are not always involved in the self-association process.
Recently, Ding et al. (18) obtained a proteinase-K-resistant domain of the VSV Pind protein (residues 107 to 177) which was crystallized as a homodimer, and the authors predicted that this domain mediate tetramerization of Pind protein by possible interdimer interactions (19). However, by direct mutation and biochemical analyses, we have mapped the self-association domain within residues 161 to 210; thus, only a series of 17 amino acids overlaps between the two predicted self-association domains. Thus, it appears that the additional amino acids which contain an important beta-sheet element may hold the subunits together. It is interesting that the Pind mutant Δ138-160, which had a major portion of amino acids 107 to 177 deleted, efficiently self-associates and is active in transcription in vivo. It is possible that during the proteolysis of Pind, residues 178 to 210 were cleaved from the stable domain (residues 107 to 177) and were undetectable by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining. Therefore, the oligomerization domain appears to be longer than as described by Ding et al. (19), and residues 107 to 177 are possibly required for stability to aid in crystallization. Supporting this contention is the fact that we failed to express Pind mutants ΔDomain I, Δ109-137, and 109-210 in yeast or HeLa cells. However, Pind mutant 76-210 was expressed well in yeast and HeLa cells, suggesting that residues 76 to 137 are important for Pind stabilization in yeast or mammalian cells, consistent with the observation that residues 107 to 177 can be stably expressed in Escherichia coli (19). Alternatively, one could envision rearrangements of the central domain that would be possible. Further studies are clearly needed to establish the precise amino acid boundaries of the self-association domain.
Recently, a number of insertion and deletion mutations were introduced into the hinge region of Pind protein and their effects in transcription and replication were studied (13). It was shown that amino acid residues 191 to 210 were absolutely required for VSV transcription and replication in vivo, consistent with our finding that this region is critically important for the self-association of the Pind protein.
In summary, we have mapped the functional self-association domain (residues 161 to 210) of Pind protein within the hinge region by using in vitro and in vivo approaches. Furthermore, our studies demonstrate an essential role of P protein self-association in the transcription of VSV genome RNA.
Acknowledgments
We thank Denis Gerlier for critically reading the manuscript.
This work was supported by a grant from the NIH (AI26585) to A.K.B.
REFERENCES
- 1.Asenjo, A., and N. Villanueva. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett. 467:279-284. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore, D., A. S. Huang, and M. Stampfer. 1970. Ribonucleic acid synthesis of vesicular stomatitis virus. II. An RNA polymerase in the virion. Proc. Natl. Acad. Sci. USA 66:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, A. K., S. A. Moyer, and D. P. Rhodes. 1974. Studies on the in vitro adenylation of RNA by vesicular stomatitis virus. Virology 61:547-558. [DOI] [PubMed] [Google Scholar]
- 4.Barik, S., and A. K. Banerjee. 1991. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in E. coli: analysis of phosphorylation status versus transcriptional activity. J. Virol. 65:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik, S., and A. K. Banerjee. 1992. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc. Natl. Acad. Sci. USA 89:6570-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barik, S., and A. K. Banerjee. 1992. Sequential phosphorylation of the phosphoprotein of vesicular stomatitis virus by cellular and viral protein kinases is essential for transcription activation. J. Virol. 66:1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canter, D. M., and J. Perrault. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376-386. [DOI] [PubMed] [Google Scholar]
- 8.Castagne, N., A. Barbier, J. Bernard, H. Rezaei, J. C. Huet, C. Henry, B. Da Costa, and J. F. Eleouet. 2004. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain. J. Gen. Virol. 85:1643-1653. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. L., T. Das, and A. K. Banerjee. 1997. Phosphorylation states of vesicular stomatitis virus P protein in vitro and in vivo. Virology 228:200-212. [DOI] [PubMed] [Google Scholar]
- 10.Chen, M., J. C. Cortay, I. R. Logan, V. Sapountzi, C. N. Robson, and D. Gerlier. 2005. Inhibition of ubiquitination and stabilization of human ubiquitin E3 ligase PIRH2 by measles virus phosphoprotein. J. Virol. 79:11824-11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary, S. K., A. G. Malur, Y. Huo, B. P. De, and A. K. Banerjee. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373-382. [DOI] [PubMed] [Google Scholar]
- 12.Curran, J., R. Boeck, N. Lin-Marq, A. Lupas, and D. Kolakofsky. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139-149. [DOI] [PubMed] [Google Scholar]
- 13.Das, S. C., and A. K. Pattnaik. 2005. Role of the hypervariable hinge region of phosphoprotein P of vesicular stomatitis virus in viral RNA synthesis and assembly of infectious virus particles. J. Virol. 79:8101-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, T., A. K. Gupta, P. W. Sims, C. A. Gelfand, J. E. Jentoft, and A. K. Banerjee. 1995. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA polymerase of vesicular stomatitis virus. J. Biol. Chem. 270:24100-24107. [DOI] [PubMed] [Google Scholar]
- 15.Das, T., A. K. Pattnaik, A. M. Takacs, T. Li, L. N. Hwang, and A. K. Banerjee. 1997. Basic amino acid residues at the carboxyl-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology 238:103-114. [DOI] [PubMed] [Google Scholar]
- 16.Davis, N. L., H. Arnheiter, and G. W. Wertz. 1986. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 59:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De, B. P., and A. K. Banerjee. 1985. Requirements and function of vesicular stomatitis virus L and NS proteins in the transcription process in vitro. Biochem. Biophys. Res. Commun. 126:40-49. [DOI] [PubMed] [Google Scholar]
- 18.Ding, H., T. J. Green, and M. Luo. 2004. Crystallization and preliminary X-ray analysis of a proteinase-K-resistant domain within the phosphoprotein of vesicular stomatitis virus (Indiana). Acta Crystallogr. D 60:2087-2090. [DOI] [PubMed] [Google Scholar]
- 19.Ding, H., T. J. Green, S. Y. Lu, and M. Luo. 2005. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80:2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson, S. U., and R. R. Wagner. 1973. L protein requirements for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 12:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emerson, S. U., and Y. H. Yu. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 15:1348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao, Y., and J. Lenard. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerize (L) and template. J. Virol. 69:7718-7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, Y., and J. Lernard. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase II. EMBO J. 14:1240-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gigant, B., F. Iseni, Y. Gaudin, M. Knossow, and D. Blondel. 2000. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J. Gen. Virol. 81:1757-1761. [DOI] [PubMed] [Google Scholar]
- 28.Gill, D. S., D. Chattopadhyay, and A. K. Banerjee. 1986. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc. Natl. Acad. Sci. USA 83:8873-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grdzelishvili, V., S. Smallwood, D. Tower, R. Hall, D. Hunt, and S. A. Moyer. 2005. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 79:7327-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 31.Hwang, L. N., N. Englund, T. Das, A. K. Banerjee, and A. Pattnaik. 1999. Optimal replication activity of vesicular stomatitis virus RNA polymerase requires phosphorylation of a residue(s) at carboxy-terminal domain II of its accessory subunit, phosphoprotein P. J. Virol. 73:5613-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, T., and A. K. Pattnaik. 1997. Replication signal in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232:248-259. [DOI] [PubMed] [Google Scholar]
- 33.Lupas, A., M. V. Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 34.Masters, P. S., and A. K. Banerjee. 1988. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N or vesicular stomatitis virus. J. Virol. 62:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller, P., N. Pariente, H. D. Klenk, and S. Becker. 2005. Homo-self-association of Marburg virus VP35 is essential for its function in replication and transcription. J. Virol. 79:14876-14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naito, S., and A. Ishihama. 1976. Function and structure of RNA polymerase from vesicular stomatitis virus. J. Biol. Chem. 251:4307-4314. [PubMed] [Google Scholar]
- 37.Nishio, M., M. Tsurudome, M. Ito, N. Watanabe, M. Kawano, H. Komada, and Y. Ito. 1997. Human parainfluenza virus type 2 phosphoprotein: mapping of monoclonal antibody epitopes and location of the multimerization domain. J. Gen. Virol. 78:1303-1308. [DOI] [PubMed] [Google Scholar]
- 38.Pattnaik, A. K., L. Hwang, T. Li, N. Englund, M. Mathur, T. Das, and A. K. Banerjee. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71:8167-8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peluso, R. W., and S. A. Moyer. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369-376. [DOI] [PubMed] [Google Scholar]
- 40.Raha, T., E. Samal, A. Majumdar, S. Basak, D. Chattopadhyay, and D. J. Chattopadhyay. 2000. N-terminal region of P protein of Chandipura virus is responsible for phosphorylation mediated homodimerization. Protein Eng. 13:437-444. [DOI] [PubMed] [Google Scholar]
- 41.Rahaman, A., N. Srinivasan, N. Shamala, and M. S. Shaila. 2004. Phosphoprotein of the rinderpest virus forms a tetramer through a coiled coil region important for biological function. A structural insight. J. Biol. Chem. 279:23606-23614. [DOI] [PubMed] [Google Scholar]
- 42.Rose, J. K., and M. A. Whitt. 2001. Rhabdoviridae: the viruses and their replication, p. 1221-1244. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins. Philadelphia, Pa.
- 43.Shaji, D., and M. S. Shaila. 1999. Domains of rinderpest virus phosphoprotein involved in interaction with itself and the nucleocapsid protein. Virology 258:415-424. [DOI] [PubMed] [Google Scholar]
- 44.Spadafora, D., D. M. Canter, R. L. Jackson, and J. Perrault. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but not essential for transcription or replication. J. Virol. 70:4538-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stillman, E. A., J. K. Rose, and M. A. Whitt. 1995. Replication and amplification of novel vesicular stomatitis virus minigenome encoding viral structural proteins. J. Virol. 69:2946-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takacs, A. M., S. Barik, T. Das, and A. K. Banerjee. 1992. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J. Virol. 66:5842-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takacs, A. M., T. Das, and A. K. Banerjee. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. USA 90:10375-10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarbouriech, N., J. Curran, C. Ebel, R. W. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]
- 49.Tarbouriech, N., J. Curran, R. W. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]