Abstract
Recent studies indicate that human immunodeficiency virus type 1 (HIV-1) recombines at exceedingly high rates, approximately 1 order of magnitude more frequently than simple gammaretroviruses such as murine leukemia virus and spleen necrosis virus. We hypothesize that this high frequency of genetic recombination is a common feature of primate lentiviruses. Alternatively, it is possible that HIV-1 is unique among primate lentiviruses in possessing high recombination rates. Among other primate lentiviruses, only the molecular mechanisms of HIV-2 replication have been extensively studied. There are reported differences between the replication mechanisms of HIV-1 and those of HIV-2, such as preferences for RNA packaging in cis and properties of reverse transcriptase and RNase H activities. These biological disparities could lead to differences in recombination rates between the two viruses. Currently, HIV-1 is the only primate lentivirus in which recombination rates have been measured. To test our hypothesis, we established recombination systems to measure the recombination rates of two other primate lentiviruses, HIV-2 and simian immunodeficiency virus from African green monkeys (SIVagm), in one round of viral replication. We determined that, for markers separated by 588, 288, and 90 bp, HIV-2 recombined at rates of 7.4%, 5.5%, and 2.4%, respectively, whereas SIVagm recombined at rates of 7.8%, 5.6%, and 2.7%, respectively. These high recombination rates are within the same range as the previously measured HIV-1 recombination rates. Taken together, our results indicate that HIV-1, HIV-2, and SIVagm all possess high recombination frequencies; hence, the high recombination potential is most likely a common feature of primate lentivirus replication.
Primate lentiviruses consist of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency viruses (SIVs) isolated from at least 30 different nonhuman primate species in sub-Saharan Africa (52, 54, 57). African primates are the natural hosts of SIVs; however, cross-species transmission can occur, allowing SIVs to infect and adapt to other hosts. HIV-1 and HIV-2 are introduced into human populations by such cross-species transmission of SIVs. Phylogenetic analyses indicate that HIV-1 was derived from the SIV that normally infects the chimpanzee, Pan troglodytes troglodytes (SIVcpz) (19, 55), whereas HIV-2 was derived from the SIV that normally infects African sooty mangabeys, Cercocebus torquatus atys (SIVsm) (22, 40).
Frequent recombination events have occurred in the evolution of primate lentiviruses both recently and in the distant past, because mosaic genome structures have been observed at all levels of primate lentivirus classification. Currently, most primate lentiviruses can be assigned to one of the six approximately equidistant phylogenetic lineages (26, 52), including (i) SIVcpz from chimpanzees (Pan troglodytes troglodytes) together with HIV-1 (19, 33), (ii) SIVsm from sooty mangabeys (Cercocebus torquatus atys) together with HIV-2 and SIVmac from four distinct species of macaques (Macaca spp.) (9, 29, 45), (iii) SIVagm from African green monkeys (Chlorocebus spp.) (44), (iv) SIVlhoest from L'Hoest monkeys (Cercopithecus lhoesti lhoesti) (27) together with SIVsun from sun-tailed monkeys (Cercopithecus lhoesti solatus) (5) and SIVmnd from mandrills (Mandrillus sphinx) (60), (v) SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis) (28), and (vi) SIVcol from a guereza colobus (Colobus guereza) (13). However, several other SIV strains—including SIVmnd2 from wild mandrills (Mandrillus sphinx) in Cameroon (59), SIVrcm from red-capped mangabeys (Cercocebus torquatus torquatus) in Nigeria (6), SIVgsn from greater spot-nosed monkeys (Cercopithecus nictitans) in Cameroon (12, 14), SIVsab from sabaeus monkeys (Cercopithecus sabaeus) (36), and SIVdrl from drills (Mandrillus leucophaeus) (13)—cannot be classified into one of the six lentivirus lineages because of their mosaic genomes. However, it was recently suggested that SIVgsn, SIVmon from mona monkeys (Cercopithecus mona), and SIVmus from mustached monkeys (Cercopithecus cephus) represent a new primate lentivirus lineage (12). In addition, careful phylogenetic analyses also revealed the existence of mosaic genomes in the six so-called pure lineages (52). All of these studies suggest that recombination has occurred among diverse SIV lineages early in the evolution of primate lentiviruses. However, identification of pure lineages or recombinants is mainly a function of chronological findings (14). For example, SIVcpz, the progenitor of HIV-1, was initially characterized as a pure lineage, and the mosaic genomes of SIVgsn and SIVrcm were thought to be recombinants of SIVcpz (6, 14) and other SIV strains. However, it was recently demonstrated after extensive phylogenetic analysis that SIVcpz is most likely a recombinant of SIVgsn and SIVrcm (3). SIVcpz is most closely related to SIVrcm in the 5′ region of the genome and to SIVgsn in the 3′ half of the genome.
In addition to recombination between different SIVs, there is ample evidence that recombination occurs frequently within populations of the same virus. Mosaic genomes have been found in HIV-1 (19), HIV-2 (21), SIVsm (54), and SIVlhoest (4). Sequence analyses of SIVsm isolates revealed extensive recombination among SIVsm viruses (54). Additionally, of the more than 70,000 HIV-1 sequences in the Los Alamos National Laboratory database, approximately 8% display mosaic genome structures (http://www.hiv.lanl.gov). Furthermore, epidemic intersubtype recombinant strains of HIV-1 have been observed in various geographic areas, and individuals infected with more than one genetically distinct HIV-1 often also harbor hybrid viruses in their viral populations (15, 20, 41, 43, 51, 63).
Consistent with phylogenetic analyses indicating frequent recombination during the evolution of primate lentiviruses and HIV-1, frequent HIV-1 recombination has also been observed in cell culture systems (8, 10, 11, 18, 35, 46, 49, 50). Similarly, SIVmac recombination was detected in inoculated rhesus macaques (39, 61). Interestingly, it was shown that HIV-1 recombines at a much higher frequency (approximately 10 times) than murine leukemia virus (MLV) and spleen necrosis virus (SNV) in one round of viral replication using the same target sequence assays (1, 31, 46, 49). Currently, the mechanistic differences that caused the disparity of the recombination rates in these viruses remain to be fully elucidated.
Based on the high HIV-1 recombination rates and the frequently observed mosaic genomes in other primate lentiviruses, we hypothesize that high genetic recombination potential is a common feature among primate lentiviruses but not gammaretroviruses. Alternatively, it is also possible that each primate lentivirus possesses a different recombination potential due to the biological features of the virus, and HIV-1 is unique in its ability to recombine extremely frequently. There are at least two factors that can affect the observed recombination rate: the frequency of copackaging of two different RNAs into the same virion (heterozygous formation) and the ability of reverse transcriptase (RT) to switch between the two copackaged RNAs during DNA synthesis (template switching). Although recombination can occur during the reverse transcription of any virus, only heterozygous virions yield genotypically different recombinants (31). Therefore, the frequency of heterozygous virion formation dictates the observed recombination rate. HIV-1 and HIV-2 have different preferences for genomic RNA packaging: HIV-2 Gag preferentially packages RNA from which it was translated (cis packaging) (24, 37), whereas HIV-1 Gag does not have the same preference (42). The differences in RNA selection could affect the frequency of heterozygous formation, thereby altering the observed recombination rates. The frequency of RT switching from one RNA template to the other RNA depends on the balance between polymerase and RNase H activities, as proposed by the dynamic copy choice model (34). The RNase H activity of HIV-2 was also shown to be much lower than that of HIV-1 in vitro (56), although a more recent study indicated that they are comparable (48). If HIV-1 and HIV-2 differ in the balance of polymerase and RNase H activities in RT, then the RT molecules of these two viruses may switch templates at different frequencies and alter the observed recombination rates. Therefore, there are sufficient differences between HIV-1 and HIV-2 replication that could lead to different recombination rates for the two viruses.
The natural host of SIVagm is the African green monkey. Many studies have focused on the pathogenicity of the virus (44); however, very little is known about the molecular mechanisms of SIVagm replication including the preferences of RNA packaging and the balance between polymerase and RNase H activities (44). Therefore, it has been entirely unclear whether SIVagm has a recombination rate similar to that of HIV-1.
Previously, we used a flow cytometry-based system to measure HIV-1 recombination rates. Recombination rates between markers separated by 103, 288, and 588 bp were 1.4%, 3.8%, and 6.9%, respectively. In this report, to examine whether recombination potential varies among different primate lentiviruses, we established systems to measure the recombination rates of HIV-2 and SIVagm, each representing a distinct phylogenetic lineage of primate lentiviruses, in one round of viral replication. Our results show that both HIV-2 and SIVagm recombined at high rates, within the same range as that of HIV-1. Taken together, our results indicate that three primate lentiviruses of different lineages all recombine at very high frequencies; therefore, the high recombination potential is most likely a common feature of primate lentivirus replication.
MATERIALS AND METHODS
Plasmid construction.
Plasmids were constructed with standard molecular cloning techniques (53). The general structures of all constructs were verified by restriction digestion, and PCR-amplified regions were further characterized by DNA sequencing to avoid inadvertent mutations. HIV-2 vectors were derived from pROD12 (a gift from Keith Peden), which contains the full-length, infectious HIV-2 molecular clone. pML2DEL3 (a gift from Maria Leavitt) was generated from pROD12 by introducing a 0.8-kb inactivating deletion in env and a start codon deletion plus a linker insertion in nef. All of the vectors used in the recombination studies contain two of the three marker genes: the mouse heat-stable antigen (HSA) gene (HSA) (37) or the mouse Thy1.2 gene (Thy) (23), along with the green fluorescent protein (GFP) gene (GFP) (7). The internal ribosomal entry site (IRES) from encephalomyocarditis virus was used to express the downstream marker gene in one transcript. DNA fragments containing either HSA-IRES-GFP or Thy-IRES-GFP were isolated from pON-H0, pON-T1, pON-T3, or pON-T6 (49) and inserted into pML2DEL3 to generate pHIV2-H0G, pHIV2-T100G, pHIV2-T300G, or pHIV2-T600G, respectively. The GFP genes of pHIV2-H0G, pHIV2-T100G, pHIV2-T300G, and pHIV2-T600G contain inactivating mutations 15, 105, 303, and 603 bp downstream from the GFP start codon, respectively. Additionally, an inactivating mutation (4-bp insertion) was introduced into the vpr gene of the aforementioned HIV-2 vectors used in recombination studies to abolish the cell-arresting activity of Vpr protein and to facilitate the generation of provirus-containing cell lines.
SIVagm vectors were derived from pTan (a gift from Feng Gao) (58), which contains a full-length, infectious molecular clone derived from the Tan-1 strain of SIVagm (36). All major open reading frames in pTan are functional, except that vpr contains an in-frame stop codon. Vector pTan1-XMS (a gift from William Fu) was generated by inserting a 19-nucleotide linker into the XhoI site of pTan to inactivate env. Vector pTan1-nef was generated from pTan1-XMS by replacing the 5′ portion of nef from the start codon to the BsiwI site with a linker sequence. DNA fragments containing either HSA-IRES-GFP or Thy-IRES-GFP were isolated from pON-H0, pON-T1, pON-T3, or pON-T6 and inserted into pTan1-nef to generate pTan-H0G, pTan-T100G, pTan-T300G, or pTan-T600G, respectively. The GFP genes in pTan-H0G, pTan-T100G, pTan-T300G, and pTan-T600G contain inactivating mutations 15, 105, 303, and 603 bp, respectively, downstream from the GFP start codon.
Plasmid pIIINL(AD8)env was derived from the AD8 strain of HIV-1, which uses CCR5 as a coreceptor (a gift from Eric Freed). Viruses pseudotyped with AD8 Env were used to infect Hut/CCR5 cells to measure recombination rates. Plasmid pHCMV-G expresses the vesicular stomatitis virus G glycoprotein (VSV-G) (25); VSV-G pseudotyped viruses were used to infect 293T cells to generate producer cell lines.
Cells, transfections, and infections.
293T cells are a human embryonic kidney cell line containing simian virus 40 large T antigen (16, 47). Hut/CCR5 cells were derived from Hut78, a human T-cell line, and express CCR5 (62). The 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% calf serum, penicillin (50 U/ml), and streptomycin (50 U/ml). Hut/CCR5 cells were maintained in RPMI medium supplemented with 10% fetal calf serum and antibiotics. All cultured cells were maintained in a humidified 37°C incubator with 5% CO2.
To generate vector-derived viruses, 293T cells were transiently transfected with the HIV-1 vector and envelope-expressing plasmid by the calcium phosphate method (53), using the MBS mammalian transfection kit (Stratagene). Viral supernatants were harvested 24 h later and clarified through a 0.45-μm-pore-size filter to remove cellular debris and then used immediately or stored at −80°C prior to infection.
For infection of 293T cells, 106 cells were plated in a 100-mm-diameter dish and infected 24 h later. Serial dilutions were generated from each viral stock and used to infect target cells in the presence of Polybrene at a final concentration of 10 μg/ml. Viruses were removed 2 h later, and fresh medium was added to the cells; at 48 h postinfection, cells were processed and flow cytometry analyses were performed. For infection of Hut/CCR5 cells, 2.5 × 105 cells in 1 ml of medium were plated in a six-well plate, 1 ml of viruses was added, and infected cells were processed and analyzed at 72 h postinfection.
Antibody staining and flow cytometry analyses.
Cells were stained with phycoerythrin-conjugated anti-HSA antibody (BD Biosciences) and allophycocyanin-conjugated anti-Thy-1 antibody (eBioscience). Cells for flow cytometry analyses were fixed with 2% paraformaldehyde, whereas cells for sorting were resuspended in phosphate-buffered saline containing 2% calf serum. Flow cytometry analyses were performed on a FACSCalibur system, and cell sorting was performed on a FACSVantage SE system with the FACSDiVa digital option (BD Biosciences). Data from flow cytometry were analyzed with Flowjo software (Tree Star).
Calculation of recombination rates.
Multiplicity of infection (MOI) was calculated from the number of infected cells obtained by flow cytometry as previously described (49). Recombination rates were calculated by dividing the GFP MOI by the total infection MOI. The percentage of the theoretical maximum measurable recombination rate (TMMRR) was calculated as recombination rate/0.125 (49). Modeling was done via polynomial linear regression as in a previous paper (49), and the 95% confidence band was established by adding two estimated standard errors of estimate to the predicted percentage of theoretical maximum measurable rate at each marker distance.
RESULTS
System used to measure HIV-2 recombination rates.
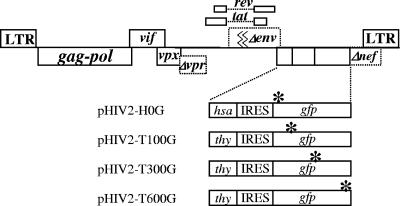
A series of Rod-12-based HIV-2 vectors were generated to contain all functional viral genes except env, vpr, and nef. The env gene was inactivated to better regulate the virus replication cycle; the vpr gene was inactivated to abolish the cell cycle arrest ability of Vpr, thereby facilitating the establishment of producer cell lines. Each vector contained two marker genes, which were inserted into the nef reading frame. One vector, pHIV2-H0G, contained a functional HSA gene and a nonfunctional GFP gene with an inactivating mutation 15 bp downstream of the GFP start codon. Another set of vectors, pHIV2-T100G, pHIV2-T300G, and pHIV2-T600G, each contained a functional Thy and a nonfunctional GFP gene; the inactivating mutation was located at 105, 303, and 603 bp, respectively, downstream of the GFP start codon (Fig. 1).
FIG. 1.
General structure of HIV-2 vectors used to measure recombination rates. All vectors share the same structure but differ in the marker genes carried. Asterisks indicate inactivating mutations in GFP. LTR, long terminal repeat.
We generated producer cells containing the two parental vectors by sequentially infecting 293T cells with VSV-G pseudotyped viruses derived from each vector. Infections were performed at a low MOI (0.05 to 0.1) to ensure that the majority of the infected cells contained one of each parental vector. Dually infected cells (cells expressing both HSA and Thy) were enriched to more than 95% by cell sorting. In each round of cell sorting, at least 5 × 105 cells were sorted to ensure that the final cell pool represented a large number of independently infected cells. As a result, each producer cell line is a pool of dually infected cells containing a minimum of 1 × 105 independent infection events.
To measure the recombination rate, producer cells were transfected with pIIINL(AD8) that expressed CCR5-tropic HIV-1 Env. Viruses harvested from transfected producer cells were used to infect the human T-cell line Hut/CCR5, which was then processed and analyzed by flow cytometry. Cells infected by the vector viruses expressed HSA (HSA+) or Thy (Thy+) or both markers if dually infected; however, cells infected by parental viruses would not express GFP (GFP−) because both vectors carry an inactivated GFP gene. Cells expressing GFP (GFP+) resulted from infection by recombinant proviruses that had reconstituted the functional GFP during reverse transcription. Because the virus producer cells do not express CD4 and the vectors do not carry a functional env gene, reinfection could not occur in producer or target cells; thus, this assay measured recombination that occurred during one round of virus replication.
HIV-2 recombination rates for markers separated by 90, 288, and 588 bp.
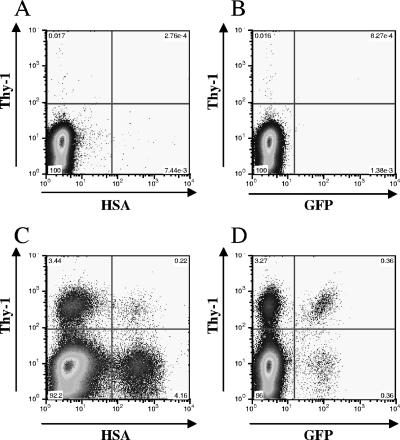
To measure HIV-2 recombination rates at three different genetic distances, we generated a total of six producer cell lines, two for each marker distance. Helper plasmids were transfected into producer cell lines, viruses were harvested and used to infect Hut/CCR5 cells, and the infected cells were analyzed by flow cytometry. Representative flow cytometry analyses of mock-infected and infected Hut/CCR5 cells are shown in Fig. 2. Mock-infected cells had very few HSA+, Thy+, or GFP+ cells (Fig. 2A and B); in contrast, HSA+, Thy+, or GFP+ cell populations could be easily detected in infected cells (Fig. 2C and D). Additionally, recombinants containing GFP+ cells were detected in both the Thy+ and Thy−/HSA+ cells (Fig. 2D and data not shown). Data generated from three independent experiments are summarized in Table 1. The numbers of total infected cells and GFP+ cells were converted to total infection MOIs and GFP MOIs, respectively, and the recombination rate was calculated by comparing the GFP MOI with the infection MOI.
FIG. 2.
Representative flow cytometry analyses of mock-infected and infected target cells. (A and B) Mock-infected Hut/CCR5 target cells stained with anti-HSA and anti-Thy-1 antibodies. (C and D) Hut/CCR5 target cells infected with virus harvested from a producer cell line harboring both HIV2-H0G and HIV2-T600G; cells were stained with anti-HSA and anti-Thy-1 antibodies. Expression levels of marker genes are indicated on the x and y axes.
TABLE 1.
HIV-2 recombination rates in Hut/CCR5 cells
| Expt no. | Marker distance (bp) | No. of cells
|
MOI
|
Recombination ratea (%) | % of TMMRRb | |||
|---|---|---|---|---|---|---|---|---|
| Total | Infected | GFP+ | Infection | GFP | ||||
| 1 | 90 | 361,134 | 16,382 | 405 | 0.052 | 0.00112 | 2.2 | 17.2 |
| 288 | 421,556 | 18,611 | 1,030 | 0.045 | 0.00244 | 5.4 | 43.4 | |
| 588 | 238,995 | 14,164 | 1,010 | 0.061 | 0.00424 | 7.0 | 55.6 | |
| 2 | 90 | 320,226 | 13,996 | 332 | 0.045 | 0.00103 | 2.3 | 18.3 |
| 288 | 326,315 | 14,756 | 725 | 0.046 | 0.00222 | 4.8 | 38.6 | |
| 588 | 409,408 | 19,490 | 1,361 | 0.049 | 0.00333 | 6.8 | 54.4 | |
| 3 | 90 | 669,949 | 7,659 | 197 | 0.011 | 0.00029 | 2.6 | 21.1 |
| 288 | 531,655 | 7,533 | 463 | 0.014 | 0.00087 | 6.2 | 49.7 | |
| 588 | 570,511 | 13,037 | 1,086 | 0.023 | 0.00191 | 8.3 | 66.4 | |
Means ± standard deviations for markers separated by the indicated distances were as follows: 90 bp, 2.4% ± 0.2%; 288 bp, 5.5% ± 0.6%; 588 bp, 7.4% ± 0.7%.
Means ± standard deviations for markers separated by the indicated distances were as follows: 90 bp, 18.9% ± 1.6%; 288 bp, 43.9% ± 4.5%; 588 bp, 58.8% ± 5.4%.
In this system, the highest theoretical frequency of GFP+ events among all infection events was 12.5%. If the RNAs from the two parental viruses were expressed at similar levels and copackaging of RNA was random, then 50% of the virions were heterozygotes and could yield observable recombinants. If recombination between two markers reached the maximum rate and the two markers randomly assorted, four genotypes could be generated at equal frequencies: the two parental genotypes, the recombinant with two mutations in GFP, and the recombinant with wild-type GFP. Of these four genotypes, only the recombinant with wild-type GFP was able to confer GFP+; thus, the highest GFP+ events were 12.5% of the total infection events (50%/4 = 12.5%). Because the theoretical maximum measurable GFP+ events in this system were 12.5% instead of 100%, we also converted the recombination rate into the percentage of TMMRR by multiplying the recombination rate by 8 (100%/12.5%). As shown in Table 1, the HIV-2 recombination rates of 2.4%, 5.5%, and 7.4% for markers separated by 90, 288, and 588 bp, respectively, correspond to 18.9%, 43.9%, and 58.8% of the TMMRR, respectively.
SIVagm recombination rates for markers separated by 90, 288, and 588 bp.
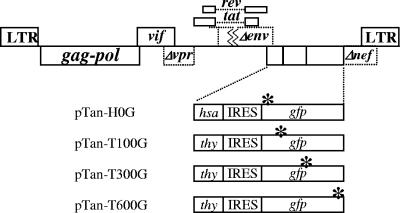
To measure SIVagm recombination rates, we established a system similar to that used in the HIV-2 study. We generated several vectors based on pTan, which contains the full-length infectious SIVagm molecular clone with functional major open reading frames except for vpr, which has an in-frame stop codon (58). We modified pTan by inactivating env with a frameshift mutation and by inserting either the HSA-IRES-GFP or Thy-IRES-GFP DNA fragment into the nef reading frame (Fig. 3). The same inactivating mutations as those in the aforementioned HIV-2 vectors were introduced into the SIVagm-derived vectors; the vector names reflect the encoded markers and the positions of the inactivating mutations in GFP (Fig. 1). By using different pairs of the SIVagm-based vectors, we measured the recombination rates at three marker distances.
FIG. 3.
General structure of SIVagm vectors used to measure recombination rates. All vectors share the same structure but differ in the marker genes carried. Asterisks indicate inactivating mutations in GFP. LTR, long terminal repeat.
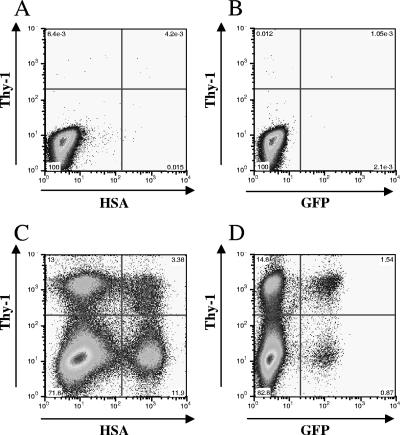
To measure the recombination rate at each genetic distance, we generated two independent producer cell lines doubly infected with a pair of vector viruses. We used the same experimental procedures as those described to measure the HIV-2 recombination rates. Representative flow cytometry analyses of the target Hut/CCR5 cells are shown in Fig. 4. As expected, very few cells that were positive for any of the three markers were detected in the mock-infected cells (Fig. 4A and B), whereas HSA+, Thy+, or GFP+ cell populations could be readily detected in infected cells (Fig. 4C and D). Data from three independent experiments are summarized in Table 2. SIVagm recombination rates were calculated using the same methods as those used to calculate HIV-2 recombination rates. As shown in Table 2, the recombination rates for SIVagm were 2.7%, 5.6%, and 7.8% for markers separated by 90, 288, and 588 bp, respectively, which correspond to 21.9%, 44.9%, and 62.4% of the TMMRR, respectively.
FIG. 4.
Representative flow cytometry analyses of mock-infected and infected target cells. (A and B) Mock-infected Hut/CCR5 target cells stained with anti-HSA and anti-Thy-1 antibodies. (C and D) Hut/CCR5 target cells infected with virus harvested from a producer cell line harboring both Tan-H0G and Tan-T600G.
TABLE 2.
SIVagm recombination rates in Hut/CCR cells
| Expt no. | Marker distance (bp) | No. of cells
|
MOI
|
% of TMMRRb | ||||
|---|---|---|---|---|---|---|---|---|
| Total | Infected | GFP+ | Infection | GFP | GFP/infectiona | |||
| 1 | 90 | 240,821 | 71,193 | 2,335 | 0.350 | 0.0097 | 2.8 | 22.2 |
| 288 | 285,101 | 52,246 | 3,070 | 0.202 | 0.0108 | 5.3 | 42.8 | |
| 588 | 261,476 | 100,252 | 9,822 | 0.484 | 0.0383 | 7.9 | 63.3 | |
| 2 | 90 | 198,984 | 70,998 | 2,485 | 0.441 | 0.0125 | 2.8 | 22.7 |
| 288 | 45,436 | 12,328 | 801 | 0.317 | 0.0178 | 5.6 | 44.9 | |
| 588 | 295,208 | 123,450 | 12,298 | 0.542 | 0.0425 | 7.8 | 62.7 | |
| 3 | 90 | 225,247 | 40,055 | 1,149 | 0.196 | 0.0051 | 2.6 | 20.8 |
| 288 | 198,666 | 58,191 | 4,004 | 0.347 | 0.0204 | 5.9 | 47.0 | |
| 588 | 229,025 | 64,002 | 5,634 | 0.328 | 0.0249 | 7.6 | 60.7 | |
Means ± standard deviations for markers separated by the indicated distances were as follows: 90 bp, 2.7 ± 0.1; 288 bp, 5.6 ± 0.2; 588 bp, 7.8 ± 0.1.
Means ± standard deviations for markers separated by the indicated distances were as follows: 90 bp, 21.9% ± 0.8%; 288 bp, 44.9% ± 1.7%; 588 bp, 62.4% ± 1.3%.
Comparison of HIV-1, HIV-2, and SIVagm recombination rates.
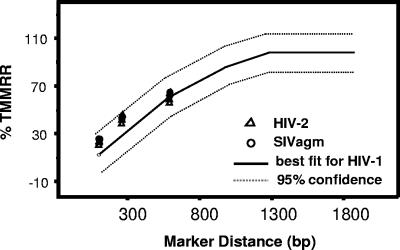
We previously established a quadratic regression model of the relationship between marker distances and recombination rates in HIV-1 using 41 data points collected from two different studies (49). This model had an outstanding fit to the data, with an R2 value of 0.984, and only one data point was outside the 95% confidence band of the model. A modified version of the model is shown in Fig. 5; this model illustrates the finding that recombination rates increased with marker distances when the markers were separated by shorter distances; when marker distances increased to more than 1 kb, recombination rates began to reach a plateau. Because HIV-1 recombination rates did not reach a plateau at marker distances lower than 588 bp, we measured the HIV-2 and SIVagm recombination rates at these shorter marker distances. To compare the recombination rates of HIV-2 and SIVagm with those of HIV-1, we plotted the HIV-2 and SIVagm recombination rates over marker distances in the existing HIV-1 model. As shown in Fig. 5, the measured recombination rates of HIV-2 and SIVagm at the marker distance of 588 bp were almost identical to that of HIV-1, although the measured recombination rates of HIV-2 and SIVagm at marker distances of 90 and 288 bp appeared to be slightly higher than those of HIV-1. All data points from HIV-2 and SIVagm recombination studies fell inside the 95% confidence band of the HIV-1 recombination model, indicating that HIV-2 and SIVagm recombined at levels similar to those of HIV-1.
FIG. 5.
Comparison of HIV-2, SIVagm, and HIV-1 recombination rates. The previously described model of the relationship between HIV-1 recombination rate and marker distance is shown as a solid line, and its 95% confidence bands are shown as dashed lines. The 95% confidence band was computed by adding two estimated standard errors of estimate to predicted scores. The measured HIV-2 and SIVagm recombination rates at three marker distances (from Tables 1 and 2) are indicated by triangles and circles, respectively.
DISCUSSION
HIV-1 recombines at exceedingly high frequencies; to examine whether other primate lentiviruses also have a high recombination potential, we measured the recombination rates of HIV-2 and SIVagm. We observed that both HIV-2 and SIVagm recombined at high frequencies; at a marker distance of 588 bp, both viruses recombined at frequencies that were approximately 60% of the theoretical maximum measurable rate. Together with our previous HIV-1 work, our results indicate high recombination rates in three distinct primate lentiviruses representing three major evolutionary lineages of currently known primate lentiviruses. These results suggest that the high recombination potential is a conserved feature in primate lentivirus evolution. Therefore, recombination can be used as a powerful tool to reassort variation in the primate lentiviral genome to increase diversity in the viral population and to enhance the evolutionary capacity of the virus.
The frequency of heterozygous virion formation affects the measured recombination rate. Although all retroviruses package two copies of RNAs into a virion, many aspects of RNA packaging are unknown. It is currently still an open issue whether one dimeric viral RNA or two monomeric viral RNAs are packaged by Gag during virus assembly. It is also unclear how often RNAs from two proviruses are copackaged in various retroviral systems, and whether RNA packaging mechanisms affect the heterozygous virion formation. From our previous study, we concluded that heterozygous virions are formed very efficiently during HIV-1 replication; with two highly homologous viruses, heterozygous virions are formed at the frequency expected from random copackaging of RNA (49, 50). This conclusion was derived from the observation that HIV-1 recombination rates can reach the theoretical maximum measurable rate at a marker distance of 1.3 kb but do not exceed this theoretical rate with further increase in marker distances (50) (Fig. 5). We previously proposed that HIV-1 Gag packages dimeric RNA, based on our recombination and other studies, because mismatch sequences in the dimer initiation signal can significantly reduce the recombination rate (10).
The proposed mechanisms of genomic RNA packaging in HIV-2 are different from those of HIV-1 (24, 37). The cis-acting packaging signal for HIV-2 is upstream of the major splice donor and is present on both spliced and full-length RNA transcripts. It has been proposed that, to ensure preferential packaging of full-length RNA, HIV-2 Gag proteins have a strong preference for packaging the RNA from which they were translated (24). This model implies that HIV-2 Gag RNA recognition occurs before RNA dimerization. Therefore, it is likely that two copies of monomeric viral RNAs are selected and packaged; RNA dimerization occurs during or after virion assembly. Despite these differences in RNA packaging mechanisms between HIV-1 and HIV-2, copackaging of RNAs from two highly homologous viruses appears to be frequent for HIV-2. Similarly, RNA copackaging also occurs in SIVagm, although little is known about the molecular mechanisms of SIVagm RNA packaging. The high recombination frequency displayed by HIV-2 and SIVagm suggests that the formation of heterozygous virions approaches the level expected from random copackaging of viral RNA for these viruses.
Interestingly, viral RNA may not be copackaged randomly in other viruses such as some of the gammaretroviruses. The recombination rates measured in three primate lentiviruses are approximately 10 times higher than those from two gammaretroviruses, MLV and SNV (49, 50; also results described in this paper). In our earlier studies, we found that although SNV and MLV have lower overall recombination rates, recombinants often have multiple crossovers (1, 2, 30-32). Our results indicated that in MLV and SNV, recombination occurs only in certain viral populations; however, viruses within these populations undergo very frequent recombination (1, 2, 30). We postulated two possible mechanisms that could have caused this phenomenon: either the reverse transcription complex in the recombining viruses had different structures to allow RT better access to both RNAs, or the RNA copackaging was not random and heterozygous viruses were formed far less than expected from random RNA assortment (30). Later, it was observed that MLV and HIV-1 have comparable template switching frequencies but different recombination rates (46). It was also shown that heterodimeric RNA occurred less frequently in MLV than HIV-1; however, the estimated two- to threefold difference in heterozygous formation does not entirely account for all of the differences in a 10-fold discrepancy in recombination rate (17). It was also concluded that cotransfected MLV vectors recombine more frequently than MLV vectors introduced by separate transfections; it was proposed that cotransfected vectors were integrated at nearby locations, and thus RNAs can traffic together and form heterozygous viruses more frequently (38). A more recent paper indicated that although the overall MLV recombination occurs at a rate lower than that of HIV-1, recombination occurs very frequently in viruses derived from heterozygous virions (64). Together, all of these results point to the idea that although MLV is perfectly capable of frequent template switching, there is a mechanistic limitation(s) to prevent observable recombination in the entire MLV population. The limitation(s) most likely includes nonrandom RNA copackaging to decrease the formation of heterozygous viruses; it is unclear at this time whether other unknown factors also play important roles as well.
In this study, we conclude that primate lentiviruses recombine frequently and at a rate much higher than those of the known simple retroviruses. The high recombination potential of the primate lentiviruses can expedite the generation of broad genetic diversity in the viral population. This broad genetic diversity allows the emergence of variants upon the change of selection pressure and facilitates adaptation of the virus. Therefore, the high recombination potential could provide a strong advantage for primate lentiviruses. Understanding viral replication mechanisms and the ability of the virus to generate genetic diversity may shed light on the pathogenesis of these viruses.
Acknowledgments
We thank Vinay K. Pathak for intellectual input and helpful discussions throughout this project; Anne Arthur for expert editorial help; and Keith Peden, Feng Gao, and Eric Freed for gifts of plasmids.
This research was supported by the Intramural Research program of the NIH, National Cancer Institute, Center for Cancer Research.
REFERENCES
- 1.Anderson, J. A., E. H. Bowman, and W. S. Hu. 1998. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J. Virol. 72:1195-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. A., V. K. Pathak, and W. S. Hu. 2000. Effect of the murine leukemia virus extended packaging signal on the rates and locations of retroviral recombination. J. Virol. 74:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 4.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J.-P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. S. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., T. D. Rhodes, and W.-S. Hu. 2005. Comparison of the genetic recombination rates of human immunodeficiency virus type 1 in macrophages and T cells. J. Virol. 79:9337-9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Z., P. Telfer, A. Gettie, P. Reed, L. Zhang, D. D. Ho, and P. A. Marx. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin, M. P. S., T. D. Rhodes, J. Chen, W. Fu, and W.-S. Hu. 2005. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. USA 102:9002-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel, F., M. D. Hoggan, R. L. Willey, K. Strebel, M. A. Martin, and R. Repaske. 1989. Genetic recombination of human immunodeficiency virus. J. Virol. 63:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courgnaud, V., B. Abela, X. Pourrut, E. Mpoudi-Ngole, S. Loul, E. Delaporte, and M. Peeters. 2003. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different Cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77:12523-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A.-M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz, R. S., E. C. Sabino, A. Mayer, J. W. Mosley, and M. P. Busch. 1995. Dual human immunodeficiency virus type 1 infection and recombination in a dually exposed transfusion recipient. J. Virol. 69:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flynn, J. A., W. An, S. R. King, and A. Telesnitsky. 2004. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 78:12129-12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galetto, R., A. Moumen, V. Giacomoni, M. Veron, P. Charneau, and M. Negroni. 2004. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 279:36625-36632. [DOI] [PubMed] [Google Scholar]
- 19.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 20.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, F., L. Yue, A. T. White, P. G. Pappas, J. Barchue, A. P. Hanson, B. M. Greene, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 23.Giguere, V., K.-I. Isobe, and F. Grosveld. 1985. Structure of the murine Thy-1 gene. EMBO J. 4:2017-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, S. D. C., J. F. Allen, and A. M. L. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guibinga, G. H., A. Miyanohara, J. D. Esko, and T. Friedmann. 2002. Cell surface heparan sulfate is a receptor for attachment of envelope protein-free retrovirus-like particles and VSV-G pseudotyped MLV-derived retrovirus vectors to target cells. Mol. Ther. 5:538-546. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch, V. M., G. A. Dapolito, S. Goldstein, H. McClure, P. Emau, P. N. Fultz, M. Isahakia, R. Lenroot, G. Myers, and P. R. Johnson. 1993. A distinct African lentivirus from Sykes' monkeys. J. Virol. 67:1517-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch, V. M., R. A. Olmsted, M. Murphey-Corb, R. H. Purcell, and P. R. Johnson. 1989. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339:389-392. [DOI] [PubMed] [Google Scholar]
- 30.Hu, W. S., E. H. Bowman, K. A. Delviks, and V. K. Pathak. 1997. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J. Virol. 71:6028-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu, W. S., and H. M. Temin. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. USA 87:1556-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 33.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 34.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jetzt, A. E., H. Yu, G. J. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2000. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 74:1234-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kharytonchyk, S., A. Kireyeva, A. Osipovich, and I. Fomin. 2005. Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the same chromosomal site. Retrovirology 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, E.-Y., M. Busch, K. Abel, L. Fritts, P. Bustamante, J. Stanton, D. Lu, S. Wu, J. Glowczwskie, T. Rourke, D. Bogdan, M. Piatak, Jr., J. D. Lifson, R. C. Desrosiers, S. Wolinsky, and C. J. Miller. 2005. Retroviral recombination in vivo: viral replication patterns and genetic structure of simian immunodeficiency virus (SIV) populations in rhesus macaques after simultaneous or sequential intravaginal inoculation with SIVmac239Δvpx/Δvpr and SIVmac239Δnef. J. Virol. 79:4886-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemey, P., O. G. Pybus, B. Wang, N. K. Saksena, M. Salemi, and A.-M. Vandamme. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. USA 100:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCutchan, F. E., P. A. Hegerich, T. P. Brennan, P. Phanuphak, P. Singharaj, A. Jugsudee, P. W. Berman, A. M. Gray, A. K. Fowler, and D. S. Burke. 1992. Genetic variants of HIV-1 in Thailand. AIDS Res. Hum. Retrovir. 8:1887-1895. [DOI] [PubMed] [Google Scholar]
- 44.Muller, M. C., and F. Barre-Sinoussi. 2003. SIVagm: genetic and biological features associated with replication. Front. Biosci. 8:1170-1185. [DOI] [PubMed] [Google Scholar]
- 45.Novembre, F. J., V. M. Hirsch, H. M. McClure, P. N. Fultz, and P. R. Johnson. 1992. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology 186:783-787. [DOI] [PubMed] [Google Scholar]
- 46.Onafuwa, A., W. An, N. D. Robson, and A. Telesnitsky. 2003. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 77:4577-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Post, K., J. Guo, K. J. Howard, M. D. Powell, J. T. Miller, A. Hizi, S. F. J. Le Grice, and J. G. Levin. 2003. Human immunodeficiency virus type 2 reverse transcriptase activity in model systems that mimic steps in reverse transcription. J. Virol. 77:7623-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes, T., O. Nikolaitchik, J. Chen, D. Powell, and W. S. Hu. 2005. Genetic recombination of human immunodeficiency virus type I in one round of viral replication: effects of genetic distance, target cells, accessory genes, and lack of high negative interference in crossover events. J. Virol. 79:1666-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes, T., H. Wargo, and W. S. Hu. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saksena, N. K., B. Wang, Y. C. Ge, S. H. Xiang, D. E. Dwyer, and A. L. Cunningham. 1997. Coinfection and genetic recombination between HIV-1 strains: possible biological implications in Australia and South East Asia. Ann. Acad. Med. Singapore 26:121-127. [PubMed] [Google Scholar]
- 52.Salemi, M., T. De Oliveira, V. Courgnaud, V. Moulton, B. Holland, S. Cassol, W. M. Switzer, and A.-M. Vandamme. 2003. Mosaic genomes of the six major primate lentivirus lineages revealed by phylogenetic analyses. J. Virol. 77:7202-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Santiago, M. L., F. Range, B. F. Keele, Y. Li, E. Bailes, F. Bibollet-Ruche, C. Fruteau, R. Noe, M. Peeters, J. F. Y. Brookfield, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2005. Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai forest, Cote d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J. Virol. 79:12515-12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S.-J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 56.Sevilya, Z., S. Loya, N. Adir, and A. Hizi. 2003. The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is modulated by residue 294 of the small subunit. Nucleic Acids Res. 31:1481-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharp, P. M., G. M. Shaw, and B. H. Hahn. 2005. Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79:3891-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soares, M. A., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 59.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J.-C. Plantier, F. Gao, K. Abernethy, L. J. T. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsujimoto, H., A. Hasegawa, N. Maki, M. Fukasawa, T. Miura, S. Speidel, R. W. Cooper, E. N. Moriyama, T. Gojobori, and M. Hayami. 1989. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341:539-541. [DOI] [PubMed] [Google Scholar]
- 61.Wooley, D. P., R. A. Smith, S. Czajak, and R. C. Desrosiers. 1997. Direct demonstration of retroviral recombination in a rhesus monkey. J. Virol. 71:9650-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, L., T. D. Martin, R. Vazeux, D. Unutmaz, and V. N. KewalRamani. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J. Virol. 76:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, T., N. Wang, A. Carr, S. Wolinsky, and D. D. Ho. 1995. Evidence for coinfection by multiple strains of human immunodeficiency virus type 1 subtype B in an acute seroconvertor. J. Virol. 69:1324-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhuang, J., S. Mukherjee, Y. Ron, and J. P. Dougherty. 2006. High rate of genetic recombination in murine leukemia virus: implications for influencing proviral ploidy. J. Virol. 80:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]