Abstract
Foot-and-mouth disease virus (FMDV) can use a number of integrins as receptors to initiate infection. Attachment to the integrin is mediated by a highly conserved arginine-glycine-aspartic acid (RGD) tripeptide located on the GH loop of VP1. Other residues of this loop are also conserved and may contribute to integrin binding. In this study we have used a 17-mer peptide, whose sequence corresponds to the GH loop of VP1 of type O FMDV, as a competitor of integrin-mediated virus binding and infection. Alanine substitution through this peptide identified the leucines at the first and fourth positions following RGD (RGD+1 and RGD+4 sites) as key for inhibition of virus binding and infection mediated by αvβ6 or αvβ8 but not for inhibition of virus binding to αvβ3. We also show that FMDV peptides containing either methionine or arginine at the RGD+1 site, which reflects the natural sequence variation seen across the FMDV serotypes, are effective inhibitors for αvβ6. In contrast, although RGDM-containing peptides were effective for αvβ8, RGDR-containing peptides were not. These observations were confirmed by showing that a virus containing an RGDR motif uses αvβ8 less efficiently than αvβ6 as a receptor for infection. Finally, evidence is presented that shows αvβ3 to be a poor receptor for infection by type O FMDV. Taken together, our data suggest that the integrin binding loop of FMDV has most likely evolved for binding to αvβ6 with a higher affinity than to αvβ3 and αvβ8.
Foot-and-mouth disease virus (FMDV) is the etiological agent of foot-and-mouth disease, a severe vesicular disease of cloven-hoofed animals, including cattle, sheep, goats, and pigs. The virus exists as seven serotypes, which are members of the genus Aphthovirus of the family Picornaviridae. The type O and A viruses have the broadest geographical distribution and occur in many parts of the world, including Africa, southern Asia, the Far East, and South America (25).
The mature virus particle consists of 60 copies (each) of four virus-encoded structural proteins, VP1 to VP4. These proteins form an icosahedral capsid that encloses a single-stranded positive-sense RNA genome. A major structural feature of the capsid is a surface-exposed, conformationally flexible loop: the GH loop of VP1 (1, 30). Synthetic peptides corresponding to this loop inhibit FMDV binding and infection of cells in culture (4, 17, 29, 50), are highly immunogenic, and induce high levels of neutralizing antibody (8, 41, 42). In all of the FMD viruses studied, the VP1 GH loop is structurally disordered (1, 13, 27). For the type O viruses this has been attributed, in part, to the presence of a disulfide bond tethering one end of the loop to VP2. Crystallographic analysis of chemically reduced virus (44), in which this disulfide was broken, resulted in the loop adopting predominantly one conformation. In this structure the loop lies in a small depression on the surface of the capsid and is formed by a short region of β-strand, followed by an arginine-glycine-aspartic acid (RGD) tripeptide in an open conformation prior to a 310 helix. This loop structure was conserved in a type C FMDV VP1 GH-loop peptide, in complex with the Fab fragment of two different neutralizing antibodies raised against the same virus (19, 52, 53).
The RGD motif is highly conserved in field strains of FMDV and mediates cell attachment by binding to integrin receptors. To date, four integrins (αvβ1, αvβ3, αvβ6, and αvβ8) have been reported to serve as receptors for FMDV on cultured cells (5, 20, 22, 24, 39). Virus binding to the integrin promotes clathrin-dependent uptake of the virus-integrin complex into early and recycling endosomes, where the prevailing low pH triggers capsid disassembly and transfer of the viral RNA into the cytoplasm by a currently unknown mechanism (3, 7, 40). Integrins are also believed to be the receptors used to initiate infection in animals (35, 37). Recently, we have shown that αvβ6, but not αvβ3, is constitutively expressed on the epithelial cells targeted during the acute phase of infection in cattle (37), suggesting that αvβ6, rather than αvβ3, is the receptor that determines the tissue tropism of FMDV.
Despite having an RGD, FMDV appears to be unable to use all of the RGD-dependent integrins as receptors to initiate infection, and evidence that two such integrins, α5β1 and αvβ5, serve as receptors for FMDV has been consistently negative (2, 32, 39). The factors that determine the integrin specificity of FMDV are not well defined. In addition to the RGD, other residues of the VP1 GH loop are conserved across the different FMDV serotypes, and these could play a significant role in integrin binding. In FMDV, a leucine (L or Leu) (or rarely an isoleucine) is conserved at the fourth position following RGD (RGD+4), and Leu is most commonly found in the RGD+1 position; however, arginine (R or Arg) or methionine (M or Met) can occupy this location in certain non-type-O viruses. A number of studies have implicated the residues that flank the RGD in determining the receptor specificity of FMDV. Rieder et al. (1994) and Leippert et al. (1997) concluded that the residues that follow the RGD motif influence virus binding to BHK cells (28, 43). However, virus binding to BHK cells may also be mediated by surface heparan sulfate (21, 45), and integrin binding by these viruses has not been determined. Mateu et al. (1996) showed that certain amino acid substitutions at the RGD+1 and RGD+4 sites dramatically reduced the ability of synthetic peptides, derived from the VP1 GH loop of FMDV C-S8c1, to inhibit infection of BHK cells by the same strain of virus (33). However, due to a lack of antibody reagents to hamster integrins, the identities of the integrins expressed on BHK cells have not been determined. Our studies have shown that although FMDV binding to αvβ1 and αvβ3 can be inhibited by a GRGDSP peptide, this peptide is an ineffective inhibitor of virus binding to αvβ6. These observations suggest that residues in addition to RGD are required for peptide binding to αvβ6. However, the contribution made by each residue of the VP1 GH loop to the integrin specificity of FMDV is currently unknown.
Here we show that the integrin-binding loop of FMDV is most highly adapted for binding to integrin αvβ6. Alanine-scanning substitution of a 17-mer peptide, corresponding in sequence to the VP1 GH loop of type O FMDV, showed that the residues that flank the RGD are not required for peptide binding to αvβ3. In contrast, alanine substitution at the RGD+1 and RGD+4 sites reduced the ability of the peptide to bind αvβ6 and αvβ8. Furthermore, we show that the natural sequence variation at the RGD+1 site (RGDL, RGDM, or RGDR) seen across the FMDV serotypes did not compromise the ability of the peptide to bind αvβ6. In contrast, an Arg at RGD+1 was shown to be unfavorable for peptide binding to αvβ8. These observations were supported by showing that a virus containing an RGDR motif uses αvβ8 less efficiently than αvβ6 as a receptor for infection. Finally, evidence is presented that shows αvβ3 to be a poor receptor for infection by type O FMDV.
MATERIALS AND METHODS
Cells and viruses.
BHK, MDBK, and SW480 cell lines transfected to express human integrins (SW480-αvβ6, SW480-αvβ3, and SW480-αvβ8) were cultivated as described previously (11, 23, 38, 54, 59). IBRS2 cells were cultivated in Glasgow's modified Eagle's medium supplemented with 10% adult bovine serum, 20 mM glutamine, penicillin (100 SI units/ml), and streptomycin (100 μg/ml). Primary bovine thyroid (pBTY) cells were prepared and cultivated as described previously (48). Preparation of working stocks of FMDV O1Kcad2 using pBTY cells and virus purification on sucrose gradients were as described previously (13). Working stocks of FMDV O1BFS were prepared using BHK cells. Unless otherwise stated, the multiplicity of infection (MOI) was based on the virus titer on BHK cells as described previously (24).
Peptides.
The FMDV 17-mer peptide (VPNLRGDLQVLAQKVAR) with its sequence derived from the VP1, GH loop of FMDV O1Kcad2 (henceforth known as the wild-type (WT) FMDV peptide) (RGD tripeptide is highlighted in bold) and its control RGE version were synthesized at the peptide synthesis facility at the Institute for Animal Health, Compton, United Kingdom. A number of variant peptides based on this sequence were also synthesized. These included peptides containing RGDM (L8M) or RGDR (L8R) in place of the WT RGDL motif and a series of peptides where each residue in turn of the FMDV 17-mer was replaced by an alanine (A). These peptides are referred to here as V1A, P2A, N3A, L4A, R5A, G6A, D7A, L8A, Q9A, V10A, L11A, Q13A, K14A, V15A, and R17A. Other peptides used were a 17-mer peptide (STAIRGDRAVLAAKYAN) (RGDR motif is boldfaced) with its sequence base on the VP1, GH loop of SAT-2 FMDV and variants of this peptide containing RGDL or RGDM in place of RGDR. The FMDV 12-mer peptide (VPNLRGDLQVLA) and its control RGE version were synthesized at the Oxford Centre for Molecular Science, New Chemistry Laboratory, Oxford, United Kingdom. The GRGDSP peptide and its control RGE version were purchased from Novabiochem.
Integrins and antibodies.
The purified human integrin αvβ3 was obtained from Chemicon. Recombinant αvβ6 was purified from Chinese hamster ovary cells stably transfected with human αv and β6 integrin cDNA (54). Each chain was truncated to remove the transmembrane and cytoplasmic domains, and integrin purification was carried out as previously described by Ferris et al. (2005) (16). The integrin used in this study was of the same batch used by Ferris et al. (2005), and the purified integrin separated on a sodium dodecyl sulfate-polyacrylamide gel as two clear bands which correspond to the truncated αv and β6 subunits (16).
The anti-integrin, mouse monoclonal antibodies (MAbs) used in this study were 23C6 (immunoglobulin G1 [IgG1]) and LM609 (IgG1), both anti-αvβ3; P1F6 (IgG1), anti-αvβ5; R6G9 (IgG2a), anti-β6; 10D5 (IgG2a), anti-αvβ6 (all from Chemicon); 37E1 (IgG2a), anti-αvβ8 (20); L230 (IgG1), anti-αv (23); and 6.8G6 (IgG1), anti-αvβ6 (55). The LIBS-1 MAb was obtained from Mark Ginsberg (Department of Medicine, University of California San Diego, La Jolla, Calif.). The anti-FMDV MAbs D9 (mouse IgG2a) and B2 (mouse IgG1), which recognize antigenic site 1 of type O FMDV (34), and MAb 2C2 (mouse IgG2a) (14), which recognizes the 3A protein (10), were obtained from Emiliana Brocchi (Istituto Zooprofilattico Sperimentale della Lombardia e dell'Emilia Romagna, Brescia, Italy). MAbs B2 and D9 were purified with protein A (Pierce) according to the manufacturer's instructions. MAb 2C2 was used as diluted ascites. The guinea-pig anti-type-O FMDV polyclonal antiserum was obtained from the FMDV world reference center Institute for Animal Health (Pirbright, United Kingdom).
ELISA.
The use of integrins as immobilized ligands for FMDV in an enzyme-linked immunosorbent assay (ELISA) has been previously described in detail (23). The same protocol was used here for αvβ6. Briefly, plastic 96-well plates were coated with integrin at the appropriate concentration in coating buffer (20 mM Tris [pH 7.4], 2 mM CaCl2, 1 mM MgCl2 in 0.85% saline) overnight at 4°C. One hundred microliters of purified FMDV (1 μg/ml) in binding buffer (coating buffer containing 2% bovine serum albumin) was added to the integrin-coated wells for 1 h at room temperature (rt). The wells were washed three times and incubated sequentially with a guinea-pig, anti-type-O FMDV polyclonal antiserum and a rabbit, anti-guinea-pig alkaline phosphatase conjugate (Sigma) for 45 min each at rt. Wells were washed three times, and alkaline phosphate substrate (Sigma) was added. The optical density of the wells was read at 405 nm (OD405). Competing antibodies were added to the immobilized integrin for 15 min prior to the addition of virus. Competing peptides were mixed with FMDV prior to the addition to the integrin-coated wells. The amount of integrin immobilized on the plate was determined using the anti-αv MAb L230 and a rabbit, antimouse alkaline phosphatase conjugate in the assay described above.
Flow cytometry.
Cells were harvested using cell dissociation solution (Sigma) with the exception of pBTY cells, which were harvested using limited digestion with trypsin. Following trypsinization, the pBTY cells were incubated for 10 min in cell culture medium. Integrin expression and virus binding were analyzed by flow cytometry on ice as previously described (20). Briefly, the cells (30 μl/well; 1 × 107 cells/ml) in fluorescence-activated cell sorting buffer (20 mM Tris [pH 7.4], 1 mM CaCl2, 0.5 mM MgCl2, 2% goat serum, 2% bovine serum albumin in 0.85% saline) were incubated sequentially with primary anti-integrin (10 mg/ml) and secondary goat anti-mouse isotype-specific R-phycoerythrin-conjugated antibodies (Southern Biotechnology Associates) for 30 min each. The cells were washed and resuspended in 1% paraformaldehyde. Fluorescence staining was analyzed by flow cytometry using a FACSCalibur (Becton Dickinson), counting 6,000 cells per sample. Background fluorescence was determined by omitting the primary antibody from the assay. For virus binding, the cells were incubated with purified O1Kcad2 (10 μg/ml) for 0.5 h. After washing, the cells were incubated sequentially with anti-FMDV MAb D9 (10 μg/ml) and a goat antimouse IgG2a-specific R-phycoerythrin conjugate (Southern Biotechnology Associates). Background fluorescence was determined by omitting either FMDV or MAb D9 from the assay. Both control conditions gave nearly identical results. For competition experiments, peptides or anti-integrin MAbs were added to the cells for 0.5 h prior to addition of virus. For the experiments involving competition with peptides or anti-integrin IgG1 MAbs, cell-bound virus was also detected using the MAb D9 (IgG2a). However, when the competing MAb was IgG2a, virus was detected using the MAb B2 (IgG1). For these experiments, additional controls were performed to verify that the R-phycoerythrin-conjugated secondary antibodies did not cross-react with the competing MAbs.
Infectivity assay.
For the enzyme-linked immunospot (ELISPOT) assay, cells prepared in 96-well plates were infected with FMDV for 1 h at 37°C. The cells were washed twice for 1 min each with 0.1 M citric acid, pH 5.2, to inactivate the extracellular virus. The pH was restored by washing four times with Dulbecco's modified Eagle's medium, and infection was allowed to progress for a further 4 h at 37°C. Cells were fixed in 4% paraformaldehyde for 45 min and infection quantified using an ELISPOT plate reader as described previously (7). Briefly, infected cells are permeabilized with 0.1% Triton and identified by sequential incubation with the anti-FMDV MAb 2C2 (which recognizes the nonstructural 3A protein), a goat antimouse IgG2a-specific biotin conjugate (Southern Biotechnology Associates), and a streptavidin-conjugated alkaline phosphatase. In the presence of enzyme substrate, the infected cells are stained dark blue and counted using an ELISPOT apparatus (Carl Zeiss). For competition experiments, the cells were pretreated with either the peptides or MAbs for 0.5 h at rt prior to addition of virus. In experiments using MAbs as competitors, the competing antibody was used at a concentration that could not be detected in the assay.
Modeling the FMDV GH loop/αvβ3 complex.
Residues RGD (145 to 147) from VP1 of FMDV O1BFS (Protein Data Bank [PDB] code 1FOD) (6) were superimposed on the RGDF ligand bound to αvβ3 (PDB 1L5G) (6) using the program SHP (49). The conformation of RGD motifs is closely conserved in active integrin binding ligands, hence the superimposition gave a root mean square deviation of 0.45 Å2. The rest of the FMDV loop was then rotated accordingly. The overall structure of αvβ3 is expected to be comparable to that of αvβ6, since β3 and β6 show 47% amino acid identity over the ectodomains (46).
RESULTS
FMDV binding to purified integrins αvβ3 and αvβ6.
Initially we established an ELISA to investigate FMDV binding to purified αvβ6. Virus binding to the immobilized integrin was detected using a guinea-pig anti-FMDV polyclonal antiserum and a rabbit anti-guinea-pig alkaline phosphatase conjugate (see Methods). This assay has been used previously to demonstrate authentic cation- and RGD-dependent binding of FMDV to αvβ3 (23). Figure 1A shows that binding of virus (FMDV O1Kcad2) to αvβ3 (1 μg/ml) and αvβ6 (0.25 μg/ml) is concentration dependent and saturable. However, it should be noted that the data shown in Fig. 1A were obtained 0.5 h and 2.5 h after the addition of enzyme substrate (see Methods) for αvβ6 and αvβ3, respectively. Figure 1B shows the OD405s for virus binding to αvβ3 (1 μg/ml) and αvβ6 (0.25 μg/ml) recorded at various time points after the addition of enzyme substrate. At all time points investigated, the OD405 was much greater for αvβ6 than for αvβ3, showing that a greater amount of virus had bound this integrin. These observations could be explained if αvβ6 was immobilized with a greater efficiency to the ELISA plate. To test this point, equivalent amounts of integrin (1 μg/ml) were applied to the plate, and bound integrin was detected with the anti-αv MAb L230 (see Methods). Similar OD405s for MAb L230 binding were obtained for both integrins, showing comparable amounts of integrin were immobilized (data not shown). The above observations suggest that under nearly identical ELISA conditions, a greater amount of virus binds to αvβ6 than to αvβ3.
FIG. 1.
FMDV binding to purified integrins. The integrin was immobilized on the plate, and virus (FMDV O1Kcad2; 1 μg/ml) binding was detected using an ELISA (see Methods). Panels A and B show virus binding (OD405) to integrins αvβ3 (1 μg/ml) (solid triangles) and αvβ6 (0.25 μg/ml) (solid squares). For panel A the data were collected at 2.5 h and 0.5 h after addition of the enzyme substrate for αvβ3 and αvβ6, respectively. Each point is the mean ± standard deviation for triplicate wells. Panel B shows virus binding to αvβ3 and αvβ6 as a function of time after addition of the enzyme substrate. Each point is the mean ± standard deviation for triplicate wells. Panel C shows that FMDV binding to αvβ6 is inhibited by function-blocking MAbs (10 μg/ml) to αvβ6 (10D5) or the αv subunit (L230) but not by a function-blocking MAb to αvβ5 (P1F6) or the nonblocking anti-β6 MAb (R6G9). Virus binding is expressed as a percentage of virus binding in the absence of competing antibodies. Each point is the mean ± standard deviation for triplicate wells.
Figure 1C shows that binding of virus to αvβ6 is inhibited by a function-blocking MAb to either αvβ6 (10D5) or the αv subunit (L230) but not by a function-blocking MAb to αvβ5 (P1F6) or by the nonblocking anti-β6 MAb (MAb R6G9), confirming binding was mediated primarily by αvβ6. In addition, virus binding to αvβ6 was completely inhibited by 1 mM EDTA (data not shown), an observation consistent with the known cation dependency of ligand binding to this integrin (55).
Peptide inhibition of FMDV binding to purified integrins αvβ3 and αvβ6.
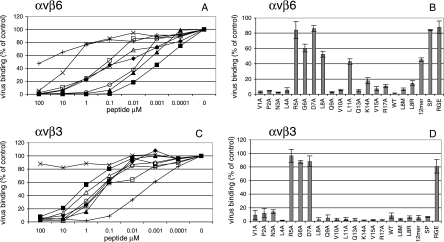
Next we determined the inhibitory effect of synthetic RGD-containing peptides on FMDV binding to the purified integrins. Figure 2 shows that virus binding to αvβ3 (Fig. 2C and D) and αvβ6 (Fig. 2A and B) is inhibited by the WT FMDV peptide but not by its non-biologically active RGE version. The 50% inhibitory concentration (IC50) of the peptide was much greater for αvβ3 (IC50 = 800 nM) than αvβ6 (IC50 = 0.3 nM), suggesting that the WT FMDV peptide has a greater affinity for αvβ6. Two other RGD-containing peptides were included in these studies: (i) a short version of the FMDV peptide truncated after the Ala residue at position 12 (henceforth known as the FMDV 12-mer), and (ii) a GRGDSP peptide. Truncation of the five C-terminal residues increased the inhibitory effect of the WT FMDV peptide on virus binding to αvβ3 (∼4-fold reduction in the IC50 [IC50 = 230 nM]; Table 1). In contrast, for αvβ6 the truncated peptide was a less-effective inhibitor than the WT FMDV peptide, as shown by a ∼70-fold increase in the IC50 (IC50 = 22 nM). These observations suggest that the C-terminal residues of the WT FMDV peptide are involved in binding to αvβ6 but not to αvβ3. This conclusion was supported by the observations that although the GRGDSP was a potent inhibitor of FMDV binding to αvβ3 (IC50 = 2.6 nM) (Fig. 2C and 2; Table 1), this peptide was a poor inhibitor for αvβ6, being equivalent to the RGE control version of the WT FMDV peptide (Fig. 2B).
FIG. 2.
Peptide inhibition of FMDV binding to purified integrins αvβ3 and αvβ6. Panels A and B and panels C and D show virus (FMDV O1Kcad2; 1 μg/ml) binding to αvβ6 and αvβ3, respectively. Virus binding is shown as the percentage of virus binding in the absence of competition. Panels A and C, solid squares show data for the WT FMDV peptide; solid diamond, FMDV 12-mer; solid triangle, L8M; open circle, L8R; X, D7A; open square, L8A; open triangle, L11A; +, GRGDSP. Panels B and D show virus binding in the presence of one peptide concentration (0.1 μM for αvβ6 and 100 μM for αvβ3), i.e., the concentration where the FMDV peptide inhibited virus binding by ∼95% (see panels A and C). Peptides: WT, WT FMDV peptide; RGE, control RGE version of the WT FMDV peptide; 12mer, the FMDV 12-mer; SP, GRGDSP; variants of the WT FMDV peptide containing amino acid substitutions are indicated (L8M, L8R, or V1A-R17A). Means and standard deviations are shown for three independent experiments, each conducted in duplicate.
TABLE 1.
Peptide inhibition of virus binding
| Peptidea | IC50 (nM) for binding of virus to integrin or cell type
|
|||||
|---|---|---|---|---|---|---|
| αvβ3b | αvβ6b | SW480-αvβ6 | pBTY | SW480-αvβ8 | IBRS2 | |
| WTc | 800 | 0.3 | 6.7 | <19 | 125 | 200 |
| L8M | 175 | 5.3 | 1.8 | ND | 480 | 460 |
| L8R | 150 | 10 | 6 | ND | 8,100 | 5,200 |
| 12-merd | 230 | 22 | 2,800 | ND | 2,400 | 3,400 |
| L8A | 70 | 100 | 2,300 | 1,000 | 10,900 | 10,800 |
| L11A | 300 | 40 | 1,700 | 310 | 3,700 | 2,000 |
| D7A | >10,000 | 2,100 | 9,000 | 7,400 | 70,000 | 51,000 |
| GRGDSP | 2.6 | 54,000 | NDe | ND | ND | ND |
Peptides containing amino acid substitutions (single-letter code) are denoted by the amino acid in the FMDV peptide and the residue number, followed by the substituted amino acid.
Purified integrin.
The FMDV peptide VPNLRGDLQVLAQKVAR. (RGD motif is boldfaced.)
The FMDV 12-mer peptide VPNLRGDLQVLA. (RGD motif is boldfaced.)
ND, not done.
Figure 2 also shows the results of competition experiments using a series of variant peptides where each residue in turn of the WT FMDV peptide was replaced by Ala. For clarity, only a selected number of peptides are shown in panels A and C, whereas panels B and D show data for all peptides but at only one concentration (i.e., the concentration where the WT FMDV peptide inhibited virus binding by ∼95%). As expected, Ala substitutions within the RGD dramatically reduced the inhibitory effect of the peptide on virus binding to both integrins (Fig. 2B and D). Alanine substitution at any other residue did not reduce the inhibitory effect of the peptide for αvβ3 (Fig. 2D; Table 1). In fact, Ala substitutions at several of these sites led to a reduction in the IC50 of the peptide (Table 1). Similarly, Ala substitution at most of the non-RGD residues did not reduce the inhibitory effect of the peptide on virus binding to αvβ6 (Fig. 2B). However, in contrast to αvβ3, Ala substitution at the RGD+1 (L8A peptide) or RGD+4 (L11A peptide) site led to a marked reduction in the inhibitory effect of the peptide for αvβ6 (Fig. 2A and B; Table 1). The IC50s of the L8A and L11A peptides were increased ∼330 and ∼130-fold, respectively, over that of the WT FMDV peptide. These data show that binding of the WT FMDV peptide to αvβ3 is dependent only on the RGD motif, whereas binding to αvβ6 requires an intact RGD and the presence of Leu residues at the RGD+1 and RGD+4 sites. However, despite containing the RGD+1 and RGD+4 leucines, the FMDV 12-mer had an IC50 similar to that of the L8A and L11A peptides, which suggests that the structure of the FMDV peptide may also influence integrin binding (see Discussion).
Although Leu is most commonly found immediately following the RGD, in some non-type-O viruses, arginine (Arg) or methionine (Met) can occupy this site. To reflect this variation, two further FMDV peptides containing either Arg or Met at the RGD+1 site were investigated. Substitution of Leu at the RGD+1 site by either Arg (L8R) or Met (L8M) led to an increase in the inhibitory effect of the FMDV peptide (∼4-fold reduction in the IC50; Table 1) on virus binding to αvβ3. In contrast, the peptides containing RGDM (IC50, 5.3 nM) or RGDR (IC50, 10 nM) appeared to have a reduced inhibitory effect on virus binding to αvβ6 (L8M, ∼16-fold, and L8R, ∼30-fold increase in the IC50 [Table 1]) compared to that of the WT FMDV peptide. However, the differences in IC50s of the WT FMDV, L8R, and L8M peptides for αvβ6 were not observed when the integrin was present at the surfaces of cells (Table 1) (see Discussion).
Peptide inhibition of FMDV binding and infection of SW480 integrin-transfected cell lines.
Next we investigated the inhibitory effect of the above peptides on FMDV binding to integrins when they are expressed on the surfaces of cells. Recently we have shown that αvβ8 is a receptor for FMDV infection (20), and this integrin was included in these studies. For these studies we used SW480 cell lines that had been stably transfected to express either human αvβ3 (SW480-αvβ3) (59), αvβ6 (SW480-αvβ6) (54), or αvβ8 (SW480-αvβ8) (11, 38).
Binding of FMDV to SW480 cells expressing either αvβ6 or αvβ8 was readily detected (Fig. 3). This binding was inhibited by >70% (data not shown) by function-blocking MAbs to αvβ6 (MAb 10D5) or αvβ8 (MAb 37E1), respectively, thus confirming our previous observations, which concluded that αvβ6 and αvβ8 are the major receptors for virus attachment on SW480-αvβ6 and SW480-αvβ8 cells (20, 24). In contrast, virus binding to SW480-αvβ3 cells was low (Fig. 3), and we were unable to carry out antibody and peptide competition studies (see below) using this cell line.
FIG. 3.
Flow cytometric analysis of FMDV binding to integrin-transfected SW480 cells. The panels show FMDV (O1Kcad2; 1 μg/ml) binding to SW480-αvβ3, SW480-αvβ6, or SW480-αvβ8. Virus binding (white histogram) was detected using the anti-FMDV MAb D9 and a goat antimouse IgG2a-specific R-phycoerythrin conjugate. Background fluorescence (shaded histogram) was determined by omitting the primary antibody. One experiment representative of three is shown; each gave similar results.
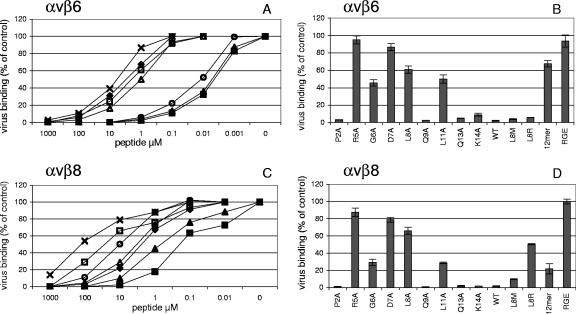
The inhibitory effects of the above peptides on FMDV binding to αvβ6 at the surfaces of SW480-αvβ6 cells were similar to those seen with the purified integrin (Fig. 4A and B) as follows: (i) an intact RGD was required for effective inhibition, since peptides containing amino acid substitutions within this motif were poor inhibitors; (ii) truncation of the five C-terminal residues (the FMDV 12-mer) lead to an increase in the IC50 of the peptide, although the effect of this truncation on the IC50 appeared much greater for SW480-αvβ6 (∼400-fold increase) than for the purified αvβ6 integrin (∼70-fold increase); and (iii) Ala substitution at the RGD+1 or RGD+4 sites resulted in an increased IC50 of the peptide (L8A, ∼350-fold, and L11A, ∼250-fold), whereas Ala substitution at any other residue did not alter its inhibitory effect. However, one noticeable difference between purified and cell-associated integrin is that with SW480-αvβ6 cells, the peptides containing either Met (L8M) or Arg (L8R) at the RGD+1 site had an inhibitory effective on virus binding similar to that of the WT FMDV peptide (containing an RGDL) (Fig. 4A and B; Table 1). As stated above, this is in contrast with the observations made using these peptides in competition experiments with purified αvβ6 (see Discussion).
FIG. 4.
Peptide inhibition of FMDV binding to integrin-transfected SW480 cells. Panels A and B and panels C and D show virus binding to SW480-αvβ6 and SW480-αvβ8, respectively. Virus binding is shown as the percentage of virus binding in the absence of competition. In panels A and C, solid squares show WT FMDV peptide; solid diamond, FMDV 12-mer; solid triangle, L8M; open circle, L8R; X, D7A; open square, L8A; open triangle, L11A. Panels B and D show virus binding in the presence of one peptide concentration (1 μM for SW480-αvβ6 or 10 μM for SW480-αvβ8), i.e., the concentration where the WT FMDV peptide inhibited virus binding by ∼95% (see panels A and C). Peptides: WT, WT FMDV peptide; RGE, control RGE version of the WT FMDV peptide; 12mer, FMDV 12-mer; variants of the WT FMDV peptide containing amino acid substitutions are indicated (L8M, L8R, or P2A-K14A). Means (n = 3) and standard deviations are shown for one experiment representative of two. Each gave nearly identical results.
The WT FMDV peptide also inhibited FMDV binding to αvβ8 on SW480-αvβ8 cells, and this inhibition was dependent on an intact RGD (Fig. 4C and D; Table 1). Although we have not determined the precise level of integrin expression on SW480-αvβ6 and SW480-αvβ8 cells, some clues regarding the relative inhibitory efficiency of the WT FMDV peptide for αvβ6 and αvβ8 can be gained by comparing the amount of virus binding with the IC50s. The IC50 of the WT FMDV peptide was ∼18 times greater for SW480-αvβ8 (IC50 = 125 nM) than for SW480-αvβ6 (IC50 = 6.7 nM), even though the two sets of cells bound similar amounts of virus (Table 1; Fig. 3). These observations suggest that the WT FMDV peptide has a greater affinity for αvβ6 over αvβ8.
Similarly to the case with SW480-αvβ6, truncation of the five C-terminal residues increased the IC50 of the peptide (by ∼20-fold) for virus binding to SW480-αvβ8 cells, as did Ala substitution at either the RGD+1 (∼90-fold) or RGD+4 (∼30-fold) site. These data suggest that the C-terminal residues (in particular the Leu residues at the RGD+1 and RGD+4 sites) are also required for binding of the FMDV peptide to αvβ8. Also similar to the case with SW480-αvβ6 was the observation that the RGDM-containing peptide had an inhibitory effect similar to that of the WT FMDV peptide on virus binding to SW480-αvβ8. However, in contrast to SW480-αvβ6, inclusion of Arg at the RGD+1 site appeared less favorable for binding to αvβ8, since the peptide containing an RGDR motif was a poor inhibitor of virus binding to this integrin (Fig. 4C and D; Table 1).
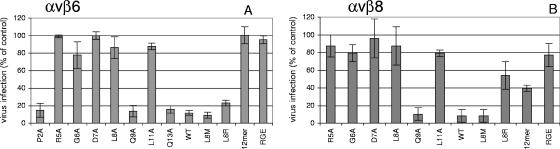
Next we investigated the ability of the above peptides to inhibit infection of the integrin-transfected cells (Fig. 5). SW480-αvβ3, SW480-αvβ6, and SW480-αvβ8 cells were infected with FMDV O1Kcad2 at an MOI of ∼1, and infection was quantified using an antibody to the FMDV 3A proteins (a marker for virus replication) in the ELISPOT assay (see Methods). At this MOI, ∼40% of the SW480-αvβ6 and ∼30% of the SW480-αvβ8 cells were infected, whereas the number of SW480-αvβ3 cells infected was low (<1%). Competition experiments (data not shown) using function-blocking MAbs to αvβ6 (MAb 10D5) and αvβ8 (MAb 37E1) confirmed our previous observations that these integrins are used as receptors to initiate infection on SW480-αvβ6 and SW480-αvβ8 cells, respectively. Since the number of SW480-αvβ3 cells infected was low, the effect on infection of antibody or peptide competition could not be quantified.
FIG. 5.
Peptide inhibition of FMDV infection of integrin-transfected cells. Panel A shows data for SW480-αvβ6 and panel B for SW480-αvβ8 cells. Infection is shown as the percentage of cells infected in the absence of competition and was quantified using MAb 2C2 and the ELISPOT reader (see Methods). Peptides: 1 μM (panel A) or 10 μM (panel B); WT, the WT FMDV peptide; RGE, the control RGE version of the WT FMDV peptide; 12mer, the FMDV 12-mer; variants of the WT FMDV peptide containing amino acid substitutions are indicated (L8M, L8R, or P2A-Q13A). The means (n = 3) and standard deviations are shown for one experiment representative of two. Each gave nearly identical results.
The inhibitory effect of the above peptides on FMDV binding to αvβ6 and αvβ8 was fully reflected in their ability to inhibit infection mediated by these same integrins, since under conditions where infection of SW480-αvβ6 and SW480-αvβ8 cells was inhibited by the WT FMDV peptide, (i) peptides containing amino acid substitutions within the RGD were poor inhibitors; (ii) Ala substitutions at the RGD+1 or RGD+4 sites dramatically reduced the inhibitory effect of the peptide; (iii) compared to the WT FMDV peptide, the FMDV 12-mer was a less-potent inhibitor of infection mediated by either integrin; (iv) the peptide containing an RGDM motif inhibited infection mediated by both αvβ6 and αvβ8; and (v) although the peptide containing an RGDR motif inhibited infection mediated by αvβ6, this peptide was a poor inhibitor of infection mediated by αvβ8.
Peptide competition studies using integrins derived from the natural hosts of FMDV.
The above studies have identified the sequence requirements for binding of a type O FMDV, VP1 GH-loop peptide (the WT FMDV peptide) to integrins αvβ3, αvβ6, and αvβ8. In addition, these studies have suggested that αvβ3 is a poor receptor for infection by type O FMDV. For the above studies we used integrins of human origin. Although human and bovine integrin subunits share a high degree of amino acid sequence identity (93% for the β6 subunits and 95% for both the αv and β3 subunits), humans are not normally susceptible to FMDV. We therefore confirmed our observations with integrins derived from the natural bovine and porcine hosts. For these studies we used a number of bovine and porcine cells expressing defined integrin receptors. These were the bovine kidney cell line (MDBK) and an immortalized bovine thyroid cell line (CT23), both of which express αvβ3 (Fig. 6); a porcine kidney cell line (IBRS2), which expresses αvβ8 (Fig. 6); and primary bovine thyroid cells (pBTY). Primary BTY cells are normally enriched for cells expressing αvβ6 but may also contain a small number of cells expressing αvβ3 and αvβ8 (Fig. 6).
FIG. 6.
Flow cytometric analysis of FMDV binding and integrin expression on IBRS2, MDBK, and pBTY cells. Expression of integrins αvβ3, αvβ6, and αvβ8 and FMDV (O1Kcad2; 1 μg/ml) binding to pBTY, IBRS2, MDBK, and CT23 cells are shown. Virus binding (white histogram) was detected using the anti-FMDV MAb D9 and a goat antimouse IgG2a-specific R-phycoerythrin conjugate. Background fluorescence (shaded histogram) was determined by omitting the anti-FMDV antibody. Integrin expression (white histogram) was detected using MAb 23C6 (αvβ3), MAb 10D5 (αvβ6), or MAb 37E1 (αvβ8). Background fluorescence (shaded histogram) was determined by omitting the anti-integrin antibody. For CT23 cells, virus binding is also shown in the presence of Mn2+ ions (black histogram). One experiment representative of three is shown, and each gave similar results.
FMDV binding was detected with MDBK cells (Fig. 6); however, this binding was not inhibited by function-blocking MAbs (23C6 and LM609; 10 μg/ml) to αvβ3 (data not shown). Similar to our observations with SW480-αvβ3 cells, virus binding (Fig. 6) to CT23 cells was low. Ligand binding to αvβ3 is regulated by changes in the activation state of the integrin (see Discussion); therefore, a possible explanation for the poor utilization of αvβ3 as a receptor for FMDV attachment on MDBK and CT23 cells is that αvβ3 may be primarily in an inactive or low-affinity state. Ligand binding (including FMDV) to αvβ3 is enhanced following integrin activation by manganese (Mn2+) ions (23, 47, 51). Inclusion of Mn2+ (1 mM) in the binding buffer led to an increase in the amount of virus binding to CT23 cells (Fig. 6, black histogram) but not to MDBK cells (data not shown). However, similar to the case with MDBK cells, this binding was not inhibited by preincubation of the cells with the anti-αvβ3, function-blocking MAb 23C6 or LM609. These data show that although FMDV binds to MDBK and CT23 cells, it could not be concluded that αvβ3 was being used as a receptor for virus attachment.
Using the ELISPOT assay, infection of MDBK and CT23 cells by FMDV O1Kcad2 was poor (<1% of the cells were infected). Similarly, infection of these cells was poor in the presence of 1 mM Mn2+. To confirm the low susceptibility of these cells to FMDV, the time of infection was extended to 24 h. Under these conditions the infectivity of both cell lines remained poor. To confirm that the low infectivity was not due to an inability of these cells to support intracellular virus replication, CT23 and MDBK cells were infected with FMDV O1BFS. This virus binds heparan sulfate and uses heparan sulfate proteoglycans as alternative receptors for cell entry without the mediation of integrins (21, 45). Infection by O1BFS resulted in an extensive cytopathic effect at ∼8 h postinfection (data not shown), indicating that the failure of these cells to support infection by the O1Kcad2 virus was due not to a general defect in intracellular virus replication but to a failure of the virus to enter the cell.
FMDV binding to pBTY and IBRS2 cells was detected (Fig. 6), and this binding was inhibited (∼70% inhibition) by function-blocking MAbs to either αvβ6 (MAb 6.8G6; 10 μg/ml) or αvβ8 (MAb 37E1; 10 μg/ml), respectively (data not shown). Virus binding to pBTY and IBRS2 cells was not inhibited by function-blocking MAbs to αvβ3 (MAbs 23C6 and LM609), showing that this integrin is not a major receptor for virus attachment on these cells. Consistent with the above observations, MAb 6.8G6 (but not MAb 23C6) inhibited infection of pBTY cells by >80%, whereas MAb 37E1 (but not MAbs 23C6 or 6.8G6) inhibited infection of IBRS2 cells by ∼55%. Taken together, these observations confirm αvβ6 and αvβ8 as the major receptors used by FMDV for virus attachment and infection of pBTY and IBRS2 cells, respectively.
We also determined the ability of a number of the above peptides to inhibit FMDV binding (Fig. 7) and infection (Fig. 8) of pBTY (mediated by αvβ6) and IBRS2 cells (mediated by αvβ8). These studies confirmed the above observations made with the integrin-transfected SW480 cell lines, since under conditions where the WT FMDV peptide was an effective inhibitor of virus binding and infection for pBTY and IBRS2 cells, (i) peptides containing amino acid substitutions within the RGD were poor inhibitors, (ii) Ala substitution at the RGD+1 or RGD+4 site reduced the inhibitory effects of the peptide, and (iii) although the peptide containing an RGDM motif was an effective inhibitor of virus binding and infection of IBRS2 cells (via αvβ8), the RGDR peptide was not (Fig. 7 and 8; Table 1).
FIG. 7.
Peptide inhibition of FMDV binding to IBRS2 and pBTY cells. Panels A and B and panels C and D show virus binding to pBTY and IBRS2 cells, respectively. Virus binding is shown as the percentage of virus bound in the absence of competition. In panels A and C, the solid square represents the WT FMDV peptide; solid diamond, FMDV 12-mer (not in panel A); solid triangle, L8M (not in panel A); open circle, L8R (not in panel A); X, D7A; open square, L8A; open triangle, L11A. Panels B and D show virus binding in the presence of one peptide concentrations (1 μM for pBTY cells and 10 μM for IBRS2 cells), i.e., the concentration where the WT FMDV peptide inhibited virus binding by ∼95% (see panels A and C). Peptides: WT, the WT FMDV peptide; RGE, the control RGE version of the WT FMDV peptide; 12mer, the FMDV 12-mer; variants of the WT FMDV peptide containing amino acid substitutions are indicated (L8M, L8R or P2A-K14A). Means (n = 3) and standard deviations are shown for one experiment representative of two; each gave nearly identical results.
FIG. 8.
Peptide inhibition of infection of pBTY and IBRS2 cells by FMDV. Panels A and B show data for pBTY and IBRS2 cells, respectively. Infection is shown as the percentage of infected cells in the absence of competing peptides and was quantified using MAb 2C2 and the ELISPOT reader (see Methods). Peptides: 1 μM (panel A) or 10 μM (panel B); WT, the WT FMDV peptide; RGE, the control RGE version of the WT FMDV peptide; 12mer, the FMDV 12-mer; variants of the WT FMDV peptide containing amino acid substitutions are indicated (L8M, L8R, or P2A-L11A). Means (n = 3) and standard deviations are shown for one experiment representative of two; each gave nearly identical results.
FMDV binding to αvβ3 does not enhance MAb LIBS-1 binding.
As stated above, a possible explanation for the poor utilization of αvβ3 as a receptor for FMDV attachment on MDBK, CT23, and SW480-αvβ3 cells is that the integrin may be in an inactive or low-affinity state. Ligand binding to αvβ3 can be detected using so-called ligand-inducing binding site (LIBS) antibodies, which recognize epitopes that are exposed within the integrin ectodomain upon ligation of the integrin to RGD-containing ligands and peptides (9, 18). Figure 9 shows the binding of one such β3-specific antibody, MAb LIBS-1, to MDBK and SW480-β3 cells. For both cell lines, a small amount of LIBS-1 binding was detected on the mock-treated cells (Fig. 9). This binding was not detected with mock-transfected SW480 cells which do not express αvβ3 (data not shown). LIBS-1 binding to MDBK and SW480-αvβ3 cells was enhanced in the presence of the FMDV 12-mer peptide, showing that the peptide had bound to αvβ3 (Fig. 9). This enhancement was not seen in the presence of the RGE version of the peptide (Fig. 9), demonstrating the RGD dependency of the enhanced LIBS-1 binding. Similar results were obtained using the GRGDSP and GRGESP peptides (data not shown). We also determined the level of LIBS-1 binding to SW480-αvβ3 and MDBK cells in the presence of FMDV (data not shown); these studies showed that LIBS-1 binding was not enhanced above the resting level on incubation of the cells with virus. These data can be interpreted to show that FMDV had not bound to αvβ3, since the LIBS-1 epitope resides away from the RGD-binding pocket, and hence the binding of virus would not be expected to block binding of the LIBS-1 antibody.
FIG. 9.
Flow cytometric analysis of MAb LIBS-1 binding to αvβ3-expressing cells. MAb LIBS-1 binding is shown for MDBK and SW480-αvβ3 cells. The black histogram shows LIBS-1 binding in the presence of the RGE version of the FMDV 12-mer peptide. The white histogram shows LIBS-1 binding in the absence of peptide. These histograms are virtually identical, and the white histogram is obscured on the figure. Background (gray histogram) was determined in the absence of the LIBS-1 MAb. The striped histogram shows LIBS-1 binding in the presence of the RGD-containing FMDV 12-mer peptide. One experiment representative of two is shown; each gave nearly identical results.
Peptide inhibition using a VP1 GH loop peptide derived from FMDV SAT-2.
The above studies have shown that an Arg at the RGD+1 site is unfavorable for binding of the WT FMDV peptide to αvβ8. However, these studies used a peptide with a sequence derived from a GH loop of a type O virus which does not normally have an RGDR configuration. It is possible, therefore, that RGDR-containing peptides could be effective inhibitors of virus binding to αvβ8 when the RGDR is located within a sequence derived from an FMDV (e.g., SAT-2) that normally has this motif as part of its VP1 GH loop.
Figure 10 shows that FMDV binding and infection of SW480-αvβ6 cells are effectively inhibited by an RGDR-containing peptide with its sequence derived from a SAT-2 virus (STAIRGDRAVLAAKYAN). Figure 10 also shows that this peptide is a poor inhibitor of virus binding and infection of SW480-αvβ8 cells. Further, Fig. 10 shows that the ability of the SAT-2 peptide to inhibit virus binding and infection of SW480-αvβ8 is enhanced by substitution of the Arg immediately following the RGD by either Met or Leu. These observations support our conclusions that RGDR-containing peptides are poor inhibitors for αvβ8.
FIG. 10.
Inhibition of FMDV (O1Kcad2) binding and infection of SW480-αvβ6 and SW480-αvβ8 cells by RGDR-containing peptides. Panel A shows peptide (1 μM) inhibition of virus binding to SW480-αvβ6 (black boxes) and SW480-αvβ8 (gray boxes). Virus binding is shown as the percentage of binding in the absence of competition. Panel B shows peptide (10 μM) inhibition of infection of SW480-αvβ6 (black boxes) and SW480-αvβ8 (gray boxes). Infection is shown as the percent infection in the absence of competition. Peptides: SAT2-R is the WT SAT-2 peptide containing an RGDR motif; SAT2-M, the SAT-2 peptide containing RGDM; SAT2-L, the SAT-2 peptide containing an RGDL. Means (n = 3) and standard deviations are shown for one experiment representative of two; each gave nearly identical results.
The above observations suggest that FMD viruses that have an RGDR motif would use αvβ8 less efficiently as a receptor to initiate infection than αvβ6. To investigate this possibility, we compared the infectivities of two related viruses (FMDV O1Kcad2 and FMDV O1Kf480) for pBTY and IBRS2 cells. FMDV O1Kf480 is a MAb escape mutant of FMDV O1Kcad2, and these viruses have identical capsid sequences except for a single residue change immediately following the RGD motif; in O1Kcad2 this residue is Leu, whereas in O1Kf480 it is Arg (12). Primary BTY and IBRS2 cells were infected with each virus at the same MOI (MOI = 0.5; the MOI was determined by titration of each virus on pBTY cells) and infection quantified using the ELISPOT assay (see Methods). As expected, nearly identical numbers of pBTY cells were infected with either virus. In contrast, the number of IBRS2 cells infected by O1Kf480 was ∼50% (n = 3; data not shown) reduced from the number of cells infected with O1Kcad2, confirming that infection mediated by αvβ8 was less efficient for a virus containing an RGDR motif.
DISCUSSION
FMDV has been reported to use a number of RGD-binding integrins as receptors to initiate infection. Virus binding to the integrin is mediated by a highly conserved RGD motif located on a surface-exposed loop (the GH loop of VP1) of the capsid. In addition to the RGD, other residues of the VP1 GH loop are conserved (see the introduction), and these could play a role in integrin binding.
Here we have determined the sequence requirements of a synthetic peptide (the WT FMDV peptide) corresponding to the VP1 GH loop of type O FMDV for binding to integrins αvβ3, αvβ6, and αvβ8. This study has shown that the Leu residues at the RGD+1 and RGD+4 sites (which are conserved in type O FMDV) make a major contribution to peptide binding to both αvβ6 and αvβ8. Peptides containing Ala substitutions at either of these sites were poor inhibitors of virus binding and infection mediated by these integrins. These observations were confirmed using purified αvβ6 and αvβ6- and αvβ8-expressing cells, which included cells derived from the natural bovine and porcine hosts of FMDV. In contrast, Ala substitutions at these residues did not reduce the inhibitory effect of the peptide for virus binding to purified αvβ3. In fact, Ala substitutions at all of the non-RGD residues (including the RGD+1 and RGD+4 sites) increased the inhibitory effect of the peptide for αvβ3, albeit by a small amount, as did truncation of the five C-terminal peptide residues (FMDV 12-mer), suggesting that the sequence of the VP1 GH loop of type O viruses may not be optimal for binding this integrin.
The FMDV 12-mer peptide was shown to be a poor inhibitor of FMDV binding to αvβ6 and αvβ8 despite this peptide containing the RGD+1 and RGD+4 leucines. This suggests that the structure of the peptide may also influence integrin binding. The structures of chemically reduced virus (30) and of FMDV-derived peptides complexed with two different Fab fragments (19, 52, 53) show that the RGD+1 and RGD+4 leucines form a hydrophobic face of the 310 helix (30, 33, 52). The structure-based modeling (Fig. 11) of the FMDV peptide with αvβ3 predicts that the leucines are predisposed to interact with the β-chain ligand-binding domain (the βA domain) (57); the RGD+1 Leu most likely interacts with helix α1 and the RGD+4 Leu with the so-called “specificity loop” (57, 58). In these regions, the residues postulated to interact with the FMDV loop show only 25% identity between integrin species, and this may explain how this region could confer specificity of FMDV binding. It is likely that the WT FMDV peptide used in this study adopts a conformation closer to that of the virus, since it incorporates the entire 310 helix at its 3′ end. Truncation of the peptide after residue 12 (as in the FMDV 12-mer) would be expected to disrupt this helix and hence peptide-integrin binding. The question remains, though, of how the model of the WT FMDV peptide with αvβ3 can accommodate the FMDV sequence variation at the RGD+1 site. One possibility is that the hydrophobic stalk of Leu may be the key recognition element, and Met or Arg could fulfill the same specificity role.
FIG. 11.
Structure-based modeling of the FMDV peptide with integrin αvβ3. Panel a shows the FMDV VP1 GH loop of serotype O1BFS (residues 135 to 157) (in blue cartoon) with the RGD motif (residues 145 to 147; red sticks) superimposed on the RGDF ligand (green sticks) as bound to the integrin αvβ3 in the crystal structure (PDB code 1L5G). The position of the leucines at RGD+1 and RGD+4 are denoted in orange. Panel B shows the FMDV VP1 GH loop (as in panel A) in the context of αvβ3. The loop is predicted to bind in the crevice between the αv β-propeller domain (yellow/green ribbon) and the ligand-binding domain of β3 (βA) (purple/pink ribbon). In the model, the orientation of the RGD is such that the Arg would contact the αv β-propeller domain and the Asp the βA domain. Panel c shows a closeup of panel B with the side chains of Leu (RGD+1 and RGD+4) shown as orange sticks projecting from the same face of the 310 helix. In this orientation they interact solely with the α1 helix (Leu RGD+1) and the “specificity loop” (Leu RGD+4) of the βA domain.
The normal sequence variation seen at the RGD+1 site across the different FMDV serotypes (see the introduction) did not compromise the ability of the WT FMDV peptide to bind αvβ6, since peptides containing RGDL, RGDM, or RGDR had similar inhibitory effects on virus binding and infection mediated by this integrin. Our initial studies suggested that inclusion of Arg or Met immediately after the RGD reduced the inhibitory effect of the peptide for virus binding to purified αvβ6, albeit by a small amount; however, these differences were not seen when the peptides were used as inhibitors of virus binding to αvβ6 on the surfaces of cells (SW480-αvβ6). Recently we have shown that recombinant αvβ6, lacking the entire β6 cytoplasmic domain, was able to support FMDV binding (20, 36); however, the β6 truncated integrin did not appear to assemble correctly at the cell surface and had a relaxed specificity for RGD-containing peptides. Specifically, a GRGDSP peptide was able to inhibit virus binding to the β6 truncated integrin but not to WT αvβ6. The purified αvβ6 used in the ELISA in the current study lacks the transmembrane and cytoplasmic domain of both α and β chains and therefore may display a relaxed peptide-binding specificity. With these observations in mind, we believe our data obtained using αvβ6-expressing cells to be more reliable.
The FMDV peptide containing an RGDM motif was also an effective inhibitor of virus binding and infection mediated by αvβ8. However, in contrast to the case with αvβ6, inclusion of an Arg and the RGD+1 site was unfavorable for αvβ8, since the RGDR-containing FMDV peptide was a poor inhibitor of virus binding and infection mediated by this integrin. These observations were confirmed using a second series of peptides corresponding in sequence to the VP1 GH loop of SAT-2 FMDV, which normally contains an RGDR motif within this loop. The WT SAT-2 peptide (containing an RGDR motif) was shown to be a poor inhibitor of virus binding and infection mediated by αvβ8, whereas variants of this peptide, containing either RGDM or RGDL, were effective in these roles. Consistent with these observations, a virus (FMDV O1Kf480) containing an RGDR motif was shown to have a reduced infectivity for IBRS2 cells (mediated by αvβ8) compared with that of O1Kcad2, which contains an RGDL motif.
The above observations suggest that inclusion of an Arg at the RGD+1 site is unfavorable for infection mediated by αvβ8 and, hence, suggest that the integrin binding loop of FMDV is most highly adapted for virus binding to αvβ6. A number of other observations support this conclusion: (i) the IC50 of the WT FMDV peptide for inhibition of virus binding to αvβ6 was ∼18-fold lower than that for αvβ8, suggesting that the peptide has a higher binding affinity for αvβ6; (ii) at high concentrations, a GRGDSP peptide has been shown to partially inhibit FMDV binding to SW480-αvβ8 but not to SW480-αvβ6 cells, which suggests that the C-terminal residues of the VP1 GH loop are essential for binding to αvβ6 but not for αvβ8; (iii) a related picornavirus, coxsackievirus A9, has been shown to use αvβ6 as a receptor for infection (56) and has an RGDMSTL sequence as part of its integrin binding loop; (iv) the RGDLXXL motif (where X represents any amino acid) was highly represented in a phage peptide library panned on αvβ6 (26); and (v) a ligand mimetic MAb specific for αvβ6 has been recently been described that contains an RGDRXXL motif as part of CDR-H3 (complementarity determining region-heavy chain 3) (55); this antibody does not bind the closely related integrin αvβ8, presumably due to the presence of the Arg at the RGD+1 site.
This study has shown that MDBK, CT23, and SW480-αvβ3 have a low susceptibility to FMDV O1Kcad2 despite these cells expressing αvβ3. For SW480-αvβ3 this can be explained by the failure of the virus to bind these cells. This explanation does not account for the low susceptibility of MDBK and CT23 cells, since virus binding was detected. This binding was not inhibited by function-blocking antibodies to αvβ3, showing that it was not mediated by this integrin. However, virus binding to MDBK cells was inhibited by the WT FMDV peptide but not by its RGE version, suggesting that integrins may mediate virus attachment. Nevertheless, we did not detect expression of αvβ6 or αvβ8 on MDBK cells by flow cytometry, suggesting that virus binding to MDBK cells was not mediated by these integrins (see Fig. 6), and presently we do not know the identity of the attachment receptor. Similarly, we do not know where the block to infection occurs, but it is likely to result from a defect in cell entry and not intracellular virus replication, since MDBK and CT23 cells were susceptible to FMDV O1BFS, a virus that uses heparan sulfate proteoglycans as its receptors.
Ligand binding to αvβ3 is regulated by conformational changes in the integrin ectodomain in a process called integrin activation (31). In the bent or closed conformation, the RGD-binding pocket is inaccessible to ligands such as vitronectin and fibronectin (51). Ligand binding is facilitated by conversion of the integrin to an extended or open conformation in which the RGD-binding pocket becomes exposed (9, 18). This conversion can be followed using anti-LIBS antibodies that recognize epitopes that are hidden in the bent conformation but become accessible to antibody on integrin activation. The bent conformation is accessible to small RGD-containing peptides, however, and the binding of such peptides triggers conversion to the open conformation and hence the binding of LIBS antibodies (51). Our studies using the LIBS-1 antibody have shown that on MDBK and SW480-αvβ3 cells, αvβ3 was primarily in the bent conformation, since incubation of these cells with RGD-containing peptides led to a large increase in the expression of the LIBS-1 epitope. However, this observation is unlikely to explain the failure of FMDV to bind αvβ3, since binding of virus to these cells in the presence of Mn2+ ions was not inhibited by function-blocking antibodies to this integrin, and infection of CT23, MDBK, and SW480-αvβ3 cells was not enhanced by Mn2+ ions, which promote integrin activation and ligand binding. Nevertheless, despite being a poor receptor for virus attachment when expressed on cells, FMDV was found to bind immobilized αvβ3 in the ELISA, although, under nearly identical conditions, not as efficiently as αvβ6. This would suggest that FMDV may have a lower affinity for αvβ3, which could explain the apparent discrepancy between virus binding to purified αvβ3 and to that present on cells; in the ELISA, the integrin is immobilized at a high concentration and virus binding is more a measure of the avidity of the interaction, whereas binding to the integrin at the cell surface is more a measure of the binding affinity. A low affinity for αvβ3 may also explain the observations made by Duque et al. (2004), who showed that soluble αvβ3 was unable to inhibit FMDV infection of BHK cells (15).
In conclusion, these studies have shown that the residues at the RGD+1 and RGD+4 sites make a major contribution to the specificity of the integrin-binding loop of FMDV and have established αvβ6 as being the integrin species to which FMDV appears to be most highly adapted. Furthermore, our data suggest that the structure of the VP1 GH loop downstream of RGD is required to maintain the contacting residues in the correct orientation for integrin binding. In addition, these studies have shown that αvβ3 is a poor receptor for infection by type O FMDV. This information has increased our understanding of how FMDV selects its integrin receptors, provides the key to understanding how FMDV targets epithelial cells in vivo, and is essential for the future development of a common, nonimmune control strategy for all FMDV serotypes based on antiviral agents which specifically block virus attachment to αvβ6.
Acknowledgments
We thank Sheila Wilsden for pBTYs and IBRS2s, Larry Hunt for peptide synthesis, Weixian Lu for biotechnological assistance, Emiliana Brocchi for MAb 2C2, Mark Ginsberg for LIBS-1, and BIOGEN Idec for MAb 6.8G6.
This work was supported by the BBSRC (201/C15845) and DEFRA (SE2717).
REFERENCES
- 1.Acharya, R., E. Fry, D. Stuart, G. Fox, D. Rowlands, and F. Brown. 1989. The three-dimensional structure of foot-and-mouth disease virus at 2.9 A resolution. Nature 337:709-716. [DOI] [PubMed] [Google Scholar]
- 2.Baranowski, E., C. M. Ruiz-Jarabo, N. Sevilla, D. Andreu, E. Beck, and E. Domingo. 2000. Cell recognition by foot-and-mouth disease virus that lacks the RGD integrin-binding motif: flexibility in aphthovirus receptor usage. J. Virol. 74:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxt, B. 1987. Effect of lysosomotropic compounds on early events in foot-and-mouth disease virus replication. Virus Res. 7:257-271. [DOI] [PubMed] [Google Scholar]
- 4.Baxt, B., and Y. Becker. 1990. The effect of peptides containing the arginine-glycine-aspartic acid sequence on the adsorption of foot-and-mouth disease virus to tissue culture cells. Virus Genes. 4:73-83. [DOI] [PubMed] [Google Scholar]
- 5.Berinstein, A., M. Roivainen, T. Hovi, P. W. Mason, and B. Baxt. 1995. Antibodies to the vitronectin receptor (integrin alpha V beta 3) inhibit binding and infection of foot-and-mouth disease virus to cultured cells. J. Virol. 69:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berryman, S., S. Clark, P. Monaghan, and T. Jackson. 2005. Early events in integrin αvβ6-mediated cell entry of foot-and-mouth disease virus. J. Virol. 79:8519-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bittle, J. L., R. A. Houghten, H. Alexander, T. M. Shinnick, J. G. Sutcliffe, R. A. Lerner, D. J. Rowlands, and F. Brown. 1982. Protection against foot-and-mouth disease by immunization with a chemically synthesized peptide predicted from the viral nucleotide sequence. Nature 298:30-33. [DOI] [PubMed] [Google Scholar]
- 9.Blystone, S. D., M. P. Williams, S. E. Slater, and E. J. Brown. 1997. Requirement of integrin beta3 tyrosine 747 for beta3 tyrosine phosphorylation and regulation of alphavbeta3 avidity. J. Biol. Chem. 272:28757-28761. [DOI] [PubMed] [Google Scholar]
- 10.Bruderer, U., H. Swam, B. Haas, N. Visser, E. Brocchi, S. Grazioli, J. J. Esterhuysen, W. Vosloo, M. Forsyth, N. Aggarwal, et al. 2004. Differentiating infection from vaccination in foot-and-mouth-disease: evaluation of an ELISA based on recombinant 3ABC. Vet. Microbiol. 101:187-197. [DOI] [PubMed]
- 11.Cambier, S., D. Z. Mu, D. O'Connell, K. Boylen, W. Travis, W. H. Liu, V. C. Broaddus, and S. L. Nishimura. 2000. A role for the integrin alphavbeta8 in the negative regulation of epithelial cell growth. Cancer Res. 60:7084-7093. [PubMed] [Google Scholar]
- 12.Crowther, J. R., S. Farias, W. C. Carpenter, and A. R. Samuel. 1993. Identification of a fifth neutralizable site on type O foot-and-mouth disease virus following characterization of single and quintuple monoclonal antibody escape mutants. J. Gen. Virol. 74:1547-1553. [DOI] [PubMed] [Google Scholar]
- 13.Curry, S., E. Fry, W. Blakemore, R. Abu-Ghazaleh, T. Jackson, A. King, S. Lea, J. Newman, D. Rowlands, and D. Stuart. 1996. Perturbations in the surface structure of A22 Iraq foot-and-mouth disease virus accompanying coupled changes in host cell specificity and antigenicity. Structure 4:135-145. [DOI] [PubMed] [Google Scholar]
- 14.De Diego, M., E. Brocchi, D. Mackay, and F. De Simone. 1997. The non-structural polyprotein 3ABC of foot-and-mouth disease virus as a diagnostic antigen in ELISA to differentiate infected from vaccinated cattle. Arch. Virol. 142:2021-2033. [DOI] [PubMed] [Google Scholar]
- 15.Duque, H., M. LaRocco, W. T. Golde, and B. Baxt. 2004. Interactions of foot-and-mouth disease virus with soluble bovine αVβ3 and αVβ6 integrins. J. Virol. 78:9773-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris, N. P., N. G. Abrescia, D. I. Stuart, T. Jackson, A. Burman, D. P. King, and D. J. Paton. 2005. Utility of recombinant integrin alpha v beta 6 as a capture reagent in immunoassays for the diagnosis of foot-and-mouth disease. J. Virol. Methods 127:69-79. [DOI] [PubMed] [Google Scholar]
- 17.Fox, G., N. R. Parry, P. V. Barnett, B. McGinn, D. J. Rowlands, and F. Brown. 1989. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70:625-637. [DOI] [PubMed] [Google Scholar]
- 18.Frelinger, A. L., III, I. Cohen, E. F. Plow, M. A. Smith, J. Roberts, S. C. Lam, and M. H. Ginsberg. 1990. Selective inhibition of integrin function by antibodies specific for ligand-occupied receptor conformers. J. Biol. Chem. 265:6346-6352. [PubMed] [Google Scholar]
- 19.Hewat, E. A., N. Verdaguer, I. Fita, W. Blakemore, S. Brookes, A. King, J. Newman, E. Domingo, M. G. Mateu, and D. I. Stuart. 1997. Structure of the complex of an Fab fragment of a neutralizing antibody with foot-and-mouth disease virus: positioning of a highly mobile antigenic loop. EMBO J. 16:1492-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, T., S. Clark, S. Berryman, A. Burman, S. Cambier, D. Mu, S. Nishimura, and A. M. King. 2004. Integrin αvβ8 functions as a receptor for foot-and-mouth disease virus: role of the beta-chain cytodomain in integrin-mediated infection. J. Virol. 78:4533-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, T., F. M. Ellard, R. A. Ghazaleh, S. M. Brookes, W. E. Blakemore, A. H. Corteyn, D. I. Stuart, J. W. Newman, and A. M. King. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J. Virol. 70:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, T., A. P. Mould, D. Sheppard, and A. M. King. 2002. Integrin αvβ1 is a receptor for foot-and-mouth disease virus. J. Virol. 76:935-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, T., A. Sharma, R. A. Ghazaleh, W. E. Blakemore, F. M. Ellard, D. L. Simmons, J. W. Newman, D. I. Stuart, and A. M. King. 1997. Arginine-glycine-aspartic acid-specific binding by foot-and-mouth disease viruses to the purified integrin αvβ3 in vitro. J. Virol. 71:8357-8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, T., D. Sheppard, M. Denyer, W. Blakemore, and A. M. King. 2000. The epithelial integrin αvβ6 is a receptor for foot-and-mouth disease virus. J. Virol. 74:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles, N. J., and A. R. Samuel. 2003. Molecular epidemiology of foot-and-mouth disease virus. Virus Res. 91:65-80. [DOI] [PubMed] [Google Scholar]
- 26.Kraft, S., B. Diefenbach, R. Mehta, A. Jonczyk, G. A. Luckenbach, and S. L. Goodman. 1999. Definition of an unexpected ligand recognition motif for alphav beta6 integrin. J. Biol. Chem. 274:1979-1985. [DOI] [PubMed] [Google Scholar]
- 27.Lea, S., J. Hernandez, W. Blakemore, E. Brocchi, S. Curry, E. Domingo, E. Fry, R. Abu-Ghazaleh, A. King, J. Newman, et al. 1994. The structure and antigenicity of a type C foot-and-mouth disease virus. Structure 2:123-139. [DOI] [PubMed] [Google Scholar]
- 28.Leippert, M., E. Beck, F. Weiland, and E. Pfaff. 1997. Point mutations within the βG-βH loop of foot-and-mouth disease virus O1K affect virus attachment to target cells. J. Virol. 71:1046-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liebermann, H., R. Dolling, D. Schmidt, and G. Thalmann. 1991. RGD-containing peptides of VP1 of foot-and-mouth disease virus (FMDV) prevent virus infection in vitro. Acta Virol. 35:90-93. [PubMed] [Google Scholar]
- 30.Logan, D., R. Abu-Ghazaleh, W. Blakemore, S. Curry, T. Jackson, A. King, S. Lea, R. Lewis, J. Newman, N. Parry, et al. 1993. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature 362:566-568. [DOI] [PubMed] [Google Scholar]
- 31.Luo, B. H., J. Takagi, and T. A. Springer. 2004. Locking the beta3 integrin I-like domain into high and low affinity conformations with disulfides. J. Biol. Chem. 279:10215-10221. [DOI] [PubMed] [Google Scholar]
- 32.Mason, P. W., B. Baxt, F. Brown, J. Harber, A. Murdin, and E. Wimmer. 1993. Antibody-complexed foot-and-mouth disease virus, but not poliovirus, can infect normally insusceptible cells via the Fc receptor. Virology 192:568-577. [DOI] [PubMed] [Google Scholar]
- 33.Mateu, M. G., M. L. Valero, D. Andreu, and E. Domingo. 1996. Systematic replacement of amino acid residues within an Arg-Gly-Asp-containing loop of foot-and-mouth disease virus and effect on cell recognition. J. Biol. Chem. 271:12814-12819. [DOI] [PubMed] [Google Scholar]
- 34.McCahon, D., J. R. Crowther, G. J. Belsham, J. D. Kitson, M. Duchesne, P. Have, R. H. Meloen, D. O. Morgan, and F. De Simone. 1989. Evidence for at least four antigenic sites on type O foot-and-mouth disease virus involved in neutralization; identification by single and multiple site monoclonal antibody-resistant mutants. J. Gen. Virol. 70:639-645. [DOI] [PubMed] [Google Scholar]
- 35.McKenna, T. S., J. Lubroth, E. Rieder, B. Baxt, and P. W. Mason. 1995. Receptor binding site-deleted foot-and-mouth disease (FMD) virus protects cattle from FMD. J. Virol. 69:5787-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, L. C., W. Blakemore, D. Sheppard, A. Atakilit, A. M. King, and T. Jackson. 2001. Role of the cytoplasmic domain of the β-subunit of integrin αvβ6 in infection by foot-and-mouth disease virus. J. Virol. 75:4158-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monaghan, P., S. Gold, J. Simpson, Z. Zhang, P. H. Weinrab, S. M. Violette, S. Alexandersen, and T. Jackson. 2005. The alpha(v)beta6 integrin receptor for FMDV is expressed constitutively on the epithelial cells targeted in cattle. J. Gen. Virol. 86:2769-2780. [DOI] [PubMed] [Google Scholar]
- 38.Mu, D., S. Cambier, L. Fjellbirkeland, J. L. Baron, J. S. Munger, H. Kawakatsu, D. Sheppard, V. C. Broaddus, and S. L. Nishimura. 2002. The integrin alpha(v)beta8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-beta1. J. Cell Biol. 157:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neff, S., D. Sa-Carvalho, E. Rieder, P. W. Mason, S. D. Blystone, E. J. Brown, and B. Baxt. 1998. Foot-and-mouth disease virus virulent for cattle utilizes the integrin αvβ3 as its receptor. J. Virol. 72:3587-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Donnell, V., M. LaRocco, H. Duque, and B. Baxt. 2005. Analysis of foot-and-mouth disease virus internalization events in cultured cells. J. Virol. 79:8506-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfaff, E., M. Mussgay, H. O. Bohm, G. E. Schulz, and H. Schaller. 1982. Antibodies against a preselected peptide recognize and neutralize foot and mouth disease virus. EMBO J. 1:869-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaff, E., H. J. Thiel, E. Beck, K. Strohmaier, and H. Schaller. 1988. Analysis of neutralizing epitopes on foot-and-mouth disease virus. J. Virol. 62:2033-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieder, E., B. Baxt, and P. W. Mason. 1994. Animal-derived antigenic variants of foot-and-mouth disease virus type A12 have low affinity for cells in culture. J. Virol. 68:5296-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowlands, D., D. Logan, R. Abu-Ghazaleh, W. Blakemore, S. Curry, T. Jackson, A. King, S. Lea, R. Lewis, J. Newman, et al. 1994. The structure of an immunodominant loop on foot and mouth disease virus, serotype O1, determined under reducing conditions. Arch. Virol. Suppl. 9:51-58. [DOI] [PubMed] [Google Scholar]
- 45.Sa-Carvalho, D., E. Rieder, B. Baxt, R. Rodarte, A. Tanuri, and P. W. Mason. 1997. Tissue culture adaptation of foot-and-mouth disease virus selects viruses that bind to heparin and are attenuated in cattle. J. Virol. 71:5115-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheppard, D., C. Rozzo, L. Starr, V. Quaranta, D. J. Erle, and R. Pytela. 1990. Complete amino acid sequence of a novel integrin beta subunit (beta 6) identified in epithelial cells using the polymerase chain reaction. J. Biol. Chem. 265:11502-11507. [PubMed] [Google Scholar]
- 47.Smith, J. W., R. S. Piotrowicz, and D. Mathis. 1994. A mechanism for divalent cation regulation of beta 3-integrins. J. Biol. Chem. 269:960-967. [PubMed] [Google Scholar]
- 48.Snowdon, W. A. 1966. Growth of foot-and mouth disease virus in monolayer cultures of calf thyroid cells. Nature 210:1079-1080. [DOI] [PubMed] [Google Scholar]
- 49.Stuart, D. I., M. Levine, H. Muirhead, and D. K. Stammers. 1979. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol Biol. 134:109-142. [DOI] [PubMed] [Google Scholar]
- 50.Surovoi, A., V. T. Ivanov, A. V. Chepurkin, V. N. Ivaniushchenkov, and N. N. Driagalin. 1988. Is the Arg-Gly-Asp sequence the site for foot-and-mouth disease virus binding with cell receptor? Bioorg. Khim. 14:965-968. (In Russian.) [PubMed] [Google Scholar]
- 51.Takagi, J., B. M. Petre, T. Walz, and T. A. Springer. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110:599-611. [DOI] [PubMed] [Google Scholar]
- 52.Verdaguer, N., M. G. Mateu, D. Andreu, E. Giralt, E. Domingo, and I. Fita. 1995. Structure of the major antigenic loop of foot-and-mouth disease virus complexed with a neutralizing antibody: direct involvement of the Arg-Gly-Asp motif in the interaction. EMBO J. 14:1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verdaguer, N., G. Schoehn, W. F. Ochoa, I. Fita, S. Brookes, A. King, E. Domingo, M. G. Mateu, D. Stuart, and E. A. Hewat. 1999. Flexibility of the major antigenic loop of foot-and-mouth disease virus bound to a Fab fragment of a neutralising antibody: structure and neutralisation. Virology 255:260-268. [DOI] [PubMed] [Google Scholar]
- 54.Weinacker, A., A. Chen, M. Agrez, R. I. Cone, S. Nishimura, E. Wayner, R. Pytela, and D. Sheppard. 1994. Role of the integrin alpha v beta 6 in cell attachment to fibronectin. Heterologous expression of intact and secreted forms of the receptor. J. Biol. Chem. 269:6940-6948. [PubMed] [Google Scholar]
- 55.Weinreb, P. H., K. J. Simon, P. Rayhorn, W. J. Yang, D. R. Leone, B. M. Dolinski, B. R. Pearse, Y. Yokota, H. Kawakatsu, A. Atakilit, D. Sheppard, and S. M. Violette. 2004. Function-blocking integrin alphavbeta6 monoclonal antibodies: distinct ligand-mimetic and nonligand-mimetic classes. J. Biol. Chem. 279:17875-17887. [DOI] [PubMed] [Google Scholar]
- 56.Williams, C. H., T. Kajander, T. Hyypia, T. Jackson, D. Sheppard, and G. Stanway. 2004. Integrin αvβ6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78:6967-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong, J.-P., T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D. L. Scott, A. Joachimiak, S. L. Goodman, and M. A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin alpha vbeta 3. Science 294:339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong, J. P., T. Stehle, R. Zhang, A. Joachimiak, M. Frech, S. L. Goodman, and M. A. Arnaout. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296:151-155. [DOI] [PubMed] [Google Scholar]
- 59.Yokosaki, Y., H. Monis, J. Chen, and D. Sheppard. 1996. Differential effects of the integrins alpha9beta1, alphavbeta3, and alphavbeta6 on cell proliferative responses to tenascin. Roles of the beta subunit extracellular and cytoplasmic domains. J. Biol. Chem. 271:24144-24150. [DOI] [PubMed] [Google Scholar]