Abstract
The replication of many viruses is absolutely dependent on proteolytic cleavage. Infected cells also use this biological mechanism to induce programmed cell death in response to viral infection. Specific inhibitors for both viral and cellular proteases are therefore of vital importance. We have recently shown that the general caspase inhibitor zVAD.fmk inhibits not only caspases, but also the 2Apro of human rhinoviruses (HRVs) (L. Deszcz, J. Seipelt, E. Vassilieva, A. Roetzer, and E. Kuechler, FEBS Lett. 560:51-55, 2004). Here, we describe a derivative of zVAD.fmk that inhibits HRV2 2Apro but that has no effect on caspase 9. This gain in specificity was achieved by replacing the aspartic acid of zVAD.fmk with methionine to generate zVAM.fmk. Methionine was chosen because an oligopeptide with methionine at the P1 position was a much better substrate than an oligopeptide with an alanine residue, which is found at the P1 position of the wild-type HRV2 2Apro cleavage site. zVAM.fmk inhibits the replication of HRV type 2 (HRV2), HRV14, and HRV16. In contrast to zVAD.fmk, however, zVAM.fmk did not inhibit apoptosis induced by puromycin in HeLa cells. zVAM.fmk inhibited in vitro the intermolecular cleavage of eukaryotic initiation factor 4GI (eIF4GI) by HRV2 2Apro at nanomolar concentrations. However, much higher concentrations of zVAM.fmk were required to inhibit HRV14 2Apro cleavage of eIF4GI. In contrast, intramolecular self-processing of HRV14 2Apro was much more susceptible to inhibition by zVAM.fmk than that of HRV2 2Apro, suggesting that zVAM.fmk inhibits HRV2 and HRV14 replication by targeting different reactions of the same proteinase.
Proteolytic processing of a large precursor protein is a central event in the life cycles of many viruses (17). This proteolysis is generally performed by virus-encoded proteinases. Thus, the reaction represents an attractive target for therapeutic intervention (8). The successful use of human immunodeficiency virus proteinase inhibitors to combat AIDS is a prime example (34). Specific inhibition of viral proteolysis in cell culture can also shed light on mechanisms of viral replication and the interplay of viruses and cells, as viral proteinases can also process cellular proteins (7).
Picornaviruses possess a single-stranded RNA genome of positive sense, which is translated on release into the cytoplasm into a large polyprotein. In enteroviruses (such as poliovirus and coxsackieviruses) and human rhinoviruses (HRVs), the polyprotein is processed mainly by the virus-encoded, chymotrypsin-like cysteine proteinase 3Cpro (13). However, the primary cleavage event, separating the structural protein precursor from the nonstructural one, is performed by 2Apro, also a chymotrypsin-like cysteine proteinase (31). 2Apro cleaves the viral polyprotein between the C terminus of VP1 and its own N terminus (31). The structures of 3Cpro and 2Apro are similar, the main difference being the absence of 4 β-strands in the N-terminal domain of 2Apro (25). Nevertheless, the specificities of the two proteins differ substantially (25).
Both 2Apro and 3Cpro from most picornaviruses have also been shown to cleave cellular proteins (25). The 2Apros of enteroviruses and human rhinoviruses have been shown to cleave the two homologues of the host protein eukaryotic initiation factor 4G (eIF4G), eIF4GI, and eIF4GII (12). This cleavage leads to an inability of the infected cell to translate its own capped mRNA, a phenomenon known as host cell shutoff (18). Initiation of protein synthesis from viral mRNA is unaffected, as it does not occur by a cap-dependent mechanism; instead, initiation of translation requires an internal ribosomal entry site (2). Another protein of the cellular translation apparatus, poly(A) binding protein (PABP), has also been shown to be cleaved by both picornavirus 2Apro and 3Cpro (15, 16).
Specific inhibitors of human rhinovirus 3Cpro have been developed and have been tested for inhibition of rhinovirus replication in the clinic (21). Certain inhibitors of human rhinovirus 3Cpro have been shown to be active against 2Apro (32). However, inhibitors specific for enterovirus or rhinovirus 2Apro alone are not available. Recent genetic evidence has shown that targeting the 2Apro self-processing reactions of these viruses might be an excellent strategy to inhibit their replication (5).
As a starting point to develop a compound specific for 2Apro, we used the inhibitor zVAD.fmk [benzyloxycarbonyl-Val-Ala-Asp (OMe)-fluoromethylketone], a general caspase inhibitor that is active in vitro against HRV and coxsackievirus serotype B4 (CVB4) 2Apro and which can also inhibit replication of these viruses in cell culture (6). As can be seen from its full name, zVAD.fmk is synthesized as the methyl ester of the aspartic acid residue. In contrast to the nonmethylated form, the methylated form can pass through the cell membrane; once inside the cell, zVAD.fmk is demethylated by endogenous esterases (20, 27). The methylated form is active against 2Apro, whereas the nonmethylated form inhibits caspases. The inhibition of caspases limits the use of zVAD.fmk to specifically investigate events in HRV replication. Furthermore, zVAD.fmk must be added repeatedly to cells infected with HRV to ensure a sufficiently high concentration of the methylated form of the inhibitor (6).
Why does only the methylated form of zVAD.fmk inhibit 2Apro? Both HRV2 and CVB4 2Apro can accept residues such as alanine, phenylalanine, and tyrosine at the P1 position (28, 29). (The nomenclature Pn-P1-P1′-Pn′ of a substrate or inhibitor is that of Berger and Schechter [3], with the scissile peptide bond lying between P1 and P1′). This preference of HRV2 2Apro for hydrophobic residues at P1 is mimicked by the methylated aspartic residue in zVAD.fmk. In contrast, aspartic acid is not accepted at P1 by HRV2 2Apro, which explains why the nonmethylated form is not active.
To improve the properties of zVAD.fmk, it was necessary to replace the methylated aspartic acid residue with a hydrophobic residue, which cannot be modified by demethylation and which in consequence cannot target caspases. Sommergruber et al. (29) showed that an oligopeptide with methionine at the P1 position was cleaved fivefold more efficiently than the corresponding wild-type peptide. We hypothesized, therefore, that the inhibitor zVAM.fmk (benzyloxycarbonyl-Val-Ala-Met-fluoromethylketone), in which the aspartic acid at P1 is replaced by methionine, should possess the required characteristics outlined above. Both zVAM.fmk and zVAD.fmk, an irreversible fluoromethylketone inhibitor, are tripeptides that mimic the structure of the three amino acids from positions P3 to P1 of the substrate. It is likely that they attach covalently to the active-site nucleophile.
This report describes the properties of zVAM.fmk and compares them to those of zVAD.fmk.
MATERIALS AND METHODS
Media, reagents, and chemicals.
zVAD.fmk was purchased from BD Pharmingen and zVAM.fmk from MP Biomedicals (Aurora, Ohio). The identity and purity of zVAM.fmk was confirmed by mass spectrometry. Anti-caspase 9 antibodies were from Alexis (San Diego, CA). Polyclonal rabbit anti-eIF4GI anti-peptide no. 7 antiserum was provided by R. Rhoads (Louisiana State University, Shreveport). Enhanced-chemiluminescence Western blotting detection reagents were from Pierce (Rockford, IL), and polyvinylpyrrolidone was from Calbiochem (Darmstadt, Germany).
Tissue culture.
Human cervix carcinoma HeLa cells (strain Ohio; European Collection of Cell Cultures, Salisbury, United Kingdom), which are particularly suited for high-yield growth of rhinoviruses, were used throughout. The cells were grown in Dulbecco modified Eagle's medium (Gibco, Eggersheim, Germany) supplemented with 10% heat-inactivated fetal calf serum (Gibco), 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C in a humidified atmosphere with 5% CO2.
Virus preparation and infection.
HRV type 2 (HRV2) and HRV14 (American Type Culture Collection) were grown in HeLa suspension cultures (strain Ohio) and purified as described previously (26). Virus titers were determined by end point dilution tests and are given as 50% tissue culture infective dose (TCID50) according to the method of Reed and Muench (23). Cells were infected with HRV14 at 100 TCID50/cell at 80 to 90% confluence in minimal essential medium (MEM) (Gibco) containing 2% fetal calf serum (Gibco), 2 mM l-glutamine, 30 mM MgCl2, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Thirty minutes postinfection (p.i.), noninternalized virus was removed by washing the cells three times with MEM, and incubation continued in Dulbecco modified Eagle's medium at 34°C.
Proliferation assay.
Cell viability was determined by using the Cell Titer 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI). Briefly, 2 × 106 cells were seeded into a 96-well plate. The next day, the cells were infected with HRV14 at 100 TCID50/cell in the absence or presence of the indicated concentration of inhibitor. Cell viability was determined 12 h p.i. by adding tetrazolium to each well, followed by incubation for 2 h at 37°C and measurement of the absorption at 492 nm in a Labsystem Multiscan RC plate reader. Each experiment was repeated three times.
35S labeling of host cell proteins.
HeLa cells were infected with HRV2 or HRV14 as described above in Dulbecco modified Eagle's medium (lacking l-methionine and l-cysteine) in the absence or presence of 200 μM zVAM.fmk. Five hours postinfection, [35S]methionine/cysteine (10 mCi/ml; Hartmann Analytic) was added to a final concentration of 100 μCi/ml; after three more hours of incubation, the cells were solubilized directly in Laemmli sample buffer. Protein extracts were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by autoradiography.
Western blotting.
Equal amounts of proteins of total cell extracts were separated by SDS-PAGE and electrotransferred onto nitrocellulose membranes (PROTRAN; Schleicher & Schuell BioScience GmbH, Dassel, Germany). The membranes were incubated overnight in phosphate-buffered saline containing 1% nonfat milk powder, 1% polyvinylpyrrolidone, and 0.1% Tween 20 at 4°C with the relevant antiserum. The antisera used in this study were a polyclonal antiserum against the N terminus of eIF4GI (anti-eIF4G-peptide 7 antiserum) (19) and a polyclonal anti-caspase 9 (1:1,000) antiserum. The immunoreactive proteins were visualized by using horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G (Bio-Rad, Hercules, CA), followed by chemiluminescence detection using a system from Pierce (Rockford, IL).
Treatment of cells with inhibitors.
Stock solutions of zVAD.fmk and zVAM.fmk (100 mM in dimethyl sulfoxide[DMSO]) were diluted in MEM in order to keep the final DMSO concentration below 0.2%. To inhibit viral-protease activity, zVAD.fmk was added at the concentrations indicated to the cells at the time of infection. Subsequently, every 4 h, the medium was removed and replaced with new medium containing zVAD.fmk at the same concentration. To inhibit viral-protease activity with zVAM.fmk, the inhibitor was added to the indicated concentration 30 min before infection. After removal of the medium at 1 h p.i., zVAM.fmk was again added to the same concentration. No further addition of zVAM.fmk was necessary. For TCID50 measurements, infected cells were incubated for a total of 12 h.
Inhibition of picornavirus 2Apro by zVAM.fmk and zVAD.fmk.
To detect in vivo inhibition, HeLa cells were infected with HRV2 or HRV14 in the presence of 200 μM zVAM.fmk; 6 h p.i., the cells were solubilized directly in Laemmli sample buffer, and protein extracts were separated by 6% SDS-PAGE and analyzed by immunoblotting. The detection of inhibition of eIF4GI cleavage in vitro was performed at 30°C in buffer A (100 mM NaCl, 25 mM Tris-HCl, 1 mM EDTA, pH 7.6) in a total volume of 20 μl. For each cleavage reaction, 9 μg of HeLa cell cytoplasmic protein extracts was incubated for 1 h at 30°C with 0.33 ng of HRV2 2Apro, 600 ng of HRV14 2Apro, or 11 ng of CVB4 2Apro in the absence and presence of different concentrations of zVAD.fmk or zVAM.fmk. The inhibitors were added to the reaction mixture 5 min before the protein extracts. The reactions were terminated by direct addition of Laemmli sample buffer. Proteins were separated by 6% SDS-PAGE and probed by Western blotting for the state of eIF4GI.
Inhibition of the self-processing cleavage reaction of 2Apro.
The plasmid pHRV2 VP1-2Apro has been described previously (11). pHRV14 VP1-2Apro contains HRV14 nucleotides 2321 to 3630. The 2Apro coding sequence is followed by two stop codons in each case. Plasmid preparation, synthesis of RNA, and in vitro translation in rabbit reticulocyte lysates were as described previously (10, 11). To examine the effects of inhibitors on self-processing, inhibitors were added to the indicated concentrations 10 min after the start of protein synthesis. Protein synthesis was then stopped at the indicated times. Control reaction mixtures contained DMSO only.
RESULTS
The ability of zVAD.fmk to inhibit the activity of HRV2 2Apro lies in the long hydrophobic side chain of the methylated aspartic acid residue, which can be bound by the deep S1 pocket present on the proteinase. Given that an oligopeptide with methionine in place of alanine at P1 in the wild-type cleavage sequence is an excellent substrate for HRV2 2Apro (29), we reasoned that replacing the methylated aspartic acid residue in zVAD.fmk with methionine to give zVAM.fmk would retain the inhibitory properties against HRV2 2Apro but exclude the affinity of the compound for caspases. In addition, this inhibitor would not be subject to loss through degradation by methylases.
zVAM.fmk affects rhinovirus replication and protects HRV-infected cells.
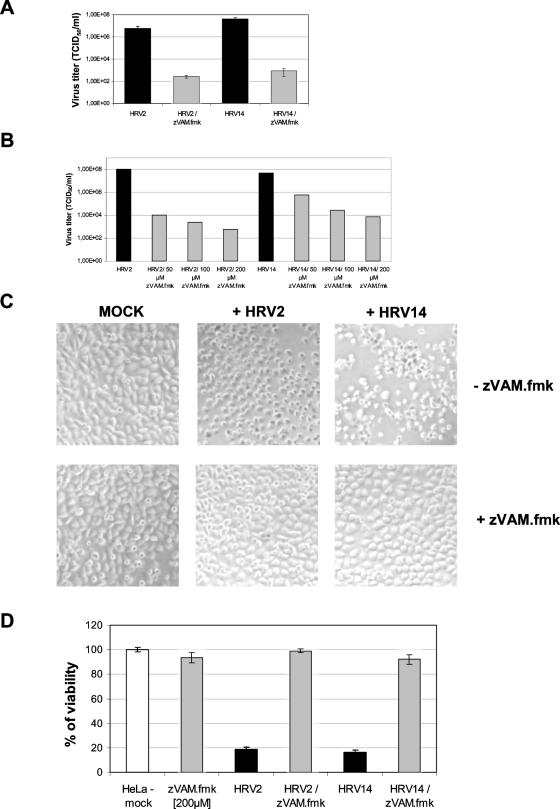
To investigate whether zVAM.fmk can inhibit HRV replication, we chose the serotypes HRV2 and HRV14. These belong to the genetic A and B groups, as well as the minor and the major receptor groups, of HRVs, respectively (24). To enable zVAM.fmk to enter the cells, the compound was added to the cells 30 min before infection with virus. HeLa cells were infected with one of the two rhinovirus serotypes (100 TCID50/cell) in the absence or presence of 200 μM zVAM.fmk, and total virus titers were determined 12 h p.i. As indicated in Fig. 1A, the presence of zVAM.fmk in the culture medium during HRV infection reduced multiplication of both virus serotypes by more than 1,000-fold. The titers observed at 12 h p.i. in the presence of zVAM.fmk were very similar to those obtained in the presence of 10 mM guanidine (data not shown). This indicates that the 12-h p.i. titers in the presence of zVAM.fmk represented the virus carried over from the initial infection. We believe, therefore, that little, if any, virus replication takes place in the presence of zVAM.fmk.
FIG. 1.
zVAM.fmk inhibits HRV replication. (A) HeLa cells were infected with 100 TCID50 units per cell in the absence (black bars) or in the presence (gray bars) of 200 μM zVAM.fmk, and the virus titers were determined 12 h p.i. Means and standard deviations were calculated from three separate experiments. (B) HeLa cells were infected with 100 TCID50 units per cell in the presence of the indicated concentrations of zVAM.fmk, and the virus titers were determined 12 h p.i. (C) HeLa cells were infected as described above at 90% confluence in the absence or presence of 200 μM zVAM.fmk, and phase-contrast photographs were taken 12 h p.i at ×2,000 magnification. (D) HeLa cells were preincubated for 30 min with 200 μM zVAM.fmk before infection with HRV2 or HRV14 (100 TCID50 units per cell) as indicated. Cell viability was determined 24 h p.i. with the proliferation assay described in Materials and Methods. Cellular metabolic activity is shown as a percentage of the control (mock-infected cells = 100%). The results were calculated from three separate experiments.
We also tested lower concentrations of zVAM.fmk (50 μM and 100 μM); however, under our standard conditions of infection (100 TCID50/cell), replication of HRV2 and HRV14 was only partially inhibited (Fig. 1B). We deemed 200 μM zVAM.fmk to be optimal, as at this concentration, it showed the strongest HRV inhibition without being toxic to the HeLa cells.
When using zVAD.fmk, it was necessary to add the inhibitor several times to the medium during infection to maintain inhibition (6). However, this was not necessary for zVAM.fmk. Additional supplementation of the compound every 4 h did not reduce the final virus titer below that seen when it was added only once prior to infection (data not shown).
We wanted to confirm that zVAM.fmk could inhibit other HRV serotypes. We therefore examined HRV16, which belongs to the major receptor group and to HRV genetic group A (24). We found that HRV16 replication was also reduced by about 1,000-fold in the presence of zVAM.fmk (data not shown).
Productive infection of picornaviruses leads to the well-documented cytopathic effect. This consists of numerous membranous vesicles, increased plasma membrane permeability, distortion and displacement of the nuclei, partial condensation of chromatin (pyknosis), rounding up of the infected cells, and eventually their lysis. A clearly visible cytopathic effect is induced in HeLa cells by HRV2 and HRV14 after 12 h under our standard conditions of infection (Fig. 1C). As zVAM.fmk significantly reduces HRV replication, the cytopathic effect does not occur at 12 h p.i. (Fig. 1C), with the host cells remaining viable for at least another 12 h. To examine the influence of zVAM.fmk on cell death induced by HRV2 or HRV14, we examined viability (Fig. 1D) by measuring mitochondrial activity. The strong cytopathic effect caused by both serotypes was reflected in the low viability observed. In contrast, cells infected in the presence of 200 μM zVAM.fmk showed almost no loss of viability compared to mock-infected cells. This indicates that zVAM.fmk can prevent infected HeLa cells from undergoing HRV-induced cell death.
zVAM.fmk does not inhibit caspases.
Programmed cell death plays an important role in the pathophysiology of viral infection. In response to certain human rhinovirus and enterovirus infections, host cells enter apoptosis (4, 9, 14, 30). We showed that HRV14 infection induces apoptosis in HeLa cells and that the process is predominantly induced via the intrinsic pathway due to extensive cytochrome c release from the mitochondria. This leads to the autocatalytic activation of apical caspase 9 (6a), a key event that sets in motion the caspase cascade.
The results in Fig. 1 showed that zVAM.fmk, like zVAD.fmk, inhibits HRV replication and prevents loss of cell viability. We have shown that zVAD.fmk inhibited both cellular caspases and HRV2 2Apro. We therefore wished to investigate the effects of zVAM.fmk on these two enzymes.
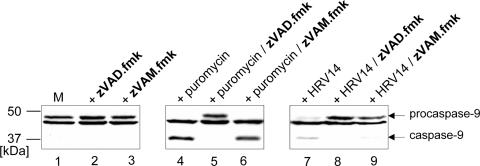
To test whether zVAM.fmk could inhibit a caspase, HeLa cells were treated with puromycin (a known inducer of apoptosis) in the absence or presence of 200 μM zVAM.fmk or 200 μM zVAD.fmk. We then investigated activation of caspase 9 by immunoblotting. Figure 2 shows that, 10 h after treatment of HeLa cells with puromycin, procaspase 9 was efficiently cleaved into its active form. In the presence of zVAD.fmk, this cleavage was completely abrogated (Fig. 2, compare lanes 4 and 5). However, the newly synthesized analog zVAM.fmk did not block caspase 9 activation (Fig. 2, lane 6). Indeed, in the presence of zVAM.fmk, caspase 9 processing was as efficient as in its absence. This indicates that, in contrast to zVAD.fmk, zVAM.fmk does not inhibit the formation of caspase 9.
FIG. 2.
Effects of zVAM.fmk and zVAD.fmk on procaspase 9 activation. HeLa cells were mock infected (lane M), treated with 100 μM puromycin for 10 h, or infected with HRV14 (100 TCID50/cell) in the absence or presence of 200 μM zVAD.fmk or 200 μM zVAM.fmk as indicated. The compounds were added to the medium 1 h prior to the addition of puromycin or HRV14. Protein extracts were prepared by lysis in sample buffer, separated by 12.5% SDS-PAGE, and probed by Western blotting for the state of caspase 9. The positions of the uncleaved form of caspase 9 and the two intermediate cleavage products detected by the polyclonal anti-caspase 9 antibody are marked. The band located between procaspase 9 and the cleavage product is a nonspecifically cross-reacting protein.
We then investigated the effects of the inhibitors on cleavage of procaspase 9, which is induced during HRV14 replication in HeLa cells (Fig. 2, lane 7). The procaspase 9 cleavage induced by HRV14 was almost completely abrogated by both zVAD.fmk and zVAM.fmk, in contrast to that induced by puromycin (Fig. 2, lanes 8 and 9). The ability of zVAM.fmk to inhibit HRV14- but not puromycin-induced caspase activation strongly suggests that zVAM.fmk inhibits apoptosis induced by HRV replication only.
zVAM.fmk prevents the shutoff of host protein synthesis induced by HRV infection.
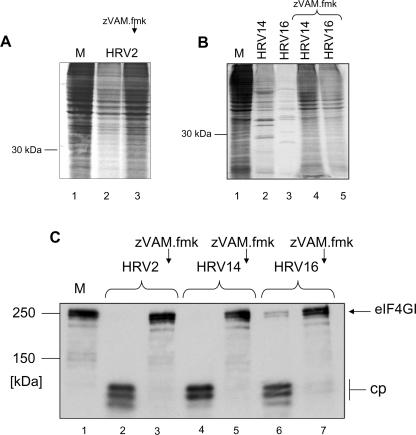
To investigate which step in virus replication was inhibited by zVAM.fmk, we examined the effect of zVAM.fmk on the host cell shutoff of protein synthesis from cellular mRNAs. We infected HeLa cells with HRV2, HRV14, and HRV16 in the absence or presence of 200 μM zVAM.fmk. Newly synthesized proteins were labeled with [35S]methionine. HRV replication significantly reduced viral-protein synthesis in the host cell (Fig. 3A, lane 2, and B, lanes 2 and 3), and viral proteins were clearly visible for HRV14 and HRV16 (Fig. 3B, lanes 2 and 3). Despite the reduction in cellular-protein synthesis, viral proteins were not evident with HRV2 (Fig. 3A, lane 2); in our hands, obtaining complete host cell shutoff with visible viral proteins is often difficult with this serotype.
FIG. 3.
(A and B) Effect of zVAM.fmk on host cell shutoff. HeLa cells were mock infected (M) or infected with HRV2, HRV14, or HRV16 for 8 h in the absence or presence of 200 μM zVAM.fmk as indicated. The compound was added 30 min before infection. [35S]methionine (0.5 μCi) was added 5 h p.i. Infection was terminated by direct addition of Laemmli sample buffer to the cells. Proteins were separated by 12.5% SDS-PAGE, and 35S-labeled proteins were detected by autoradiography. (C) In vivo inhibition of HRV2, HRV14, and HRV16 2Apro activities by zVAM.fmk. HeLa cells were mock infected (lane M) or infected with HRV2, HRV14, or HRV16 for 6 h in the absence or presence of 200 μM zVAM.fmk as indicated. The compound was added to the culture medium 30 min prior to infection. Infection was terminated by direct addition of Laemmli sample buffer to the cells. Proteins were separated by 6% SDS-PAGE, and the state of eIF4GI was probed by Western blotting. The positions of uncleaved eIF4GI and the N-terminal cleavage products (cp) detected by the anti-eIF4GI antiserum are marked.
Infection of HeLa cells with HRV2, HRV14, or HRV16 in the presence of 200 μM zVAM.fmk did not result in a reduction of host cell protein synthesis; indeed, the rate of protein synthesis was comparable to that in the mock-infected cells (Fig. 3A, compare lane 3 with lane 1, and B, compare lanes 4 and 5 with lane 1). Thus, HRV infection in the presence of 200 μM zVAM.fmk does not result in the shutoff of protein synthesis from capped mRNAs.
The high levels of protein synthesis from cellular mRNAs in the HRV-infected cells in the presence of zVAM.fmk suggested that the translation initiation factors eIF4GI and eIF4GII were no longer cleaved. We therefore examined the influence of zVAM.fmk on the cleavage of eIF4GI in cell culture. HeLa cells were infected for 6 h with HRV2, HRV14, or HRV16 in the presence or absence of zVAM.fmk. In the absence of inhibitor, eIF4GI was fully processed at this time (Fig. 3C, lanes 2, 4, and 6). In contrast, addition of 200 μM zVAM.fmk before infection with virus completely blocked cleavage of eIF4GI (Fig. 3B, lanes 3, 5, and 7). Thus, the lack of host cell shutoff in the presence of zVAM.fmk demonstrated by 35S labeling appeared to be due to protection of eIF4GI from cleavage.
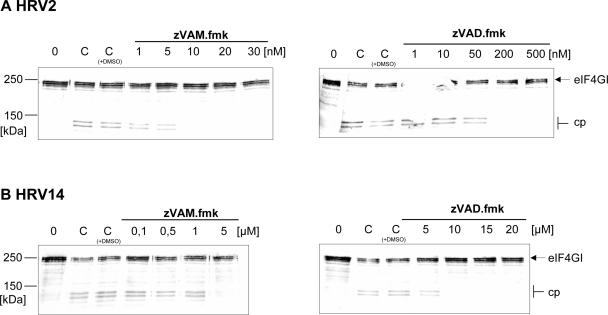
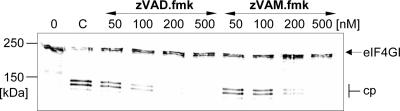
To investigate whether the protection of eIF4GI from cleavage was due to a direct inhibition of HRV2 2Apro, we examined the influence of zVAM.fmk on eIF4GI cleavage in vitro in HeLa cell extracts using recombinant HRV2 2Apro and HRV14 2Apro. The amounts of the enzymes were adjusted so that 50% of eIF4GI was cleaved at the end of the incubation (Fig. 4). Under these conditions, cleavage of eIF4GI by HRV2 2Apro was completely inhibited by the addition of 10 nM zVAM.fmk and between 50 and 200 nM zVAD.fmk. In contrast, HRV14 2Apro was inactivated by 5 μM zVAM.fmk and 10 μM zVAD.fmk (Fig. 4B). This difference in the potencies of the inhibitors toward the purified enzymes is in contrast to the inhibitors' abilities to inhibit virus replication to the same extent.
FIG. 4.
In vitro inhibition of HRV2 2Apro and HRV14 2Apro activities by zVAM.fmk and zVAD.fmk. HeLa cell protein extract (lane 0) was incubated at 30°C for 1 h with HRV2 2Apro (0.33 ng/reaction) (A) and HRV14 2Apro (600 ng/reaction) (B) in the absence (lanes C) or presence of the indicated concentrations of zVAM.fmk or zVAD.fmk. Inhibitors were added to the enzymes 5 min before addition of the extract. eIF4GI cleavage was terminated by the addition of Laemmli sample buffer. Proteins were separated by 6% SDS-PAGE and probed by Western blotting for the eIF4GI state. The positions of the uncleaved form of eIF4GI and the N-terminal cleavage products (cp) detected by the anti-eIF4GI polyclonal antiserum are marked.
Inhibition of CVB4 2Apro by zVAD.fmk and zVAM.fmk.
The amino acid sequences of the 2Apros of HRV2 and CVB4 are about 40% identical, with most of the identity lying in the C-terminal third of the molecules. However, the cleavage sites on the respective polyproteins are less similar. HRV2 2Apro cleaves (asterisk) at IITTA*GPSD, whereas CVB4 2Apro cleaves at LITT*GPYG (Table 1) (28). We wished to examine the effects of zVAM.fmk and zVAD.fmk on the activity of CVB4 2Apro assayed by cleavage of eIF4GI in HeLa extracts. Figure 5 shows that both inhibitors can inhibit CVB4 2Apro. However, a higher concentration of zVAM.fmk (more than 200 nM) is required to inhibit CVB4 2Apro than is required to inhibit HRV2 (10 nM). The different specificities of HRV2 and CVB4 2Apros are thus reflected in their different behaviors toward zVAM.fmk.
TABLE 1.
2Apro cleavage sites
| Virus | Cleavage sequence (P*P′)a |
|---|---|
| HRV2 | TRPIITTA * GPSDMYV |
| HRV14 | RKGDLKSY * GLGPRYG |
| CVB4 | ERASLITT * GPYGHQS |
| PV1b | STKDLTTY * GFGHQNK |
The cleavage sites are indicated by asterisks. The Swissprot identifiers for the amino acid sequences are POLG_HRV2, POLG_HRV14, POLG_POL1M, and POLG_CXB4J.
PV1, poliovirus type 1.
FIG. 5.
In vitro comparative inhibition of CVB4 2Apro activity by zVAD.fmk and zVAM.fmk. HeLa cell protein extract (lane 0) was incubated at 30°C for 1 h with CVB4 2Apro (11 ng/reaction) in the absence (lane C) or in the presence of zVAD.fmk or zVAM.fmk as indicated. Inhibitors were added to the enzyme 5 min before addition of the extract. eIF4GI cleavage was terminated by addition of Laemmli sample buffer. Samples were analyzed on 6% SDS-PAGE, followed by Western blotting. The positions of the uncleaved form of eIF4GI and the N-terminal cleavage products (cp) detected by the anti-eIF4GI polyclonal antibody are marked.
Inhibition of self-processing of HRV2 and HRV14 2Apros by zVAM.fmk.
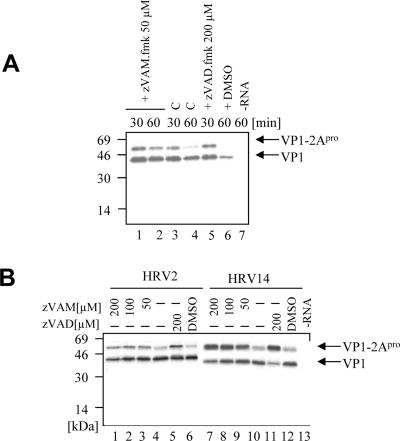
The experiments described above showed that the intermolecular cleavage activities of various 2Apros could be inhibited to various extents by zVAM.fmk. We wished to examine whether the inhibitor could also interfere with intramolecular cleavage. We therefore investigated the effects of zVAM.fmk on self-processing of the HRV2 and HRV14 2Apros. Figure 6A shows a small but measurable inhibition of HRV2 2Apro at 30 min with 50 μM zVAM.fmk (compare lanes 1 and 3). The inhibition obtained was comparable to that obtained with 200 μM zVAD.fmk (lane 5) (6). Addition of higher concentrations of zVAM.fmk did not significantly increase the inhibition of intramolecular cleavage (Fig. 6B, lanes 1 and 2). In contrast, the inhibition of HRV14 2Apro self-processing by zVAM.fmk was much more pronounced (Fig. 6B, right). Even after 60 min of translation in vitro, 50% inhibition of HRV14 2Apro cleavage was still observed at 50 μM zVAM.fmk. Lower concentrations of zVAM.fmk were not effective (data not shown). Interestingly, HRV14 2Apro was also more strongly inhibited by zVAD.fmk than was HRV2 2Apro (Fig. 6B, compare lanes 5 and 11). Furthermore, it is worth noting that neither zVAM.fmk (data not shown) nor zVAD.fmk (35) showed any activity against PV 2Apro in the self-processing reaction.
FIG. 6.
Inhibition of self-processing of HRV 2 and HRV 14 2Apro by zVAM.fmk. (A) Autoradiogram of a time course of in vitro translation of HRV2 VP1-2Apro. Ten minutes after the start of protein synthesis, zVAM.fmk was added to a final concentration of 50 μM (lanes 1 and 2). Lanes 3 and 4 show self-cleavage without an inhibitor, and lane 5 shows inhibition of self-cleavage with 200 μM zVAD.fmk. Samples were taken at the times indicated, and 2Apro self-cleavage from VP1 was examined. In lane 6, translation was performed using 0.2% DMSO only, and lane 7 shows the negative control without RNA. (B) Autoradiogram of the in vitro translation products of RNAs from HRV2 (lanes 1 to 6) and HRV14 (lanes 7 to 12) VP1-2Apros, respectively. Translation was as in panel A, with the addition of zVAM.fmk to a final concentration of 200 μM, 100 μM, or 50 μM (lanes 1 to 3 and 7 to 9). After 60 min of translation, samples were taken and analyzed as for panel A. Lanes 4 and 10 show 60-min translation of the RNAs without any inhibitor, and for comparison, lanes 5 and 11 show inhibition of self-processing by 200 μM zVAD.fmk. Translation was also performed in the presence of 0.2% DMSO only (lanes 6 and 12) and without RNA (lane 13).
These results confirm that zVAM.fmk can inhibit 2Apro self-processing. Furthermore, the data for HRV14 2Apro indicate that the effects of an inhibitor on intramolecular cleavage may differ from those in an intermolecular reaction. Indeed, these results suggest that the inhibition in vivo by zVAM.fmk of HRV14 replication is due to an effect on intramolecular processing of 2Apro. The inhibition of HRV2 replication, however, seems to occur via inhibition of the intermolecular cleavage of its 2Apro.
DISCUSSION
The ability to prevent virus replication by inhibition of viral enzymes, such as the 2Apros of HRVs and enteroviruses, is central to the understanding and treatment of viruses. No compounds are available that are specific for these 2Apro enzymes. We reported recently, however, that zVAD.fmk, a compound developed to inhibit caspases, was also active against HRV2 2Apro and could inhibit HRV2 replication (6). We set out to develop a specific inhibitor of HRV2 2Apro from zVAD.fmk by replacing the aspartic acid residue with methionine to generate zVAM.fmk. Methionine was chosen rather than alanine, which is found at the corresponding P1 position in the wild-type HRV2 cleavage sequence (Table 1), as an oligopeptide with methionine at the P1 position was cleaved five times more efficiently than the wild-type peptide. The choice of methionine was justified, as zVAM.fmk was more efficient than zVAD.fmk in inhibiting intermolecular cleavage by HRV2 2Apro (Fig. 4A, compare left and right blots). Furthermore, zVAM.fmk was effective in blocking the replication of HRV2 in cell culture (Fig. 1). In contrast to zVAD.fmk, zVAM.fmk was not active against caspase 9 and did not inhibit apoptosis of HeLa cells induced by puromycin (Fig. 1 and 2). zVAM.fmk will therefore be a useful and specific tool to examine the roles of 2Apro during HRV2 replication by allowing the inactivation of the enzyme at specific times after the start of the infectious cycle.
Although zVAM.fmk was developed using knowledge of the properties of HRV2 2Apro, it was also able to inhibit the replication of HRV14 and HRV16 in HeLa cells. The inhibitor may therefore be generally useful against HRV serotypes. Investigation of the effects of zVAM.fmk in vitro on HRV2 and HRV14 2Apros indicated that the mechanisms of action of the inhibitor on the two enzymes may differ. zVAM.fmk at nanomolar concentrations inhibits intermolecular 2Apro cleavage of eIF4GI by HRV2 2Apro. Significantly higher concentrations of zVAM.fmk, however, were required to affect the intermolecular processing of eIF4GI by HRV14 and CVB4 2Apros. zVAM.fmk could also inhibit intramolecular cleavage by 2Apro; in this reaction, zVAM.fmk was significantly more active against HRV14 2Apro than against HRV2 2Apro. These apparent differences in the modes of action of the inhibitor against the 2Apros of the two serotypes suggest that a difference in the sensitivity of viral replication of the two serotypes to the inhibitor might be visible. However, as seen in Fig. 1B, the serotypes were inhibited to similar extents at 50 and 100 μM zVAM.fmk. It is indeed possible that such differences are present, but we believe that the sensitivity of the virus replication system may not be high enough to be able to detect them. This is especially true when the concentrations required in vivo are much higher than those required in vitro.
Why is zVAM.fmk so active in inhibiting HRV14 2Apro intramolecular cleavage? The specificity of HRV14 2Apro is much less well explored than that of HRV2 2Apro. No data on the preference of the HRV14 2Apro enzyme at the P1 position is available, and the three-dimensional structure has not been solved. Wang et al. (33) reported that HRV14 2Apro cleaves a peptide corresponding to its own cleavage site poorly in vitro. An improvement in cleavage was achieved by replacing the P2′ leucine residue with proline. HRV14 2Apro was also more active on peptides resembling more closely the HRV2 2Apro cleavage site and the eIF4GI cleavage site. However, these data shed no light on P1 specificity, as the P1 amino acid was in both cases tyrosine, the residue found at P1 in the wild-type HRV14 2Apro cleavage site. Wang et al. (33) concluded that the active sites of the two 2Apros were similar but that different amino acids were responsible for substrate recognition. The results here imply further that subtle differences in the mechanism of substrate recognition during self-processing are also present.
Can inhibitors similar to zVAM.fmk that are more effective against enteroviruses be generated? The activity of zVAM.fmk against CVB4 2Apro was in between those observed for the two HRV enzymes, once again reflecting differences in the substrate binding sites. These must differ to a significant degree to take into account the residues that are present at the respective cleavage sites (Table 1). To design inhibitors with greater potency against enteroviral enzymes, use can be made of the substrate sequences. However, there is a limit to the usefulness of this information, as shown by the low activity of zVAM.fmk against HRV2 in the self-processing reaction. This indicates that the structure of the enzyme is also an important factor for the design of such specific inhibitors. As mentioned above, neither zVAM.fmk (data not shown) nor zVAD.fmk (35) showed any activity against PV 2Apro in the self-processing reaction. This result is surprising, as PV 2Apro and HRV14 2Apro, which was potently inhibited by both zVAM.fmk and zVAD.fmk in self-processing, both possess tyrosine at the P1 position and thus might be expected to have similar overall architectures at the active site. It is therefore likely that the occupancy of other residues in the inhibitor contributes to the overall spectrum of inhibition.
A high-resolution structure is available only for HRV2 2Apro (22). Modeling studies of the interactions of HRV2 2Apro with substrate indicate that the residues Cys101 and Glu102 are responsible for binding the side chain of the P1 residue. The amino acid identity between HRV2 and HRV14 is only around 40% (22). Nevertheless, the residues equivalent to Cys101 and Glu102 can be clearly identified in HRV14 2Apro and all three poliovirus types as being Ala104 and Glu105 and Ala104 and Ser105, respectively. It therefore seems likely that the amino acids present at these positions determine in some as yet undefined way how and to what extent a 2Apro is inhibited by zVAM.fmk.
In short, we have shown that inhibition of the replication of HRVs can be achieved with a substance designed for HRV2 2Apro. Recent genetic evidence has shown that the prevention of poliovirus 2Apro cleavage by mutations leads to a structural protein precursor that impairs capsid formation by correctly processed precursors (5). This may amplify the effects of specific 2Apro inhibitors. Picornavirus 2Apros therefore represent excellent targets for antiviral compounds, and further investigation appears timely in light of a recent call by the WHO for the development of antiviral compounds active against poliovirus (1).
Acknowledgments
We thank Nicole Foeger for cytoplasmic extracts, Joachim Seipelt and Andreas Roetzer for helpful discussions, Andrea Triendl for preparing HeLa cells, Caterina Sturtzel for assistance with some of the experiments, and Eva M. Schmid for plasmid pHRV14 VP1-2Apro, as well as Robert E. Rhoads for the anti-eIF4GI antiserum.
This work was supported by grants from the Austrian Science Foundation to E.K. (F-508-MED) and to T.S. (P-16189 and P-17988).
REFERENCES
- 1.Aylward, R. B., R. W. Sutter, and D. L. Heymann. 2005. Policy. OPV cessation—the final step to a “polio-free” world. Science 310:625-626. [DOI] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. R. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression, vol. 39. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 3.Berger, A., and I. Schechter. 1970. Mapping the active site of papain with the aid of peptide substrates and inhibitors. Phil. Trans. R. Soc. Lond. B 257:249-264. [DOI] [PubMed] [Google Scholar]
- 4.Carthy, C. M., B. Yanagawa, H. Luo, D. J. Granville, D. Yang, P. Cheung, C. Cheung, M. Esfandiarei, C. M. Rudin, C. B. Thompson, D. W. Hunt, and B. M. McManus. 2003. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 313:147-157. [DOI] [PubMed] [Google Scholar]
- 5.Crowder, S., and K. Kirkegaard. 2005. trans-Dominant inhibition of RNA viral replication can slow growth of drug-resistant viruses. Nat. Genet. 37:701-709. [DOI] [PubMed] [Google Scholar]
- 6.Deszcz, L., J. Seipelt, E. Vassilieva, A. Roetzer, and E. Kuechler. 2004. Antiviral activity of caspase inhibitors: effect on picornaviral 2A proteinase. FEBS Lett. 560:51-55. [DOI] [PubMed] [Google Scholar]
- 6a.Deszcz, L., E. Gaudernak, E. Kuechler, and J. Seipelt. 2005. Apoptotic events induced by human rhinovirus infection. J. Gen. Virol. 86:1379-1389. [DOI] [PubMed] [Google Scholar]
- 7.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, B., and J. Kay. 1990. Viral proteinases: weakness in strength. Biochim. Biophys. Acta 1048:1-18. [DOI] [PubMed] [Google Scholar]
- 9.Girard, S., T. Couderc, J. Destombes, D. Thiesson, F. Delpeyroux, and B. Blondel. 1999. Poliovirus induces apoptosis in the mouse central nervous system. J. Virol. 73:6066-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser, W., and T. Skern. 2000. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Lett. 480:151-155. [DOI] [PubMed] [Google Scholar]
- 11.Glaser, W., A. Triendl, and T. Skern. 2003. The processing of eIF4GI by human rhinovirus 2 2Apro: relationship to self-cleavage and role of zinc. J. Virol. 77:5021-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradi, A., Y. V. Svitkin, W. Sommergruber, H. Imataka, S. Morino, T. Skern, and N. Sonenberg. 2003. Human rhinovirus 2A proteinase cleavage sites in eukaryotic initiation factors (eIF) 4GI and eIF4GII are different. J. Virol. 77:5026-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanecak, R., B. L. Semler, C. W. Anderson, and E. Wimmer. 1982. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc. Natl. Acad. Sci. USA 79:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jelachich, M. L., and H. L. Lipton. 1996. Theiler's murine encephalomyelitis virus kills restrictive but not permissive cells by apoptosis. J. Virol. 70:6856-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerekatte, V., B. D. Keiper, C. Badorff, A. Cai, K. U. Knowlton, and R. E. Rhoads. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krausslich, H. G., and E. Wimmer. 1988. Viral proteinases. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 18.Kuechler, E., J. Seipelt, H.-D. Liebig, and W. Sommergruber. 2002. Picornavirus proteinase-mediated shutoff of host cell translation: direct cleavage of a cellular initiation factor, p. 301-311. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 19.Liebig, H.-D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, R. Rhoads, E. Kuechler, and T. Skern. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4γ and influence on in vitro translation. Biochemistry 32:7581-7588. [DOI] [PubMed] [Google Scholar]
- 20.Livingston, D. J. 1997. In vitro and in vivo studies of ICE inhibitors. J. Cell Biochem. 64:19-26. [PubMed] [Google Scholar]
- 21.Matthews, D. A., P. S. Dragovich, S. E. Webber, S. A. Fuhrman, A. K. Patick, L. S. Zalman, T. F. Hendrickson, R. A. Love, T. J. Prins, J. T. Marakovits, R. Zhou, J. Tikhe, C. E. Ford, J. W. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, M. A. Brothers, D. M. DeLisle, and S. T. Worland. 1999. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 96:11000-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen, J. F., M. M. Cherney, H. D. Liebig, T. Skern, E. Kuechler, and M. N. James. 1999. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 18:5463-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed, L. J., and H. Muench. 1938. A simple method of estimating 50 per cent end-points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 24.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 25.Skern, T., B. Hampoelz, A. Guarné, I. Fita, E. Bergmann, J. Petersen, and M. N. G. James. 2002. Structure and function of picornavirus proteinases, p. 199-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 26.Skern, T., W. Sommergruber, D. Blaas, C. Pieler, and E. Kuechler. 1984. Relationship of human rhinovirus strain 2 and poliovirus as indicated by comparison of the polymerase gene regions. Virology 136:125-132. [DOI] [PubMed] [Google Scholar]
- 27.Slee, E. A., H. Zhu, S. C. Chow, M. MacFarlane, D. W. Nicholson, and G. M. Cohen. 1996. Benzyloxycarbonyl-Val-Ala-Asp (OMe) fluoromethylketone (Z-VAD.FMK) inhibits apoptosis by blocking the processing of CPP32. Biochem. J. 315:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommergruber, W., H. Ahorn, H. Klump, J. Seipelt, A. Zoephel, F. Fessl, E. Krystek, D. Blaas, E. Kuechler, H. D. Liebig, and T. Skern. 1994. 2A proteinases of coxsackie- and rhinovirus cleave peptides derived from eIF-4γ via a common recognition motif. Virology 198:741-745. [DOI] [PubMed] [Google Scholar]
- 29.Sommergruber, W., H. Ahorn, A. Zophel, I. Maurer-Fogy, F. Fessl, G. Schnorrenberg, H. D. Liebig, D. Blaas, E. Kuechler, and T. Skern. 1992. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J. Biol. Chem. 267:22639-22644. [PubMed] [Google Scholar]
- 30.Taimen, P., H. Berghall, R. Vainionpaa, and M. Kallajoki. 2004. NuMa and nuclear lamins are cleaved during viral infection—inhibition of caspase activity prevents cleavage and rescues HeLa cells from measles virus-induced but not from rhinovirus 1B-induced cell death. Virology 320:85-98. [DOI] [PubMed] [Google Scholar]
- 31.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761-770. [DOI] [PubMed] [Google Scholar]
- 32.Wang, Q. M., R. B. Johnson, L. N. Jungheim, J. D. Cohen, and E. C. Villarreal. 1998. Dual inhibition of human rhinovirus 2A and 3C proteases by homophthalimides. Antimicrob. Agents Chemother. 42:916-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Q. M., R. B. Johnson, W. Sommergruber, and T. A. Shepherd. 1998. Development of in vitro peptide substrates for human rhinovirus-14 2A protease. Arch. Biochem. Biophys. 356:12-18. [DOI] [PubMed] [Google Scholar]
- 34.Wlodawer, A., and J. Vondrasek. 1998. Inhibitors of human immunodeficiency virus type 1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 27:249-284. [DOI] [PubMed] [Google Scholar]
- 35.Zamora, M., W. E. Marissen, and R. E. Lloyd. 2002. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J. Virol. 76:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]