Abstract
The E1 envelope protein of the alphavirus Semliki Forest virus (SFV) is a class II fusion protein that mediates low pH-triggered membrane fusion during virus infection. Like other class I and class II fusion proteins, during fusion E1 inserts into the target membrane and rearranges to form a trimeric hairpin structure. The postfusion structures of the alphavirus and flavivirus fusion proteins suggest that the “stem” region connecting the fusion protein domain III to the transmembrane domain interacts along the trimer core during the low pH-induced conformational change. However, the location of the E1 stem in the SFV particle and its rearrangement and functional importance during fusion are not known. We developed site-directed polyclonal antibodies to the N- or C-terminal regions of the SFV E1 stem and used them to study the stem during fusion. The E1 stem was hidden on neutral pH virus but became accessible after low pH-triggered dissociation of the E2/E1 heterodimer. The stem packed onto the trimer core in the postfusion conformation and became inaccessible to antibody binding. Generation of the E1 homotrimer on fusion-incompetent membranes identified an intermediate conformation in which domain III had folded back but stem packing was incomplete. Our data suggest that E1 hairpin formation occurs by the sequential packing of domain III and the stem onto the trimer core and indicate a tight correlation between stem packing and membrane merger.
Enveloped alphaviruses such as Semliki Forest virus (SFV) have an internal nucleocapsid surrounded by the viral membrane containing the transmembrane (TM) proteins E2 and E1 (reviewed in reference 46). These glycoproteins are found as heterodimers on the virus surface and form an external layer arranged with T=4 icosahedral symmetry. In the acidic environment of the endosome compartment, E1 is released from its dimeric interaction with E2, inserts into the target membrane, and forms a stable E1 homotrimer (HT) (reviewed in 22 and 25). This E1 conformational change mediates the fusion of the viral and endosomal membranes, delivering the positive-sense RNA genome into the cytosol.
The E1 and E2 proteins can be released from the virus membrane by proteolytic cleavage, producing soluble ectodomains termed E1* and E2* (23). E1* contains the E1 sequence up to residue A391 in the N-terminal portion of the “stem,” the region that connects the ectodomain to the TM anchor (see Fig. 1A). Viral E1 specifically requires cholesterol in the target membrane in order to insert into the membrane and mediate virus-membrane fusion (22). Similarly, when treated at low pH in the presence of cholesterol-containing liposomes, E1* molecules insert into liposomes and form E1* HTs (27). The E1*HT and the full-length E1HT are biochemically comparable in their resistance to trypsin digestion and to dissociation by sodium dodecyl sulfate (SDS) sample buffer at 30°C (27).
FIG. 1.
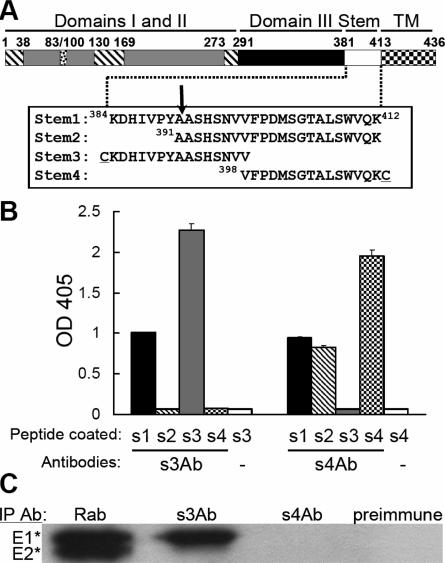
Generation and mapping of antibodies to the SFV E1 stem region. (A) Linear diagram of the primary sequence of the SFV E1 protein. Domains I (hatched), II (gray), and III (black), and the stem (white) and transmembrane (checked) regions are indicated, along with the residue numbers of the approximate domain boundaries. The fusion peptide loop (residues 83 to 100) is in domain II. The sequences of four stem peptides are shown, with an arrow indicating the protease cleavage site that produces E1*. The cysteine residues attached to the N terminus of the stem3 peptide and the C terminus of the stem4 peptide are underlined. (B) Binding of stem antibodies to stem peptides. ELISA wells were coated with the indicated stem peptides and then tested for the binding of s3Ab or s4Ab. The lanes labeled “−” represent controls in which no primary antibody was added. The data shown are the mean ± the standard deviation (SD) from triplicate wells. (C) The E1* and E2* ectodomains were immunoprecipitated as indicated with a polyclonal rabbit antibody against the SFV E1 and E2 proteins (Rab), s3Ab, s4Ab, or preimmune serum. Quantitation showed that the s3Ab immunoprecipitated 68% of the total E1* precipitated by the Rab antibody (average of two independent experiments).
The neutral pH E1 ectodomain monomer is an elongated molecule folded in three domains predominantly composed of β-strands (28, 44). Domain I is positioned in the middle of E1 and connects to domain II containing the internal fusion peptide loop at the tip of the molecule. The other side of domain I connects with domain III. Domain III has an immunoglobulin-like fold and adjoins the E1 stem region and the TM domain in the full-length E1 molecule. In the low pH-induced E1* HT conformation, domains I, II, and III essentially maintain their original folds, but domain III moves ∼37 Å toward the membrane-inserted fusion loop (15). The stem region, which is not ordered in the monomer structure, becomes highly ordered and extends along the trimer core composed of domains I and II. The remaining stem region, which was removed during the proteolytic production of E1*, is of sufficient length to complete the connection to the fused membrane in the postfusion conformation. The low pH-triggered rearrangement of E1 thus forms a structure termed a “trimer of hairpins,” in which the fusion loops and TM domains are positioned at the same side of the membrane-inserted HT.
The structures of both the neutral pH- and the low pH-induced conformations of the SFV E1 ectodomain are remarkably similar to those of the fusion proteins (termed E) from the flaviviruses dengue virus and tick-borne encephalitis virus (3, 36-38, 43, 54). The alphavirus and flavivirus fusion proteins have therefore been grouped together as “class II” virus membrane fusion proteins (28). The “class I” virus fusion proteins such as human immunodeficiency virus type 1 (HIV-1) gp41 and influenza virus HA2 also mediate fusion by rearrangement of the fusion protein to form a trimeric hairpin (17, 25). A critical step in hairpin formation and fusion is believed to involve the packing of the outer layer onto the central core. For the class I proteins, the central trimeric α-helical coiled-coil is packed by an outer layer composed of the C-terminal region of the fusion protein ectodomain (9, 17). Synthetic peptides (termed C-peptides) derived from the outer layer of several class I proteins act as potent inhibitors of the fusion reaction (reviewed in references 10 and 39). In contrast, the outer layer of the class II proteins is composed of domain III and the adjacent stem region. Similar to the class I C-peptides, it was proposed that peptides derived from the class II stem region might act as antiviral inhibitors (3, 37). The addition of exogenous domain III during the low pH-triggered fusion step blocks class II fusion protein fold-back and membrane fusion (29), strongly suggesting that blocking hairpin formation would be a viable antiviral strategy.
Despite the predicted importance of the stem of class II fusion proteins, its exact function is poorly understood. The stem of alphavirus E1 proteins has a sequence of ∼30 amino acids, most of which is not predicted to have significant secondary structure. Fitting the crystal structure of the SFV E1 ectodomain into the cryoelectron microscopic density of SFV or the alphavirus Sindbis virus suggests that the stem is very close to the viral membrane and likely to interact with E2 (44, 52). An E1-E2 interaction around the alphavirus stem region is also supported by studies of a chimeric Sindbis virus containing E1 from the alphavirus Ross River virus. Growth of this chimeric virus can be promoted by mutations in the stem region and transmembrane domain of E1 (51). Our recent studies show that the presence of the stem on exogenous domain III significantly increases its binding to the trimeric fusion protein target and enhances the potency of fusion inhibition (29). Taken together, these data suggest that the class II virus fusion protein stem region may have roles both in virus assembly and in the virus-membrane fusion reaction.
Here we set out to define the importance and features of the conformational transition of the stem region during fusion. We developed polyclonal antibodies specific to the N- or C-terminal halves of the SFV E1 stem. These reagents demonstrated that at low pH the E1 stem region converted from a strong dimer interaction with E2 to a strong interaction with the trimer core, and they revealed an intermediate conformation of the E1 HT in which domain III was folded back while stem packing was incomplete.
(The data in this study are from a thesis submitted by M. Liao in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Cells and virus.
BHK-21 cells, control cholesterol-containing C6/36 mosquito cells, and cholesterol-depleted C6/36 cells were maintained as previously described (29, 50). The SFV used in the present study was derived from the pSP6-SFV4 infectious clone (30) and propagated in BHK-21 cells. 35S-labeled SFV was prepared and gradient-purified as previously described (29).
Peptide synthesis.
The stem1 and stem2 peptides were synthesized by Genemed Synthesis, Inc. (South San Francisco, CA). The stem3 and stem4 peptides were prepared by the Laboratory for Macromolecular Analysis, Albert Einstein College of Medicine. The purity and amino acid composition of the peptides were verified by high-pressure liquid chromatography and mass spectrometry.
Preparation of the stem antibodies.
The stem3 and stem4 peptides were conjugated via their respective N- or C-terminal cysteine residues with Imject maleimide-activated mariculture keyhole limpet hemocyanin (Pierce Biotechnology, Inc., Rockford, IL) according to the manufacturer's instructions. The conjugated peptides were used to immunize New Zealand rabbits (Covance Research Products, Inc., Denver, PA). The rabbit sera were affinity purified by using stem3 or stem4 peptides coupled to UltraLink Iodoacetyl gel (Pierce). The antibodies were concentrated, and the buffer was exchanged to phosphate-buffered saline (PBS) by using Vivaspin concentrators (Vivascience, Hanover, Germany), and the protein concentrations determined by Bio-Rad protein assay. The purified antibody preparations were termed s3Ab and s4Ab.
ELISA.
Enzyme-linked immunosorbent assay (ELISA) plate wells were coated by incubation at 4°C overnight with 0.5 μM concentrations of the indicated stem peptides. The wells were then blocked by incubation with 1% bovine serum albumin (BSA) and incubated at 37°C for 1 h with 0.1 μg of s3Ab or s4Ab/ml or without antibody as a negative control. Antibody binding was detected by reacting with goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase, followed by incubation with substrate (2).
SFV-plasma membrane fusion assay.
We used a previously described fusion-infection assay (29) to follow the low pH-dependent fusion of SFV with the plasma membrane of BHK cells. In brief, virus was prebound to cells on ice and treated for 1 min at 37°C to trigger fusion of virus with the plasma membrane of the cell. Cells were cultured at 28°C overnight in the presence of 20 mM NH4Cl to prevent secondary infection, and infected cells were quantitated by immunofluorescence. SFV domain III protein containing the N-terminal His tag and C-terminal stem region was prepared and purified as previously described (29).
Immunoprecipitation analysis.
Immunoprecipitation analysis in the presence of detergent was performed essentially as previously described (12, 24). In brief, all samples were solubilized in 1% octyl-glucoside (OG) and reacted with antibody. The immune complexes were then absorbed to zysorbin, washed three times with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 8.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 [TX-100], 1% sodium deoxycholate, 0.1% SDS, 20 μg of aprotinin/ml), subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and quantified by PhosphorImager analysis using ImageQuant version 1.2 software (Molecular Dynamics, Sunnyvale, CA).
Immunoprecipitation analysis of intact virus without detergent was performed only for the experiments described in Fig. 5. 35S-labeled SFV was treated at the indicated pH at 37°C for 5 min, returned to neutral pH, reacted with the indicated antibodies in the absence of detergent, absorbed to zysorbin, washed two times with detergent-free buffer to remove unbound antibodies, and then washed three times with RIPA buffer to disrupt viruses before SDS-PAGE analysis (12). Under these conditions only the proteins specifically bound by antibody in the intact virus particle were recovered by immunoprecipitation. The total E1 present in the reaction was defined by solubilization of virus in 1% TX-100 and immunoprecipitation with rabbit polyclonal antibody against SFV E1 and E2.
FIG. 5.
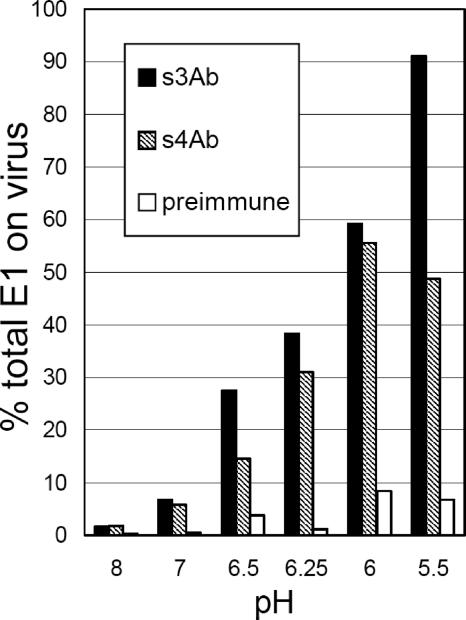
The E1 stem region on intact virus becomes exposed after low pH treatment. 35S-labeled SFV was treated at the indicated pH at 37°C for 5 min. The pH of all samples was then adjusted to pH 8, and the accessibility of the stem region was assessed by immunoprecipitation with stem antibodies as described in Materials and Methods. Antibody precipitation was expressed as a percentage of the total viral E1. The data shown are a representative example of two independent experiments.
Preparation of 35S-labeled homotrimers of the full-length E1 and E1 ectodomain.
To prepare the full-length E1 HT, 35S-labeled SFV in morpholineethanesulfonic acid buffer (pH 8.0) was mixed with 1 mM liposomes containing a 1:1:1:1.5 molar ratio of phosphatidylcholine (PC), phosphatidylethanolamine (PE), sphingomyelin (Sph), and cholesterol (Chol) (4); incubated at pH 5.5 at 37°C for 5 min; returned to pH 8.0; and solubilized in 1% TX-100 for 1 h at room temperature. The viral RNA was digested at 37°C for 10 min with 50 μg of boiled RNase A/ml, and then the viral proteins were digested with 50 μg of trypsin [TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone) treated; type XIII; Sigma, St. Louis, MO]/ml at 37°C for 15 min. Digestion was stopped by treatment with 1 mM phenylmethylsulfonyl fluoride (PMSF) and 150 μg of soybean trypsin inhibitor (Sigma)/ml for 15 min on ice. For each preparation, the isolation of full-length E1 HT was confirmed by analysis on 20% acrylamide gels and comparison with the migration of viral E1 and E1*. The mass of the full-length E1 HT was also confirmed by mass spectrometry (data not shown). SDS treatment of the full-length E1 HT was performed by heating at 95°C for 3 min in 2% SDS in PBS, followed by vigorous shaking at room temperature for 5 min. This treatment was repeated two more times, and the SDS was then diluted to <0.05% in PBS containing 1% OG before immunoprecipitation analysis.
35S-labeled E1 and E2 ectodomains (E1* and E2*) were prepared by subtilisin digestion of radiolabeled virus as previously described (13). Some p62* is always observed in these preparations. E1* homotrimers were generated by treating 35S-labeled ectodomains at pH 5.5 at 37°C for 10 min in the presence of 1 mM liposomes containing a 1:1:1:3 molar ratio of PC, PE, Sph, and Chol.
Formation of the E1 HT and the E1* HT was assessed by their characteristic resistance to dissociation by treatment with SDS sample buffer at 30°C for 3 min (13).
Liposome coflotation assay.
Membrane insertion of the E1* ectodomain was tested by treating 35S-labeled ectodomains at pH 5.5 at 37°C for 10 min in the presence of 1 mM liposomes containing a 1:1:1:3 molar ratio of PC, PE, Sph, and Chol. The samples were then adjusted to pH 8.0, and sucrose gradient flotation was used to separate the top, liposome-containing fraction from the bottom, unbound protein fraction, as previously described (1).
Virus solubilization and E2 immunodepletion.
35S-labeled SFV was solubilized with 1% TX-100 at room temperature for 30 min. An aliquot of the solubilized virus was incubated with 50 μg of boiled RNase A/ml in the presence of 5 mM PMSF at 37°C for 10 min to digest the viral RNA. The E2 was then immunodepleted by incubation on ice for 1 h with 100 μg of MAb E2-1/ml, a mouse monoclonal antibody (MAb) against E2 (24). E2-antibody complexes were removed by two serial absorptions with zysorbin, and the samples were adjusted to 0.025% TX-100 and 1% OG in PBS before further immunoprecipitation analysis. The molecular weight of the isolated E1 was verified by analysis on 20% acrylamide gels.
Low pH treatment of 35S-labeled SFV on the plasma membrane of C6/36 cells.
The conformational changes in E1 were assessed after low pH treatment in the presence of target membranes with or without cholesterol. 35S-labeled SFV was prebound to control or cholesterol-depleted C6/36 cells for 1.5 h on ice in pH 7.4 binding medium (RPMI without bicarbonate plus 0.2% BSA and 10 mM HEPES [RPMI/BSA/HEPES] supplemented with 20 mM MES and 20 mM NH4Cl). Unbound virus was removed by washing in the cold, and the cells with bound virus were treated at 37°C for 1 min with pH 5.5 medium (RPMI/BSA/HEPES plus 30 mM sodium succinate). The cells were then placed on ice, washed two times with pH 7.4 binding medium, lysed in PBS containing 1% TX-100, and digested with 50 μg of trypsin/ml at 37°C for 15 min. The digestion was stopped by the addition of 1 mM PMSF, 150 μg of soybean trypsin inhibitor/ml, 1 μg of pepstatin/ml, 50 μg of leupeptin/ml, and 100 μg of aprotinin/ml, followed by incubation on ice for ≥15 min. Samples were adjusted to 1% OG before immunoprecipitation analysis.
RESULTS
Generation and mapping of antibodies against the stem region of SFV E1.
We defined the SFV E1 stem as the sequence between lysine 384 and lysine 412 connecting domain III to the TM domain (Fig. 1A). Four peptides including various portions of this sequence were synthesized. The stem1 peptide contains the complete stem sequence; the stem2 peptide contains the stem sequence beginning with Ala391, after which is the protease cleavage site that produces E1* (15); and the stem3 peptide and stem4 peptide cover the N-terminal half and C-terminal half of the stem, respectively. The stem3 and stem4 peptides contain an N- or C-terminal cysteine residue that was coupled to a protein carrier. The coupled peptides were used to raise site-directed polyclonal antibodies termed s3Ab and s4Ab (see Materials and Methods).
The specificity of the two stem antibodies was assessed by testing binding to ELISA plates coated with the four stem peptides (Fig. 1B). As predicted, s3Ab bound efficiently to both stem1 peptide and stem3 peptide and not to stem4 peptide. Interestingly, s3Ab showed no binding to stem2 peptide, indicating that the sequence from K384 to Y390 contained essential residues for s3Ab interaction. Immunoprecipitation studies with radiolabeled E1* showed that it was efficiently precipitated by s3Ab even though it ends at residue A391 (Fig. 1C), indicating that the stem sequence 384KDHIVPYA391 is sufficient for s3Ab binding. As predicted, s4Ab showed efficient binding to the stem1, stem2, and stem4 peptides (Fig. 1B) but did not bind either stem3 peptide or E1* (Fig. 1C), which lack the sequence used for immunization.
Effects of stem antibodies and peptides on SFV fusion.
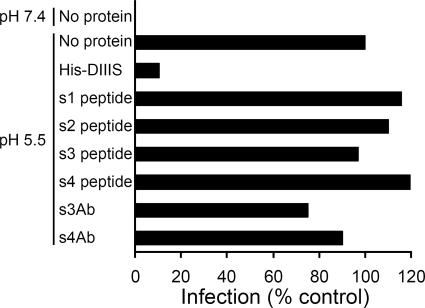
We tested the four stem peptides and two stem antibodies for their ability to inhibit the low pH-induced fusion of SFV with the plasma membrane of BHK cells. As previously described (29), SFV fused efficiently upon low pH treatment, and this activity was strongly inhibited in the presence of 0.6 μM purified domain III protein containing the complete stem region (Fig. 2). In contrast, no inhibition was observed when the fusion step was carried out in the presence of stem peptides at concentrations of 40 μg/ml (13 to 23 μM). Similarly, s3Ab and s4Ab did not inhibit SFV-plasma membrane fusion when tested at concentrations of 200 μg/ml (∼1.3 μM). In addition, neither the stem peptides nor the stem antibodies inhibited SFV-liposome fusion even when the fusion reaction was slowed by using suboptimal pH and temperature conditions (data not shown). In order to better understand these results, we analyzed the accessibility of the stem in the virus particle and during the fusion reaction.
FIG. 2.
Effect of stem peptides and stem antibodies on the fusion of SFV with BHK cells. SFV was diluted in binding medium (pH 6.5) and bound to BHK cells on ice for 90 min. The cells were then incubated at 37°C for 1 min at pH 7.4 or 5.5 in the presence of the indicated peptide or antibody. Cells infected during the low pH pulse were quantitated by immunofluorescence. The proteins were used at concentrations of 0.6 μM for His-DIIIS, 40 μg/ml for the peptides (s1 peptide, 13 μM; s2 peptide, 17 μM; s3 peptide, 23 μM; s4 peptide, 23 μM), or 200 μg/ml (∼1.3 μM) for s3Ab and s4Ab. Note that for the experiments with s3Ab and s4Ab the antibodies were also added to the virus and preincubated at room temperature for 2 h before being added to BHK cells.
Interactions between the stem antibodies and the E1 ectodomain.
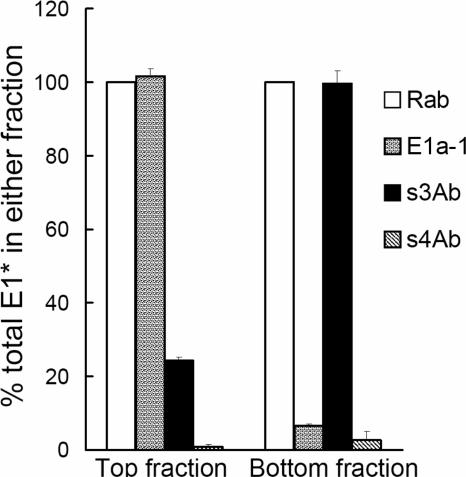
The reactivity of the stem antibodies with the E1 protein suggested that they could be useful as a tool to follow the stem region during the low pH-induced rearrangement of E1. We began this analysis of E1 conformational changes using E1*, which contains the s3Ab epitope at its C terminus. When treated at low pH in the presence of cholesterol-containing liposomes, E1* inserts into the liposomes and forms a stable homotrimer (27). The liposomes and associated E1* HT cofloat to the top of a sucrose gradient, while any remaining monomeric E1* stays in the bottom fraction (1, 27). To test antibody reactivity, immunoprecipitation of the top and bottom portions of the gradient was performed in OG, a detergent previously demonstrated to solubilize the liposomes and associated E1* while preserving the structure of the HT (14). Using this system, we demonstrated that the E1* molecules in the top fraction were efficiently immunoprecipitated by MAb E1a-1, a MAb specific for the low pH-induced conformation of E1 (Fig. 3) (24). As predicted, MAb E1a-1 showed very little reactivity against the monomeric E1* in the bottom fraction. In contrast, the s3Ab very efficiently precipitated monomeric E1* from the bottom fraction, while only ∼24% of the E1* in the top fraction reacted with s3Ab. The actual proportion of E1* molecules in the top fraction that reacted with s3Ab was probably even lower than 24%, since antibody binding to the stem of one E1* molecule would result in the precipitation of all three E1* molecules in the HT.
FIG. 3.
Binding of s3Ab is decreased after E1* trimerization. E1* was mixed with liposomes and treated at pH 5.5 to generate E1*HT. The membrane-bound E1* HTs (top fraction) and non-membrane-bound E1* monomers (bottom fraction) were then separated by floatation in sucrose gradients, solubilized in 1% OG at room temperature for 15 min, and immunoprecipitated with the indicated antibodies. For each antibody, the immunoprecipitated E1 was quantitated and expressed as a percentage of the total E1 present in the top or bottom fraction, as defined by precipitation with the Rab antibody. The data represent the mean ± the SD from three independent experiments.
The differential binding of s3Ab to monomeric and trimeric E1* suggested a significant change in the conformation or accessibility of the stem region during the formation of the E1* trimer. Comparison of the E1* monomer and HT structures (15, 28, 44) shows that the stem region recognized by s3Ab is disordered in the monomer and ordered after trimerization, with the stem interacting along the length of the trimer core. The s3Ab data were consistent with this structural information and further indicated that the stem packing onto the trimer core was tight enough to resist displacement by antibody.
Although the N-terminal stem region shows an extensive interaction with the HT core, the role of this interaction during E1* membrane insertion and trimerization was not known. To test this, E1* was prebound with different antibodies in solution at neutral pH. Samples were then treated at low pH in the presence of liposomes, and the efficiency of E1*-liposome association and SDS-resistant HT formation was evaluated (Fig. 4). E1*-membrane association was not affected by prebinding of the protein with s3Ab, showing similar or even slightly higher coflotation compared to samples treated in the absence of antibody or in the presence of the control s4Ab. Similarly, the s3Ab did not affect the formation of the SDS-resistant E1* HT (Fig. 4B). In contrast, both E1*-membrane interaction and HT formation were completely blocked by prebinding with MAb E1f, an antibody to the E1 fusion peptide loop (Fig. 4) (12). ELISA studies demonstrated that the mildly acidic pH used in Fig. 4 did not release prebound s3Ab from the stem3 peptide or E1* (data not shown). Together, these results suggest that interaction of the N-terminal stem with the trimer core is not required for stable E1*-membrane interaction or formation of the SDS-resistant HT.
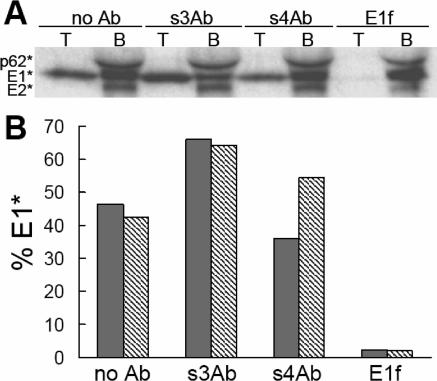
FIG. 4.
S3Ab does not inhibit E1* liposome binding or HT formation. Ectodomains were prebound with the indicated antibodies using concentrations of s3Ab and MAb E1f shown to give efficient immunoprecipitation (Fig. 1C). The samples were then treated at low pH in the presence of liposomes. An aliquot of each sample was analyzed by sucrose gradient flotation. The top liposome-containing fraction and the bottom fraction (indicated as T and B, respectively) were collected, precipitated with trichloroacetic acid, and analyzed by SDS-PAGE, as shown in panel A. The E1* in the top fraction was expressed as a percentage of the total E1* in the gradient (T + B), and results are shown as the gray bars in panel B. Another aliquot of the same samples was tested for the SDS-resistant HT using the assay described in Materials and Methods and expressed as a percentage of the total E1* (HT plus monomer), and the results are shown as the hatched bars in panel B. The data are the average of two independent experiments.
Interactions between stem antibodies and viral E1.
While extensive cryo-electron microscopy reconstruction and fitting studies have defined the arrangement of the E1 ectodomain on the virus particle (28, 32, 40, 44, 52), the stem region is not present in the neutral pH E1 structure, and its disposition on the virus and changes at low pH have not been defined. We therefore wanted to use the stem antibodies to probe the accessibility of this region on intact SFV particles.
Purified 35S-labeled SFV was incubated at 37°C for 5 min at the indicated pH, adjusted to pH 8, and then reacted with the stem antibodies in the absence of detergent (Fig. 5). A detergent wash was used at the end of the precipitation to allow quantitation of E1 that specifically bound the antibody in the intact virus particle. At pH 8 or pH 7 the E1 stem region was inaccessible for binding of either the s3Ab or s4Ab (Fig. 5). Both the N- and the C-terminal regions of the stem became increasingly accessible after exposure to decreasing pH (pH 6.5 to 5.5). Approximately 90% of the viral E1 molecules bound the s3Ab after treatment of virus at pH 5.5, while ∼50% of E1 bound the s4Ab after treatment at pH 6.0.
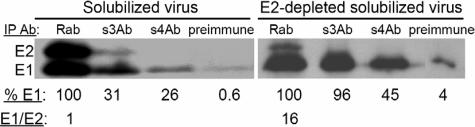
Studies of the structure of the alphavirus particle suggest that the observed inaccessibility of the E1 stem region was due to the presence of the E2 protein, which interacts closely with E1 and forms the external spike domain (see, for example, references 40 and 44). The E2/E1 dimer interaction is known to dissociate at low pH (45), and virus chimera studies suggest that E2 may interact directly with the E1 stem region (51). To test whether an E2-E1 interaction protected the stem region from antibody binding, 35S-labeled SFV was solubilized in OG and then precipitated with stem antibodies (Fig. 6, left panel). While increased immunoprecipitation was observed compared to the intact virus at neutral pH (see Fig. 5), solubilized E1 was only partially reactive with the stem antibodies (31% for s3Ab and 26% for s4Ab). This is in keeping with previous studies showing that the E2/E1 dimer is stable to treatment with OG and other nonionic detergents (18). We therefore immunodepleted the solubilized viral proteins with an antibody against E2 to enrich for monomeric E1 (55). This treatment changed the ratio of E1 to E2 from 1:1 in the solubilized virus to 16:1 in the immunodepleted sample, demonstrating efficient E2 depletion. The monomeric full-length E1 was efficiently recognized by the s3Ab (96% immunoprecipitation, Fig. 6, right panel). Thus, the stem region of full-length E1 was fully competent for s3Ab binding but was protected by the E2/E1 dimer interaction. The s4Ab also showed increased binding after E2 depletion (45% versus 26%).
FIG. 6.
The E2/E1 dimer interaction prevents the binding of stem antibodies. 35S-labeled SFV was solubilized in detergent-containing buffer and the E2 protein was removed by immunodepletion where indicated (right panel). The accessibility of the E1 stem region was tested by immunoprecipitation with the stem antibodies. For each antibody the immunoprecipitated E1 was quantitated and expressed as a percentage of the total E1 in the reaction (%E1, shown under each lane). The total E1 was defined by precipitation with the Rab antibody. The Rab sample was also used to quantitate the ratio between E1 and E2 proteins before and after immunodepletion (shown under lane 1 of each panel). The quantitative data are the average of two independent experiments.
Although both antibodies strongly reacted with stem peptides (Fig. 1B), the s4Ab showed lower binding to E1 than the s3Ab (see Fig. 6 and 7B). The C-terminal part of the stem (∼7 amino acids) was predicted to form an α-helix by several protein structure prediction servers such as Jpred (7), PSIPRED (21, 33), and HHpred (47, 48) (data not shown). The stem4 peptide might not fully present this secondary structure during antibody induction and purification, perhaps explaining why the s4Ab did not efficiently recognize the stem on viral E1. In contrast, the N-terminal part of the stem recognized by s3Ab was not predicted to form any secondary structure, and this antibody efficiently recognized both peptide and viral E1.
FIG. 7.
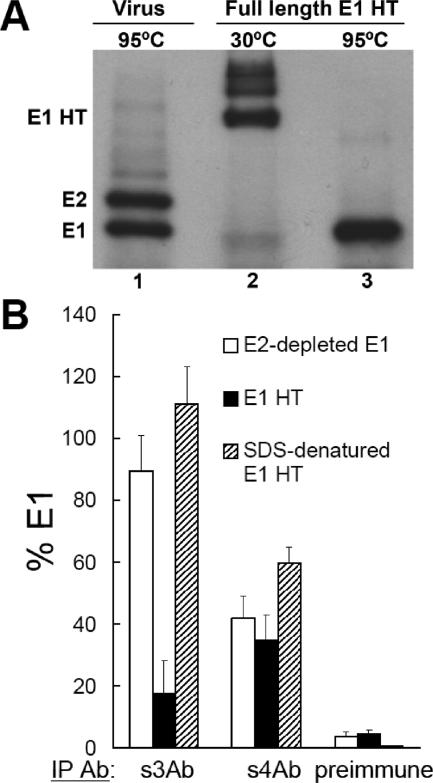
Binding of the s3Ab is decreased after trimerization of the full-length E1 protein. (A) Characterization of the E1 HT. Full-length E1 HT was generated at low pH and isolated based on its resistance to trypsin digestion. Aliquots of the samples were solubilized in SDS-sample buffer for 3 min at 30°C (lane 2) or 95°C (lane 3) and analyzed by SDS-PAGE with 11% acrylamide gels. Lane 1 shows a control of 35S-labeled SFV solubilized in SDS-sample buffer at 95°C. (B) Antibody reactivity of various E1 preparations. Monomeric E1 prepared by E2 immunodepletion (see Fig. 5), full-length E1 HT prepared as in panel A, and SDS-treated full-length E1 HT were adjusted to 1% OG and immunoprecipitated with the indicated antibodies. The reactivity with each antibody was expressed as a percentage of the total E1 precipitated by the Rab antibody. The data represent the mean ± the SD from three independent experiments.
Interactions between stem antibodies and full-length E1 HT.
Recent studies have shown differences in the stability of the full-length flavivirus E homotrimer versus the ectodomain homotrimer (49), suggesting that additional contacts are provided by stem packing. However, the conformation and accessibility of the stem region in the full-length class II fusion proteins during fusion have not been studied. To study the stem in the full-length E1 HT, 35S-labeled SFV was incubated at pH 5.5 in the presence of liposomes to induce HT formation, and the HT was isolated based on its resistance to trypsin digestion (see Materials and Methods). Isolated E1 HT was fully resistant to dissociation by SDS sample buffer at 30°C (Fig. 7A, lane 2), and dissociation by boiling in SDS sample buffer showed that only full-length E1 protein was present (Fig. 7A, lane 3). We compared the immunoreactivity of monomeric (E2-depleted) full-length E1, the E1 HT, and the E1 HT dissociated by SDS treatment (Fig. 7B). The s3Ab efficiently precipitated monomeric E1 but bound only ∼18% of the full-length E1 HT. After the E1 HT was dissociated by boiling in SDS, it was efficiently precipitated by the s3Ab, demonstrating that the residues needed for s3Ab binding were present on the isolated full-length HT. Thus, consistent with the results with E1* (see Fig. 3), the N-terminal region of the stem was fully accessible in monomeric E1, became hidden after E1 trimerization, and could be reexposed by trimer dissociation. Because of the relatively low efficiency of s4Ab binding, the accessibility of the C-terminal region of the stem remains unclear. However, the E1 protein showed comparable reactivity (∼50%) with s4Ab before and after E1 HT formation, which suggested a less drastic change than for the N-terminal stem.
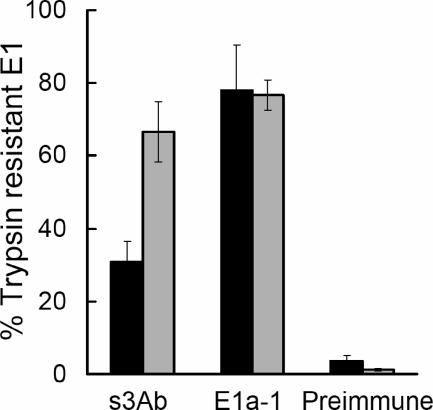
The ectodomain studies described above suggested that stem interaction was not essential for E1* membrane binding and trimerization, since these were insensitive to prebinding with s3Ab (Fig. 4). This suggests a possible role for the stem in a later stage of E1 rearrangement and membrane fusion. To test the correlation between stem packing and membrane fusion, we used the s3Ab to compare the E1 HT conformations formed under fusion-permissive versus fusion-nonpermisive conditions. 35S-labeled SFV was bound on ice to control or cholesterol-depleted C6/36 cells. The cells with bound virus were then treated at pH 5.5 for 1 min at 37°C, conditions previously shown to trigger virus fusion and infection of the control cells. Virus bound to the cholesterol-depleted cells does not fuse or infect, although the E1 HT forms and is both SDS and trypsin resistant (42, 50). After low pH treatment the cells with bound virus were lysed and the lysates digested with trypsin to isolate the full-length E1 HT. Similar to the results in Fig. 7B, the s3Ab reacted relatively inefficiently (31%) with the E1 HT formed on control cells in which membrane fusion occurred (Fig. 8). In contrast, the s3Ab reacted significantly more efficiently (67%) with the E1 HT formed on cholesterol-depleted cells in which fusion is blocked. The MAb E1a-1, an acid-conformation-specific antibody that maps to domain I (2), reacted similarly with both preparations of E1 HT, in keeping with this antibody detecting an E1 conformation correlated with the folding back of domain III (29). Thus, fusion-blocking conditions produced an intermediate E1 HT conformation in which the stem region remains more accessible to s3Ab binding, presumably because it has not yet fully packed against the trimer core.
FIG. 8.
The s3Ab identifies an intermediate HT conformation generated on fusion-inactive cholesterol-depleted target cells. 35S-labeled SFV was prebound on ice to control (black bars) or cholesterol-depleted (gray bars) C6/36 cells and treated at pH 5.5 for 1 min at 37°C to trigger the low pH-dependent conformational changes of the viral glycoproteins. The samples were then solubilized in 1% TX-100, treated with trypsin to isolate E1 HTs, and immunoprecipitated with the indicated antibodies. Reactivity with each antibody was expressed as a percentage of the total E1 precipitated by the Rab antibody. The data represent the mean ± the SD from three independent experiments.
Consistent with previous results demonstrating that the presence of cholesterol in target membranes promotes HT formation (5), the efficiency of trypsin-resistant HT production on cholesterol-depleted cells was about half of that on control cells (data not shown). In fact, a proportion of the E1a-1 reactive and SDS-resistant E1 HT formed on the cholesterol-depleted cells was trypsin sensitive and thus not tested in the experiment shown in Fig. 8. Studies that included this HT population demonstrated that it was also fully accessible to binding by the s3Ab (data not shown).
DISCUSSION
Dynamics of the E1 stem region.
Cryo-electron microscopy reconstructions and fitting of the E1 structure indicate that E2 covers much of E1 and interacts both with the E1 tip containing the fusion loop and with the stem-TM region at the other end of E1 (see, for example, references 40 and 44). We developed site-directed antibodies to the SFV E1 stem and used them to monitor this region during various stages of the virus fusion reaction. Our results showed that the stem region was inaccessible to antibody in the virus particle at neutral pH and was not exposed even in detergent-solubilized E2/E1 heterodimers. The E1 stem region became accessible to antibody binding in E2-depleted detergent-solubilized E1 or in virus particles treated at low pH, a condition that disrupts the E2-E1 interaction. Dissociation of the E2/E1 dimer is the first observable low pH-dependent step in the alphavirus fusion pathway and an important point of fusion regulation (see reference 45 and references therein). A variety of mutations in the E2 ectodomain are known to alter the pH dependence of alphavirus-membrane fusion by affecting E2/E1 dimer dissociation (16, 45, 53). Our data suggest that the stem region of E1 is also important in dimer interaction and the regulation of membrane fusion.
After the release of the E1 stem from E2 at low pH, the stem region became s3Ab inaccessible after E1 trimerization. This finding is consistent with the observed close packing of the E1 stem against the central trimer core in the E1* HT structure (15). Thus, the s3Ab revealed a dynamic series of arrangements of the stem in the virus: initial interaction with E2 on the virus particle, release from E2 at low pH, and packing as part of the outer layer of the stable trimer. These findings suggest that the sequence/structure of the stem must be optimized to balance its two distinct roles in regulatory dimer interaction versus formation of the highly stable E1 homotrimer.
The intermediate HT conformation revealed by s3Ab.
We demonstrated that the s3Ab could efficiently bind E1* at neutral pH (Fig. 1C) but that antibody binding did not inhibit the low pH-triggered interaction of E1* with liposomes or the formation of the SDS-resistant E1* HT (Fig. 4). This suggests that E1 can insert into membranes and trimerize without folding back the stem region. Comparison of the trypsin-resistant E1 trimers formed on control and cholesterol-depleted cell membranes demonstrated that the s3Ab epitope was significantly more accessible under fusion-blocking conditions (Fig. 8), suggesting that the stem does not fully pack against the trimer core in the absence of fusion. Thus, packing of the stem appears to be closely linked to the final membrane merger.
How do these results suggesting a late step in E1 hairpin formation fit with the available information on E1 rearrangement during membrane fusion? We have recently demonstrated that membrane fusion by class II viruses such as SFV and dengue virus is specifically blocked by the addition of exogenous domain III during the low pH-triggering step (29). Under these conditions, exogenous domain III stably binds to an intermediate conformation of trimeric E1, thereby preventing the fold-back of the viral domain III. The block in domain III fold-back correlates with decreased SDS resistance of the E1 trimer and with a block in the exposure of the MAb E1a-1 epitope, suggesting that before the folding-back of domain III the E1 trimer intermediate is not SDS resistant or MAb E1a-1 reactive. In the present study, the E1 HT formed on fusion-inactive cholesterol-depleted membranes was SDS and trypsin resistant and efficiently bound MAb E1a-1 but showed increased reactivity with the s3Ab compared to the HT formed on fusion-active control membranes (Fig. 8). This suggests that during E1 trimerization under cholesterol-depleted conditions the domain III folds back while the stem region does not complete its packing. Thus, the present study provides the first demonstration that the folding back of domain III and the stem can be uncoupled during the low pH-induced rearrangement of class II proteins.
The intermediate conformation of E1 revealed by the s3Ab is reminiscent of an intermediate conformation described for HIV-1 gp41, which does not form the final “trimer of hairpins” until the completion of membrane fusion (35). Interestingly, even though the data suggest that formation of the SFV trimer core and domain III fold-back occur on the cholesterol-depleted cells, the stem region did not pack along the trimer even after the HT was solubilized in detergent (Fig. 8). Since the hindrance to stem packing is presumably removed upon membrane solubilization, the stem should then readily fit into the grooves of the trimer core. It is possible that stem packing can only occur simultaneously with formation of the groove and/or that packing itself requires a low pH. Alternatively, the cholesterol-deficient target membrane may not allow complete or correct fold-back of domain III or formation of the optimal groove for stem packing.
Stages in packing of the outer layer onto the trimer core.
Our data suggest a “two-step packing” model for the membrane fusion reaction mediated by SFV E1. In such a model, the trimer core formed by domains I and II would first be bound by the folded-back domain III, followed by subsequent binding of the stem region, with the latter step being directly coupled to membrane merger.
There is evidence that the class I proteins also show two-step packing of their outer layers. For example, the C34 C-peptide of HIV-1 gp41 forms a stable six-helix bundle with a synthetic N-peptide, whereas the more C-terminal T20 C-peptide is not able to form a stable six-helix bundle (31) and is less potent at inhibiting the lipid mixing step of fusion (26). The packing of the C-terminal “leash” residues of the influenza virus HA2 ectodomain is important to complete membrane fusion but is not essential for the low pH-induced hairpin conformation (41). Thus, different parts of the outer layer region from class I fusion proteins appear to function sequentially during fusion.
This general “two-step packing” model may also be applicable to the interaction of the cellular SNARE proteins during intracellular membrane fusion events (for a review, see reference 20). The initial interaction of the N-terminal, membrane-distal portion of the v-SNARE with the trimeric t-SNARE complex is reversible and can be inhibited by exogenous peptides containing this region of the v-SNARE (34). In contrast, the C-terminal, membrane-proximal portion of the v-SNARE interacts with the t-SNARE complex after the binding of the N-terminal region of the v-SNARE becomes stable (34). Thus, different portions of the v-SNARE show sequential and perhaps functionally distinct stages of interaction.
Stem-based antiviral strategies.
Because of the general similarity of the “trimer of hairpins” of the class I and class II proteins, it was predicted that synthetic peptides containing the class II stem sequence would have fusion-blocking activity analogous to that of the class I C-peptides (3, 37). Indeed, exogenous domain III proteins containing the stem region showed significantly increased binding to E1 at low pH and a more potent inhibition of virus-membrane fusion (29), suggesting that stem interactions could significantly contribute to the driving force during membrane fusion. The lack of inhibition by the stem1-4 peptides could simply reflect a need for peptide optimization, similar to the results frequently observed with class I peptides (39). It is also not known whether a stem peptide could bind an initial trimer core versus whether binding would require the prior folding-back of domain III. If the latter case were true, the exogenous stem could be at a strong disadvantage in competing with the endogenous stem from the folded-back domain III. Recently, a synthetic peptide containing a C-terminal region of the dengue virus stem was shown to decrease infection by both dengue and West Nile viruses (19). It will be interesting to determine the effects of this stem peptide on the virus membrane fusion reaction.
Antibodies or small molecule inhibitors could be alternative methods to disrupt the interaction of the stem with the trimer core. Our data on SFV indicate that the alphavirus stem region is inaccessible to antibody in the viral particle at neutral pH. The lack of accessibility at neutral pH and the speed and endosomal location of the alphavirus fusion suggest that stem antibodies are unlikely to be useful fusion inhibitors. Small molecules have been used successfully as inhibitors of hairpin formation and membrane fusion for class I viruses such as HIV-1 (8, 11) and respiratory syncytial virus (6). A small molecule strategy might also work for the class II viruses if it could be targeted to a key stem interaction with the trimer core. The alphavirus stem region contains a number of conserved residues. It will be important to define the roles of these conserved stem residues in the dimeric interactions of E1 with E2, and in the interaction of the stem with the trimer core during fusion.
Acknowledgments
We thank Alice Guo for excellent technical assistance. We thank the members of our lab for helpful discussions and comments on the manuscript.
This study was supported by a grant to M.K. from the Public Health Service (R01 GM052929) and by Cancer Center Core Support grant NIH/NCI P30-CA13330.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, A., M. R. Klimjack, P. K. Chatterjee, and M. Kielian. 1999. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J. Virol. 73:10029-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee, P. K., C. H. Eng, and M. Kielian. 2002. Novel mutations that control the sphingolipid and cholesterol dependence of the Semliki Forest virus fusion protein. J. Virol. 76:12712-12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianci, C., D. R. Langley, D. D. Dischino, Y. Sun, K. L. Yu, A. Stanley, J. Roach, Z. Li, R. Dalterio, R. Colonno, N. A. Meanwell, and M. Krystal. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc. Natl. Acad. Sci. USA 101:15046-15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuff, J. A., and G. J. Barton. 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40:502-511. [DOI] [PubMed] [Google Scholar]
- 8.Debnath, A. K., L. Radigan, and S. Jiang. 1999. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 42:3203-3209. [DOI] [PubMed] [Google Scholar]
- 9.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Ferrer, M., T. M. Kapoor, T. Strassmaier, W. Weissenhorn, J. J. Skehel, D. Oprian, S. L. Schreiber, D. C. Wiley, and S. C. Harrison. 1999. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat. Struct. Biol. 6:953-960. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, M. Liao, L. Hammar, R. H. Cheng, and M. Kielian. 2004. Multistep regulation of membrane insertion of the fusion peptide of Semliki Forest virus. J. Virol. 78:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons, D. L., B. Reilly, A. Ahn, M.-C. Vaney, A. Vigouroux, F. A. Rey, and M. Kielian. 2004. Purification and crystallization reveal two types of interactions of the fusion protein homotrimer of Semliki Forest virus. J. Virol. 787:3514-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbons, D. L., M.-C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 16.Glomb-Reinmund, S., and M. Kielian. 1998. fus-1, a pH-shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J. Virol. 72:4281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, S. C. 2005. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 64:231-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helenius, A., and J. Kartenbeck. 1980. The effects of octylglucoside on the Semliki Forest virus membrane. Eur. J. Biochem. 106:613-618. [DOI] [PubMed] [Google Scholar]
- 19.Hrobowski, Y. M., R. F. Garry, and S. F. Michael. 2005. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell 112:519-533. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Kielian, M. 2006. Class II virus membrane fusion proteins. Virology 344:38-47. [DOI] [PubMed] [Google Scholar]
- 23.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest Virus. J. Cell Biol. 101:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian, M., and F. A. Rey. 2006. Virus membrane fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliger, Y., S. A. Gallo, S. G. Peisajovich, I. Munoz-Barroso, S. Avkin, R. Blumenthal, and Y. Shai. 2001. Mode of action of an antiviral peptide from HIV-1: inhibition at a post-lipid mixing stage. J. Biol. Chem. 276:1391-1397. [DOI] [PubMed] [Google Scholar]
- 27.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 29.Liao, M., and M. Kielian. 2005. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus-membrane fusion. J. Cell Biol. 171:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S., H. Lu, J. Niu, Y. Xu, S. Wu, and S. Jiang. 2005. Different from the HIV fusion inhibitor C34, the anti-HIV drug Fuzeon (T-20) inhibits HIV-1 entry by targeting multiple sites in gp41 and gp120. J. Biol. Chem. 280:11259-11273. [DOI] [PubMed] [Google Scholar]
- 32.Mancini, E. J., M. Clarke, B. E. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 33.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 34.Melia, T. J., T. Weber, J. A. McNew, L. E. Fisher, R. J. Johnston, F. Parlati, L. K. Mahal, T. H. Sollner, and J. E. Rothman. 2002. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J. Cell Biol. 158:929-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 38.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay, S., W. Zhang, S. Gabler, P. R. Chipman, E. G. Strauss, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, H. E., J. A. Gruenke, and J. M. White. 2003. Leash in the groove mechanism of membrane fusion. Nat. Struct. Biol. 10:1048-1053. [DOI] [PubMed] [Google Scholar]
- 42.Phalen, T., and M. Kielian. 1991. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 112:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 44.Roussel, A., J. Lescar, M.-C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Crystal structure of the Semliki Forest virus envelope protein E1 in its monomeric conformation: identification of determinants for icosahedral particle formation. Structure 14:75-86. [DOI] [PubMed] [Google Scholar]
- 45.Salminen, A., J. M. Wahlberg, M. Lobigs, P. Liljeström, and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus II: cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlesinger, S., and M. J. Schlesinger. 2001. Togaviridae: the viruses and their replication, p. 895-916. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 47.Soding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 48.Soding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stiasny, K., C. Kossl, and F. X. Heinz. 2005. Differences in the postfusion conformations of full-length and truncated class II fusion protein E of tick-borne encephalitis virus. J. Virol. 79:6511-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, J. S., E. G. Strauss, and J. H. Strauss. 1998. Molecular genetic study of the interaction of Sindbis virus E2 with Ross River virus E1 for virus budding. J. Virol. 72:1418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, X., and M. Kielian. 2004. Mutations that promote furin-independent growth of Semliki Forest virus affect p62-E1 interactions and membrane fusion. Virology 327:287-296. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziemiecki, A., and H. Garoff. 1978. Subunit composition of the membrane glycoprotein complex of Semliki Forest virus. J. Mol. Biol. 122:259-269. [DOI] [PubMed] [Google Scholar]