Abstract
Recent studies of varicella-zoster virus (VZV) DNA sequence variation, involving large numbers of globally distributed clinical isolates, suggest that this virus has diverged into at least three distinct genotypes designated European (E), Japanese (J), and mosaic (M). In the present study, we determined and analyzed the complete genomic sequences of two M VZV strains and compared them to the sequences of three E strains and two J strains retrieved from GenBank (including the Oka vaccine preparation, V-Oka). Except for a few polymorphic tandem repeat regions, the whole genome, representing approximately 125,000 nucleotides, is highly conserved, presenting a genetic similarity between the E and J genotypes of approximately 99.85%. These analyses revealed that VZV strains distinctly segregate into at least four genotypes (E, J, M1, and M2) in phylogenetic trees supported by high bootstrap values. Separate analyses of informative sites revealed that the tree topology was dependent on the region of the VZV genome used to determine the phylogeny; collectively, these results indicate the observed strain variation is likely to have resulted, at least in part, from interstrain recombination. Recombination analyses suggest that strains belonging to the M1 and M2 genotypes are mosaic recombinant strains that originated from ancestral isolates belonging to the E and J genotypes through recombination on multiple occasions. Furthermore, evidence of more recent recombination events between M1 and M2 strains is present in six segments of the VZV genome. As such, interstrain recombination in dually infected cells seems to figure prominently in the evolutionary history of VZV, a feature it has in common with other herpesviruses. In addition, we report here six novel genomic targets located in open reading frames 51 to 58 suitable for genotyping of clinical VZV isolates.
In temperate climates, varicella-zoster virus (VZV) is a ubiquitous human pathogen with tropism to skin and sensory ganglia. VZV establishes latent infections, most prominently in the dorsal root and trigeminal ganglia, resulting in a permanent association with the host that may reactivate many decades after primary infection to cause herpes zoster. Primary VZV infection is nearly always symptomatic (varicella), a normally benign childhood disease; occasionally first exposure results in more serious illness or, in rare instances, death (40). Several neurological manifestations have been described in relation to VZV reactivation, such as facial palsy, encephalitis, meningitis, and cerebral vasculitis, conditions that may occur without any dermal lesions (4, 12, 16). The combined disease burden of primary and reactivated VZV infections has motivated widespread vaccination using a live, attenuated VZV strain (V-Oka). More recently, it has been demonstrated that a high-dose formulation of V-Oka can prevent or substantially ameliorate both herpes zoster and postherpetic neuralgia in persons 60 years of age and older (33).
The genetic diversity of various human herpesviruses (cytomegalovirus, Epstein-Barr virus, herpes simplex virus type 1, human herpesvirus 7, and human herpesvirus 8) and methods for genotyping clinical isolates have been proposed by a number of laboratories (7, 11, 27, 31, 34, 35, 38, 41-43, 46). Genetic diversity among VZV strains was initially defined using either restriction enzyme mapping (1, 22) or variation in the copy number at the tandem repeat regions (6, 20, 21, 39). Phylogenetic analyses of partial VZV genome sequences have recently identified a divergence of VZV into different genotypes and recombinant strains (25, 30, 44). Loparev et al. (25) performed targeted (multiple locus) analysis of VZV polymorphisms for a large number of clinical isolates obtained from all six continents, establishing a divergence into the three genotypes E (European), J (Japanese), and M (mosaic). The M genotype could be further subdivided into the groups M1 and M2. A seminal finding of the report was that strains belonging to M1 and M2 were most common in tropical regions, whereas E and J strains tended to occur in temporate latitudes. In addition, it was demonstrated that sequence analysis of a 447-nucleotide (nt) region located in open reading frame 22 (ORF22r1) could unerringly recapitulate the genotyping results obtained from analyzing 23 single-nucleotide polymorphisms located in seven open reading frames distributed over the entire genome. These analyses implied that recombination events, which were recently found to be a common feature among wild-type isolates of another alphaherpesvirus, i.e., herpes simplex virus type 1 (5, 31), were of interest for future study as a possible mechanism for VZV strain variation (30). In the studies reported here, we determined the complete genomic sequence of two M VZV strains presented in a previous study (25). The sequences were analyzed with different phylogenetic algorithms applied on the complete genome as well as on shorter segments. The results suggest a divergence of clinical VZV isolates into at least four genotypes, designated E, J, M1, and M2, and further suggest that recombination is an important contributor to the evolution of VZV.
MATERIALS AND METHODS
VZV strains.
A total of six clinical isolates were analyzed. Strains 123 (previously named 123J) and DR (previously named Morocco 1) were selected as representative strains for the genotypes M1 and M2, respectively, and were sequenced at the National VZV Laboratory at the Centers for Disease Control and Prevention (25). Sequence data for four additional VZV strains were obtained from GenBank (BC, gi 46981409; Dumas, gi 9625875 [or 59989, older entry]; MSP, gi 46981482; and parental Oka [P-Oka], gi 26665420) and have been described previously (8, 17). The culture-attenuated vaccine preparation (V-Oka, gi 2264274) was also included in the phylogenetic analyses of complete genomic sequences. Characteristics of the VZV strains analyzed are provided in Table 1.
TABLE 1.
Characteristics of VZV strains analyzed in this study
| Strain | Origin | Geographic source | Clinical manifestation | Genotype |
|---|---|---|---|---|
| 123 | CDC | United States | Chickenpox | M1 |
| DR | CDC | Morocco | Zoster | M2 |
| BC | GenBank | Canada | Zoster | E |
| Dumas | GenBank | The Netherlands | Chickenpox | E |
| MSP | GenBank | United States | Chickenpox | E |
| P-Oka | GenBank | Japan | Chickenpox | J |
| V-Oka | GenBank | Japan | Cell culture | J |
PCR and DNA sequencing.
DNA was purified from lysates of VZV-infected cells using the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. DNA from uninfected human lung fibroblast cells was used as a negative control for all experiments. The complete DNA sequences for VZV-DR and VZV-123 were determined using the primer walking approach. Panels of sequencing primers were designed based on the published sequences for the Dumas and P-Oka strains. The 30 sets of primers were designed to amplify 2,000- to 4,000-bp overlapping regions of VZV DNA. DNA amplification reactions were performed using ThermoAce DNA polymerase preparation (Invitrogen), a high-fidelity polymerase. For DNA purification, PCR products were separated on 2% precast agarose gels (BMA, Rockland, Maine); DNA bands were excised and isolated using a gel extraction kit (QIAGEN, Valencia, Calif.). Sequencing primers were appropriately spaced (200 to 400 bp) to maximize coverage for sequencing, ensuring at least threefold redundancy for each DNA strand. Automated DNA sequencing was performed using an ABI Prism Avent 3100 genetic analyzer. Sequencing reactions of gel-purified PCR products were performed using Big Dye Terminator, version 3.0, cycle sequencing ready reaction mixture (Applied BioSystems) according to the manufacturer's instructions. Primary DNA sequence assembly and analysis were performed using Sequencher (Gene Codes Corp., Ann Arbor, Mich.), and the sequences were compared with those of VZV strains Dumas and P-Oka to identify single-nucleotide polymorphisms.
Sequence analysis.
The complete sequences were readily aligned manually thanks to the high level of conservation among all VZV genomes. The tandem repeat elements (R1 to R5) as well as hypervariable regions (i.e., loci with variable numbers of insertions or deletions of single nucleotides) were excluded prior to phylogenetic analyses due to the uncertainty regarding the phylogenetic informative degree of those regions. Phylogenetic analyses were completed using algorithms included in the Phylip package (10) and the SimPlot program (24). Owing to the limited number of complete VZV genomic sequences available, the following assumptions were made. (i) P-Oka was regarded as the representative consensus sequence for all strains of genotype J. (ii) The consensus sequence of BC, Dumas, and MSP was considered to represent the consensus sequence for all strains of genotype E. (iii) The genomic sequence for strain 123 served as the representative consensus sequence for all M1 genotype strains. (iv) The genomic sequence for strain DR was assumed to represent the consensus sequence for all M2 genotype strains. Since no outgroup sequences were included in the analyses, all phylogenetic trees generated for the study are unrooted.
Phylogenetic algorithms and methods.
All phylogenetic analyses (except for the bootscan method) were performed by the maximum likelihood method (DNAML) included in the Phylip 3.62 package (10). For simplicity, the faster neighbor-joining algorithm was selected for the boostcan method (included in the SimPlot program). To investigate whether the tree topologies were robust regarding the use of different algorithms, the maximum parsimony method (DNAPARS) was used in all analyses in parallel and a Bayesian inference algorithm (MrBayes 3.1) (23, 36) was applied on the complete genome alignment. Bootstrap replicates of the sequence alignments were constructed by using SEQBOOT. To decide a suitable evolutionary model and parameters, the complete genome alignment of all included strains was investigated by using MrModeltest 2.2 (32). The program suggested the use of the GTR+I+G (general time-reversible model using invariant sites and gamma-distributed rates at variable sites) model. The parameters were set as proposed. The simpler model, Jukes-Cantor, was applied in parallel to validate the robustness of the tree topologies regarding the choice of evolutionary model.
Phylogenetic analysis of the complete genome.
Although strains belonging to the M1 and M2 genotypes have previously been suggested as possible recombinants (25), phylogenetic analyses including all seven strains were performed on the complete genome without fragmentation. Unrooted trees were created based on 100 bootstrap replicates of the alignment.
Analysis of phylogenetically informative sites.
The four genotypes E, J, M1, and M2 can be ordered in unrooted bifurcating trees with three different topologies. In addition to the phylogenetic analysis described above, analyses of all single phylogenetically informative sites in the alignment of the consensus sequences representing E, J, M1, and M2 were performed. Each informative site, localized using the SimPlot program, supports one of the three alternative tree topologies. We investigated whether any topology was uniformly supported by a majority of the informative sites throughout the genome or if blocks of sequence were present in the genome supporting different topologies that served as evidence for recombination.
Segmentation analysis.
To investigate whether viruses classified as the M1 and M2 genotypes were derived from the E and J strains through recombination events, strains 123 and DR were separately evaluated segment by segment. Since the genome is highly conserved, with few informative sites, the SimPlot program was unable to produce reliable recombination analyses. However, using the bootscan method it was possible to locate putative recombination sites and evaluate them manually. Each predicted segment was analyzed together with the corresponding segments from the E and J strains using the maximum likelihood algorithm. In all likelihood, if M1 and M2 strains represent the culmination of recombination activity, they would have descended from different recombination events since these strains are positioned on different branches in phylogenetic trees (25). Although analyses that included both strains 123 and DR were performed for purposes of comparison, strains 123 and DR were analyzed separately. Thus, strain DR was excluded from the analyses when strain 123 was under evaluation and vice versa. Regions lacking recombination sites were excluded from the analyses. Finally, all segments with a similar phylogenetic topology were concatenated to larger segments and further reanalyzed using the maximum parsimony algorithm applied on 100 bootstrap replicates. The maximum likelihood and neighbor-joining algorithms were applied in parallel for comparison.
Nucleotide sequence accession numbers.
The complete nucleotide sequences for both 123 and DR are being deposited in GenBank (accession numbers not yet assigned).
RESULTS
Sequence data.
The VZV genome is composed of the unique long and unique short regions and consists of approximately 125,000 nucleotides. The complete genome was sequenced for strains VZV 123 and VZV DR. The sequences were compared with the published genomic sequences for the P-Oka (J), V-Oka (J), BC (E), Dumas (E), and MSP (E) strains. All seven complete genomes were comparable in total length, and all of the open reading frames described for the reference strain Dumas were present in each of the analyzed strains. The VZV genome is so highly conserved that the most distantly related genotypes examined in this study (J and M1) differed by only ∼0.2%. In a comparison between the European strain Dumas, the Japanese strain P-Oka and the mixed strain 123, 62% of all intragenic nucleotide substitutions were silent.
Phylogenetic analysis of the complete genome.
Based on analyses of selected genomic regions of clinical VZV isolates (25, 30), the VZV selected strains segregated into at least three genotypes. We performed phylogenetic analyses based on the complete genome (Fig. 1A). After aligning and cleaning the sequence data as described above (removal of repeat regions), trees were constructed from 100 bootstrap replicates. The E, J, M1, and M2 genotypes were unambiguously separated from each other, with high bootstrap values (>85%), and the complementary algorithms and evolutionary models were tested in parallel as comparison yielded the same topology and branch lengths.
FIG. 1.
(A) Phylogenetic tree based on the complete VZV genome. Bootstrap values supporting each genotype are shown. (B) Schematic view of phylogenetically informative sites (n = 49) in the VZV genome included in 18 genetic blocks. The topology supported by each informative site, the position of the site, nucleotides for the respective strain, and transversions (TV) are marked to the left. The possible tree topologies (no. 1 to 3) of the four genotypes are illustrated to the right. Nucleotide positions refer to the European strain Dumas.
Although the topology was similar to previously published results (25), the M1 and M2 branches presented here were notably longer relatively to the E and J branches. These data may be an indication that these strains diverged earlier. The genetic similarity between the most closely related genotypes, J and M2, is ∼99.89% (1.15 × 10−3 substitutions per site), while the genetic similarity between the most distantly related groups, J and M1, is ∼99.82% (1.76 × 10−3 substitutions per site). By comparison, the genetic distance between E and J is ∼99.85% (1.47 × 10−3 substitutions per site). The highest genetic divergence between strains of the same genotype was observed for genotype E (Dumas versus BC, ∼99.96% similarity or 0.39 × 10−3 substitutions per site). Previously published analyses of the genomic sequences for P-Oka and V-Oka established the high level of similarity between these viruses; our analysis established a similarity of ∼99.98% (0.15 × 10−3 substitutions per site). Similar topologies were observed regardless of whether the maximum likelihood or the neighbor-joining algorithm was used to conduct the analysis. Each phylogenetically informative site in the sequence alignment was separately compared to the three possible tree topologies of the four genotypes E, J, M1, and M2. Altogether, 49 informative sites were identified in the alignment of E, J, M1, and M2, including 41 transitions and 8 transversions.
The results indicate that the topology obtained from a phylogenetic tree based on the complete genome is not uniformly supported (Fig. 1B). Eighteen genetic blocks were identified in the VZV genome, each supporting one of three possible phylogenetic topologies. The topology described in Fig. 1A, which is represented by tree no. 2 in Fig. 1B, is only supported by 25 informative sites (n = 25) included in a total of eight blocks encompassing one block with eight informative sites, one block with five informative sites, one block with four informative sites, three blocks with two informative sites each, and two blocks with a single informative site each (Fig. 1B). The topology illustrated as tree no. 1 was supported by 11 informative sites (n = 11) included in six blocks: one block with three informative sites, three blocks with two informative sites each, and two blocks with one informative site each. Finally, the topology illustrated as tree no. 3 was supported by 13 informative sites (n = 13) included in four blocks: one block with seven informative sites, one block with four informative sites, and two blocks with one informative site each.
Recombination analysis.
The M genotype VZV strains DR and 123 were previously postulated to represent putative recombinant strains (25). We performed separate fragmentation analyses of DR and 123 to evaluate whether or not a complete genome analysis would support the hypothesis that M1 and M2 genotypes were derived through a series of recombination events between E and J genotype strains.
The sequence data were evaluated for the presence of possible recombination sites predicted by the bootscan method. Genomic segments identified through this approach revealed putative sites where viral recombination events have occurred. We performed phylogenetic analyses on each identified segment through the application of the maximum likelihood algorithm. Results from these analyses show different phylogenetic topologies as well as branch lengths.
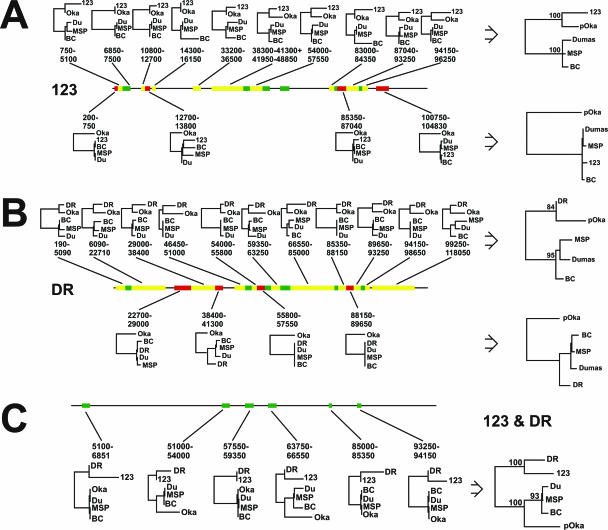
Fifteen distinct segments of interest for further evaluation were identified in our analysis of VZV genomic sequence data in the context of the DR strain (Fig. 2B). Trees based on each segment were constructed separately using the maximum likelihood algorithm. The analyses showed that 11 segments clustered DR with P-Oka and four segments clustered DR with the European strains.
FIG. 2.
(A) Fragmentation analysis of strain 123 (genotype M1). Phylogenetic trees were calculated for each fragment, and the larger trees on top and below are based on 100 bootstrap replicates of a concatenation of all segments supporting similar topologies. Nucleotide positions refer to the European strain Dumas. (B) Fragmentation analysis of strain DR (genotype M2) with phylogenetic trees based on each fragment. The larger trees on top and below are based on 100 bootstrap replicates of a concatenation of all segments supporting similar topologies. Nucleotide positions refer to the European strain Dumas. (C) Fragmentation analysis of the VZV genome with M1 and M2 genotypes included. The tree topology of the six green segments clusters M1 and M2 together and clearly separates them from the E and J genotypes. The larger tree below is based on 100 bootstrap replicates of a concatenation of all six segments. Nucleotide positions refer to the European strain Dumas.
In further support of the robustness of this segmentation pattern, all 11 segments that clustered DR with P-Oka were concatenated to a single large segment. We made the assumption that all segments with similar phylogenetic topology were descended from the same ancestral strain during recombination.
Although concatenation would be expected to yield the same topology, the results based on the extended segments were used to estimate correct branch lengths and information about the bootstrap values. The more extensive concatenated segment was reanalyzed with 100 bootstrap replicates. In addition, the four segments clustering DR with the European strains were also concatenated and reanalyzed. The results showed that the topology was supported by high bootstrap values (Fig. 2B), an observation that provides additional support for the occurrence of recombination events in the derivation of the M2 strain DR.
Fourteen segments were identified in strain 123. Again, each segment was analyzed separately using the maximum likelihood algorithm. As with the DR analysis, topology depended on which part of the genome was under investigation (Fig. 2A). Ten segments clustered 123 with P-Oka, and four segments clustered 123 with the European strains. As with strain DR, these topologies remained stable using high bootstrap values after the segments were concatenated and reanalyzed (Fig. 2A).
In addition to the separate analyses of the DR and 123 strains, we performed an additional analysis that included both DR and 123. Notably, the results revealed that six blocks clustered strains 123 and DR closely together, with wider separation from both the E and J genotypes (Fig. 2C). Analogous to the results obtained from the separate analyses of DR and 123, the topology of the six blocks was also supported when the concatenated sequences were reanalyzed by the maximum likelihood algorithm applied using 100 bootstrap replicates (Fig. 2C).
VZV genotyping.
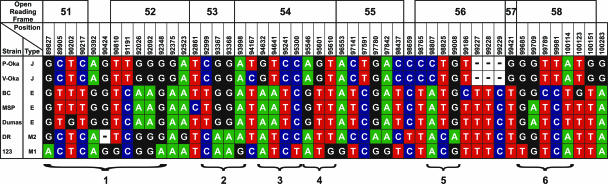
ORF22 has previously been suggested as a suitable target for genotyping of clinical VZV isolates (25). In the present study, we investigated the complete genome in an effort to uncover alternative regions that can be used to distinguish between the four major VZV genotypic groups E, J, M1, and M2. Even though these four genotypes are hitherto the only ones described, we cannot exclude that more genotypes will be revealed as more strains are sequenced, which will require additional genotyping strategies. However, although the genome is highly conserved, several regions with nucleotides specific for the four genotypes are present. The region including ORF51 to -58 contains at least six regions (Fig. 3, no. 1 to 6) that might be utilized for VZV genotyping. One region is located in ORF51 to -52, ORF53 to -54, ORF56, and ORF58, respectively, and two are located in region ORF54. Each region includes single-nucleotide polymorphisms that specifically associate with each of the four genotypes. The flanking sequences of each region have additional sites that are also likely to prove useful for VZV genotypic analysis. In addition, five sites are present in ORF51 to -58 (nt 90202, 92861, 99186, 99709, and 100123) that could be useful for the identification of stable subgroups of genotype E.
FIG. 3.
Genomic variation of a 10,456-nt stretch including ORF51 to -58 of the seven VZV strains included. Nucleotide positions refer to the European strain Dumas. Six genotyping targets are suggested where no. 4, 5, and 6 are all shorter than 430 nt.
DISCUSSION
Although the genomic stability of VZV is well established, limited genetic diversity with an intriguing correlation to geographic origin was recently documented (25, 44). Currently circulating strains of VZV can be categorized into three major genotypes, European (E), Japanese (J), and mosaic (M). Mosaic strains occur most commonly in warmer climates and are characterized by alternating E-like and J-like DNA sequences. We undertook this study to determine whether this pattern could be explained by the occurrence of recombination events. Phylogenetic analyses were performed on the complete genome alignments as well as on shorter segments. The results indicate that VZV strains in circulation comprise at least four genotypes (E, J, M1, and M2). Trees based on the complete genome alignment and supported by high bootstrap values (>99) clearly segregated these genotypes. Recombination analyses of the alignment revealed the genomes of the M1 and M2 strains contain segments that suggest an evolutionary origin from strains E and J strains. This apparent association implies that strains clustering with the M1 and M2 genotypes are recombinants derived from ancestral strains of the E and J genotypes. The M1 and M2 branches in the phylogenetic tree based on the complete genome are longer than those described in an earlier study that evaluated shorter segments of the genome (25). This observation suggests that the M1 and M2 genotypes are likely older than previously indicated and that they have evolved independently since emerging as discrete genotypes. Furthermore, six distinct segments in the alignment include informative sites that cluster the M1 and M2 genotypes into a single group separated from and intermediate between the E and J genotypes (Fig. 2C). The appearance of these segments in strains belonging to the M1 and M2 genotypes could be explained by the introduction of point mutations following the emergence of these genotypes and/or through later recombination events that cluster M1 with M2.
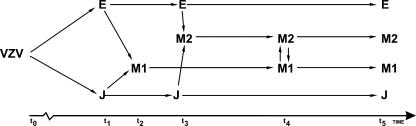
Interpretations of VZV evolution are hampered by the scarcity of point mutations. The limited numbers of fully sequenced isolates and the conservation of the genome complicate both the reconstruction of the evolutionary history and the detection of individual recombination events. Such reconstruction is complex even without these additional obstacles. Nonetheless, we were able to construct a model of the evolution of VZV based on both phylogenetic and recombination analyses (Fig. 4). We propose that the E and J genotypes have evolved from a common ancestor and that subsequent recombination between these genotypes in superinfected persons led to the emergence of at least two mosaic genotypes. Since then, the viruses have continued to evolve both independently (through point mutations) and dependently (through additional recombination events). This model of evolution is supported by the results from the informative site analysis as well as the fragmentation analyses. That said, we cannot exclude that additional hitherto unknown genotypes and recombination events may be involved in the evolutionary history of VZV. Such information could affect the proposed model and could also reveal information about the evolutionary origin of the nonspecified segments in DR and 123 (Fig. 2A and B). Nevertheless, it appears likely that recombination events occurring in dually infected persons have contributed to the evolution of VZV.
FIG. 4.
A suggested evolutionary model based on the results from the phylogenetic and recombination analyses. An ancestral VZV strain (t0) diverged into the E and J strains (t1). E and J are postulated to have recombined at least twice (t2 and t3) to form mosaic recombinant M strains. Following a period of independent evolution, the mosaic strains are postulated to have recombined at least once to establish M1 and M2 (t4). Finally, E, J, M1, and M2 have evolved independently to the present time (t5).
Recombination between different VZV strains has been described in cell culture using restriction endonuclease fragment size analysis and hybridization (9). DNA sequence and single-nucleotide polymorphism analyses have been used to detect probable recombination in several few clinical VZV isolates (30, 44), and homologous recombination has also been documented in other herpesviruses. Norberg et al. (31) recently reported that recombination events are common among clinical isolates of herpes simplex virus type 1.
Evidence for strain recombination has also been observed among clinical isolates of human herpesvirus 8 (34), cytomegalovirus (18), and Epstein-Barr virus isolates (28, 45). As such, homologous recombination appears to be a general feature of human herpesvirus evolution and an important mechanism for maintaining genetic diversity among human herpesvirus strains. An ∼34-kb DNA comparison between V-Oka and the wild-type parental ancestor P-Oka revealed that nucleotide substitutions were preferentially located in ORF62, the major transactivating protein (2). V-Oka vaccine was attenuated in vitro from a wild-type J strain, and vaccine preparations cluster phylogenetically with genotype J. The vaccine is gaining more widespread use globally and has already been introduced broadly into a number of globally distributed populations. V-Oka establishes latency and can reactivate to cause zoster, and superinfection with wild-type VZV strains has been documented frequently among vaccinated individuals (3, 13-15, 30, 37). In addition, a recent population-based study indicated that second cases of wild-type varicella occur more frequently among naturally infected persons than previously appreciated (19). These findings suggest that recombination between wild-type VZV strains and V-Oka strains is possible in both vaccine recipients and, more rarely, between wild-type viruses in persons dually infected with wild-type strains. These new opportunities for VZV recombination should provide an interesting avenue for further study.
We investigated the complete genome for isolates from each of the previously identified VZV genotypes to detect sequence variation that can be used for the development of practical VZV genotyping methods. On the basis of these analyses, we suggest ORF51 to -58 as alternative regions for genotyping of VZV clinical isolates. These regions are relatively variable and include several polymorphic sites that can be used to distinguish between the genotypes E, J, M1, and M2. We also present six examples of shorter subregions that are suitable for genotyping (no. 1 to 6 in Fig. 3). Regions 4, 5, and 6 are all shorter than 430 nucleotides and can therefore easily be amplified by a single PCR. Although the three European isolates included here are classified into the same genotype (E), we cannot exclude the possibility that as yet unidentified subgenotypes exist. At least five sites in ORF51 to -58 should be useful for the classification of variants of genotype E.
The present and previous studies (25, 30, 44) clearly demonstrate limited but informative genetic diversity among globally distributed VZV clinical strains. It is quite likely that both recombination and point mutations, in addition to polymorphic TR regions, have all played important roles in the generation of VZV variability. VZV is essentially ubiquitous, at least in temperate climates, and establishes lifelong latent infection; in addition, VZV is transmissible from cases of zoster, albeit at low efficiency. A relatively small number of individuals in regular global transit could have spread and established relatively few VZV strains in new geographic regions in recent centuries. Analyses of herpesvirus family evolution indicate that the major herpesvirus species emerged in parallel with primate evolution, probably multiple millions of years ago (26, 29). This would explain the intricate balance of power that exists between all human herpesviruses and their hosts; herpesviruses only rarely cause serious human disease, but are able to persistently infect over a human lifespan and thus expand their opportunities for further transmission. VZV may have passed through periodic evolutionary bottlenecks, which may explain the geographic separation of genotypes recently observed (25). To further explore this issue, it may be useful to analyze the evolutionary model proposed here in the context of the migration patterns of modern humans over the past 30,000 years.
In conclusion, our results suggest that the history of the VZV evolution included recombination events. Regardless of the method employed, different phylogenetic topologies were observed that were dependent on the region of the genome being investigated. We suggest that strains 123 and DR consist of genome fragments acquired from the E and J genotypes during homologous recombination. In addition, it is likely that strain 123 has acquired at least six fragments from strain DR during a more recent recombination event or vice versa.
Acknowledgments
This work was supported by grants from the National Vaccine Program office, Centers for Disease Control and Prevention, the Swedish Society of Medicine, and the LUA Foundation at Sahlgrenska University Hospital.
REFERENCES
- 1.Adams, S. G., D. E. Dohner, and L. D. Gelb. 1989. Restriction fragment differences between the genomes of the Oka varicella vaccine virus and American wild-type varicella-zoster virus. J. Med. Virol. 29:38-45. [DOI] [PubMed] [Google Scholar]
- 2.Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. [DOI] [PubMed] [Google Scholar]
- 3.Arvin, A. M. 2001. Varicella vaccine: genesis, efficacy, and attenuation. Virology 284:153-158. [DOI] [PubMed] [Google Scholar]
- 4.Bergstrom, T. 1996. Polymerase chain reaction for diagnosis of varicella zoster virus central nervous system infections without skin manifestations. Scand. J. Infect. Dis. Suppl. 100:41-45. [PubMed] [Google Scholar]
- 5.Bowden, R., H. Sakaoka, P. Donnelly, and R. Ward. 2004. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect. Genet. Evol. 4:115-123. [DOI] [PubMed] [Google Scholar]
- 6.Casey, T. A., W. T. Ruyechan, M. N. Flora, W. Reinhold, S. E. Straus, and J. Hay. 1985. Fine mapping and sequencing of a variable segment in the inverted repeat region of varicella-zoster virus DNA. J. Virol. 54:639-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package), 3.62 ed. Department of Genetics, University of Washington, Seattle.
- 11.Franti, M., J.-T. Aubin, L. Poirel, A. Gautheret-Dejean, D. Candotti, J.-M. Huraux, and H. Agut. 1998. Definition and distribution analysis of glycoprotein B gene alleles of human herpesvirus 7. J. Virol. 72:8725-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuta, Y., F. Ohtani, H. Aizawa, S. Fukuda, H. Kawabata, and T. Bergstrom. 2005. Varicella-zoster virus reactivation is an important cause of acute peripheral facial paralysis in children. Pediatr. Infect. Dis. J. 24:97-101. [DOI] [PubMed] [Google Scholar]
- 13.Galil, K., E. Fair, N. Mountcastle, P. Britz, and J. Seward. 2002. Younger age at vaccination may increase risk of varicella vaccine failure. J. Infect. Dis. 186:102-105. [DOI] [PubMed] [Google Scholar]
- 14.Galil, K., B. Lee, T. Strine, C. Carraher, A. L. Baughman, M. Eaton, J. Montero, and J. Seward. 2002. Outbreak of varicella at a day-care center despite vaccination. N. Engl. J. Med. 347:1909-1915. [DOI] [PubMed] [Google Scholar]
- 15.Gershon, A. A., S. P. Steinberg, and L. Gelb. 1984. Clinical reinfection with varicella-zoster virus. J. Infect. Dis. 149:137-142. [DOI] [PubMed] [Google Scholar]
- 16.Gnann, J. W., Jr. 2002. Varicella-zoster virus: atypical presentations and unusual complications. J. Infect. Dis. 186(Suppl. 1):S91-S98. [DOI] [PubMed] [Google Scholar]
- 17.Grose, C., S. Tyler, G. Peters, J. Hiebert, G. M. Stephens, W. T. Ruyechan, W. Jackson, J. Storlie, and G. A. Tipples. 2004. Complete DNA sequence analyses of the first two varicella-zoster virus glycoprotein E (D150N) mutant viruses found in North America: evolution of genotypes with an accelerated cell spread phenotype. J. Virol. 78:6799-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haberland, M., U. Meyer-König, and F. T. Hufert. 1999. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 80:1495-1500. [DOI] [PubMed] [Google Scholar]
- 19.Hall, S., T. Maupin, J. Seward, A. O. Jumaan, C. Peterson, G. Goldman, L. Mascola, and M. Wharton. 2002. Second varicella infections: are they more common than previously thought? Pediatrics 109:1068-1073. [DOI] [PubMed] [Google Scholar]
- 20.Hawrami, K., D. Harper, and J. Breuer. 1996. Typing of varicella zoster virus by amplification of DNA polymorphisms. J. Virol. Methods 57:169-174. [DOI] [PubMed] [Google Scholar]
- 21.Hondo, R., and Y. Yogo. 1988. Strain variation of R5 direct repeats in the right-hand portion of the long unique segment of varicella-zoster virus DNA. J. Virol. 62:2916-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hondo, R., Y. Yogo, M. Yoshida, A. Fujima, and S. Itoh. 1989. Distribution of varicella-zoster virus strains carrying a PstI-site-less mutation in Japan and DNA change responsible for the mutation. Jpn. J. Exp. Med. 59:233-237. [PubMed] [Google Scholar]
- 23.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 24.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 27.Meng, Y. X., T. J. Spira, G. J. Bhat, C. J. Birch, J. D. Druce, B. R. Edlin, R. Edwards, C. Gunthel, R. Newton, F. R. Stamey, C. Wood, and P. E. Pellett. 1999. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology 261:106-119. [DOI] [PubMed] [Google Scholar]
- 28.Midgley, R. S., N. W. Blake, Q. Y. Yao, D. Croom-Carter, S. T. Cheung, S. F. Leung, A. T. C. Chan, P. J. Johnson, D. Huang, A. B. Rickinson, and S. P. Lee. 2000. Novel intertypic recombinants of Epstein-Barr virus in the Chinese population. J. Virol. 74:1544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montague, M. G., and C. A. Hutchison III. 2000. Gene content phylogeny of herpesviruses. Proc. Natl. Acad. Sci. USA 97:5334-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norberg, P., T. Bergstrom, E. Rekabdar, M. Lindh, and J. A. Liljeqvist. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 78:10755-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nylander, J. A. A. 2004. MrModeltest, v2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 33.Oxman, M. N., M. J. Levin, G. R. Johnson, K. E. Schmader, S. E. Straus, L. D. Gelb, R. D. Arbeit, M. S. Simberkoff, A. A. Gershon, L. E. Davis, A. Weinberg, K. D. Boardman, H. M. Williams, J. H. Zhang, P. N. Peduzzi, C. E. Beisel, V. A. Morrison, J. C. Guatelli, P. A. Brooks, C. A. Kauffman, C. T. Pachucki, K. M. Neuzil, R. F. Betts, P. F. Wright, M. R. Griffin, P. Brunell, N. E. Soto, A. R. Marques, S. K. Keay, R. P. Goodman, D. J. Cotton, J. W. Gnann, Jr., J. Loutit, M. Holodniy, W. A. Keitel, G. E. Crawford, S. S. Yeh, Z. Lobo, J. F. Toney, R. N. Greenberg, P. M. Keller, R. Harbecke, A. R. Hayward, M. R. Irwin, T. C. Kyriakides, C. Y. Chan, I. S. Chan, W. W. Wang, P. W. Annunziato, and J. L. Silber. 2005. A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N. Engl. J. Med. 352:2271-2284. [DOI] [PubMed] [Google Scholar]
- 34.Poole, L. J., J.-C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rekabdar, E., P. Tunbäck, J.-Å. Liljeqvist, M. Lindh, and T. Bergström. 2002. Dichotomy of glycoprotein G gene in herpes simplex virus type 1 isolates. J. Clin. Microbiol. 40:3245-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 37.Sauerbrei, A., E. Rubtcova, P. Wutzler, D. S. Schmid, and V. N. Loparev. 2004. Genetic profile of an Oka varicella vaccine virus variant isolated from an infant with zoster. J. Clin. Microbiol. 42:5604-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepp, D. H., M. E. Match, S. M. Lipson, and R. G. Pergolizzi. 1998. A fifth human cytomegalovirus glycoprotein B genotype. Res. Virol. 149:109-114. [DOI] [PubMed] [Google Scholar]
- 39.Takayama, M., N. Takayama, N. Inoue, and Y. Kameoka. 1996. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J. Clin. Microbiol. 34:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyring, S. K. 1992. Natural history of varicella zoster virus. Semin. Dermatol. 11:211-217. [PubMed] [Google Scholar]
- 41.Umene, K., and H. Sakaoka. 1999. Evolution of herpes simplex virus type 1 under herpesviral evolutionary processes. Arch. Virol. 144:637-656. [DOI] [PubMed] [Google Scholar]
- 42.Umene, K., and H. Sakaoka. 1997. Populations of two eastern countries of Japan and Korea and with a related history share a predominant genotype of herpes simplex virus type 1. Arch. Virol. 142:1953-1961. [DOI] [PubMed] [Google Scholar]
- 43.Umene, K., and M. Yoshida. 1993. Genomic characterization of two predominant genotypes of herpes simplex virus type 1. Arch. Virol. 131:29-46. [DOI] [PubMed] [Google Scholar]
- 44.Wagenaar, T. R., V. T. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 45.Walling, D. M., A. G. Perkins, J. Webster-Cyriaque, L. Resnick, and N. Raab-Traub. 1994. The Epstein-Barr virus EBNA-2 gene in oral hairy leukoplakia: strain variation, genetic recombination, and transcriptional expression. J. Virol. 68:7918-7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimber, U., H. K. Adldinger, G. M. Lenoir, M. Vuillaume, M. V. Knebel-Doeberitz, G. Laux, C. Desgranges, P. Wittmann, U. K. Freese, U. Schneider, et al. 1986. Geographical prevalence of two types of Epstein-Barr virus. Virology 154:56-66. [DOI] [PubMed] [Google Scholar]