Abstract
Varicella-zoster virus (VZV) is a remarkably stable virus that until recently was thought to exhibit near-universal genetic homogeneity among circulating wild-type strains. In recent years, the expanding knowledge of VZV genetics has led to a number of groups proposing sequence-based typing schemes, but no study has yet examined the relationships between VZV genotypes at a full-genome level. A central hypothesis of this study is that VZV has coevolved with humankind. In this study, 11 additional full VZV genomic sequences are presented, bringing the current number of complete genomic sequences publicly available to 18. The full-genome alignment contained strains representing four distinct clades, but the possibility exists that a fifth clade comprised of African and Asian-like isolates was not represented. A consolidated VZV genotyping scheme employing the origin-associated region between reiteration region R4 and open reading frames (ORFs) 63 and 70 is described, one which accurately categorizes strains into one of four clades related to the geographic origin of the isolates. The full-genome alignment also provided evidence for recombination having occurred between the major circulating VZV clades. One Canadian clinical isolate was primarily Asian-like in origin, with most of the genome showing strong sequence identity to the Japanese-like clade B, with the exceptions being two putative recombination regions, located in ORFs 14 to 17 and ORFs 22 to 26, which showed clear similarity to the European/North American clade A. The very low rate of single-nucleotide polymorphisms scattered across the genome made full-genome sequencing the only definitive method for identifying specific VZV recombination events.
Varicella-zoster virus (VZV) is a member of the genus Varicellovirus in the Alphaherpesvirinae subfamily of the Herpesviridae. It is the causative agent of chicken pox (varicella) in children, after which it establishes latency in the sensory ganglia with the potential to reactivate at a later time to cause shingles (zoster). The sequencing of the first complete VZV genome, VZV-Dumas, by Davison and Scott (10) opened the door for genetic analysis of this extremely stable virus. The genome is comprised of ∼125 kb of linear double-stranded DNA containing approximately 71 open reading frames (ORFs). The viral structure is similar to that of other alphaherpesviruses, consisting of two unique regions, unique long and unique short, each flanked by inverted repeats; short repeats termed terminal repeat long and internal repeat long border the unique long region, while larger repeats termed terminal repeat short (TRS) and internal repeat short (IRS) border the unique short region.
A number of factors likely contribute to the overall stability of the VZV genome, one of which is the efficient proofreading activity of the DNA polymerase, which exhibits 3′ to 5′ exonuclease activity as documented in herpes simplex virus type 1 (HSV-1) (2). The synonymous and nonsynonymous mutation rates among the herpesviruses have been estimated at 1 × 10−7 and 2.7 × 10−8 mutations/site/year, respectively, based on the highly conserved gB gene (31). These mutation rates are an estimated 10 to 100 times faster than the equivalent sections of their host genomes but still very low relative to other viral families. As the alphaherpesviruses are so stable genetically with such a low rate of nucleotide substitution, recombination is thought to have played a crucial role in the evolution of different branches of the phylogenetic tree.
Initial attempts to examine genetic variation among isolates primarily used scattered restriction polymorphisms (ORF 38 PstI, ORF 54 BglI, and ORF 62 SmaI) and differences in the reiteration regions in the genome (1, 26, 27, 29, 51). A primary goal of these studies was to distinguish between circulating strains and the attenuated Oka vaccine strain, and rapid methods for differentiation of wild-type and vaccine strains have been developed based on these studies (53). In recent years, the expanding knowledge of VZV genetics has led to a number of groups proposing sequence-based typing schemes. These areas have both been the focus of recent proposals for a common worldwide VZV typing scheme, revealing evidence for geographical segregation of isolates. The development of a robust VZV genotyping scheme is useful from a molecular epidemiology perspective (linking cases/outbreaks within and between geographic regions), and phylogenetic analysis can also provide insights into VZV molecular evolution.
The increase in sequencing data available from geographically diverse isolates has enabled the identification of a large number of single-nucleotide polymorphisms (SNPs) spread across the genome, and strengthened our knowledge of the phylogenetic relationships between circulating VZV isolates. Studies on genetic variation and classification into different subgroups have been described for all other human herpesviruses, including HSV-1 (39) and HSV-2 (45), Epstein-Barr virus (47), cytomegalovirus (6, 7), and human herpesviruses 6 (9), 7 (14), and 8 (34), although none of these studies have included multiple complete genomic sequences of genetically unrelated strains in their analyses.
One such genotyping scheme for VZV was proposed by Faga et al. (13) and Wagenaar et al. (57), which amplified and sequenced 6 genes, gB (ORF 31), gE (ORF 68), gH (ORF 37), gI (ORF 67), gL (ORF 60), and IE62 (ORF 62), encompassing nearly 13 kb of the genome. Strains were then placed in a phylogenetic tree to determine relationships, with 4 major clades identified by this glycoprotein/IE62 scheme. The arbitrarily designated clade A is comprised primarily of European/North American isolates and includes the prototype VZV-Dumas, clade B is an Asian (primarily Japanese) cluster and includes VZV-Oka, clade C is an Asian-like cluster that shares some characteristics with European/North American strains, and clade D is also a European/North American cluster.
Barrett-Muir et al. (3, 4) and Quinlivan et al. (43) used heteroduplex mobility assays to study VZV isolates from the United Kingdom, Africa, Asia, and Brazil to locate a number of informative SNPs across the genome (in ORFs 1, 21, 50, and 54). Using this scattered SNP scheme, strains were assigned a genotype based on shared alleles into one of four major groups. Genotype A1/A2 strains are ubiquitous in Africa, Asia, and the Far East, genotypes B and C (containing VZV-Dumas) are chiefly European but have also been described in North America and Brazil, and genotypes J1/J2 are Japanese isolates (including VZV-Oka) that differ from genotype A strains at 2 SNPs. A third strategy proposed by Loparev et al. (30) involved sequencing a 447-bp portion of ORF 22 and, based on information from a limited number of variable SNPs, assigning strains one of three genotypes, either genotype E (European), genotype J (Japanese), or genotype M (mosaic). Genotype M is comprised of a mixture of E- and J-like alleles, and a number of variants have been observed, allowing further subclassification of strains as M1 or M2.
To our knowledge, no study has examined the relationships between VZV genotypes at a full-genome level; indeed, no study with such breadth of coverage has been carried out for any individual herpesvirus to our knowledge. In this study, we sequenced 11 full VZV genomes, bringing the number of complete genomic sequences publicly available to 18, encompassing each of the distinct VZV clades from Wagenaar et al. (57). With this newfound information, it is the goal of this study to compare and incorporate results of previous genotyping studies and formulate a consolidated VZV genotyping scheme, one which is able to distinguish between isolates from different clades as well as within an individual clade. As an important consequence of this genomic analysis, we also present evidence for recombination having occurred between the major circulating VZV clades, as Canadian strain VZV-8 from clade C contains a mixture of characteristics from European/North American clade A and Japanese clade B. Recombination events have already been demonstrated between various vaccine and wild-type pseudorabies virus (PRV, suid herpesvirus 1) strains replicating in pigs and other animals (16, 21, 22, 24). If recombination events occur during VZV infection in a frequency sufficient that they can be documented by sequencing less than 20 genomes, this observation would challenge current concepts about the inherent stability of the VZV genome. Also, this observation may have implications for immunization strategies with live attenuated human herpesvirus vaccines.
METHODS AND MATERIALS
Virus propagation and genome purification.
The strains used for genome sequencing are described in Table 1. These particular strains were chosen for sequencing to give a representative panel of Canadian and American isolates for use in facilitating identification of important SNPs for genotyping purposes, for instance, within outbreak situations. Isolates were propagated for 6 to 8 passages in cultured human melanoma cells (Mewo strain, ATCC HTB-65) in Dulbecco's modified Eagle's medium (Gibco, Burlington, Ontario, Canada) supplemented with 5% fetal bovine serum, 1% nonessential amino acids, and 1% sodium pyruvate (18). Viral nucleocapsids were isolated from infected Mewo cells exhibiting advanced cytopathic effect, and VZV genomic DNA was extracted using methods described previously (44).
TABLE 1.
Strains used in the complete genome alignment and phylogenetic analysis
| VZV strain | Sourcea | Yr isolated | Reference | Location | Accession no. |
|---|---|---|---|---|---|
| Dumas | V | Late 1970s | 10 | The Netherlands | NC001348 |
| BC | Z | 1999 | 19 | British Columbia, Canada | AY548171 |
| MSP | V | 1995 | 19 | Minnesota | AY548170 |
| SD | Unknown | 1980 | This study | South Dakota | DQ479953 |
| KEL | Z | 2002 | This study | Iowa | DQ479954 |
| 36 | V | 1998 | This study | New Brunswick, Canada | DQ479958 |
| 49 | V | 1999 | This study | New Brunswick, Canada | DQ479959 |
| 32 P5 | V | 1976 | This study | Texas | DQ479961 |
| 32 P22 | V | 1976 | This study | Texas | DQ479962 |
| 32 P72 | V | 1976 | This study | Texas | DQ479963 |
| 11 | Z | 1996 | This study | New Brunswick, Canada | DQ479955 |
| 22 | Z | 1998 | This study | New Brunswick, Canada | DQ479956 |
| 03-500 | Unknown | 2003 | This study | Alberta, Canada | DQ479957 |
| 8 | Z | 1995 | This study | New Brunswick, Canada | DQ479960 |
| pOka | V | 1970 | 17 | Japan | AB097933 |
| vOka | N/A | N/A | 17 | Japan | AB097932 |
| VarilRix | N/A | N/A | 56 | Japanb | DQ008354 |
| VariVax | N/A | N/A | 56 | Japanb | DQ008355 |
V refers to varicella, and Z refers to zoster. N/A, not applicable.
VarilRix and VariVax are attenuated vaccine strains derived from pOka.
DNA sequencing strategies.
Sequencing was performed essentially as previously described for VZV-MSP and VZV-BC (19). In summary, two general approaches were taken. One involved the construction of EcoRI-restricted VZV genomic libraries. PCR primers were designed to produce hybridization probes specific for the different VZV EcoRI fragments and were used to screen the library. In addition, a panel of custom sequencing primers were designed based on the known sequence of VZV-Dumas (10). The other approach involved a traditional shotgun sequencing strategy in which the purified viral DNA was sheared to produce a random distribution of smaller fragments (generally 1 to 3 kb in length). These were then cloned into a plasmid vector (pCR4Blunt-TOPO; Invitrogen Canada, Inc., Burlington, Ontario, Canada) to generate a recombinant library containing representative fragments spanning the entire genome. Random library clones were end sequenced using universal primers flanking the cloning site, and specific PCR products were amplified and sequenced to close gaps and resolve problem areas.
Several of the clinical isolates were found to replicate with very low titers, and as a result, it was difficult to obtain sufficient amounts of pure VZV DNA for the above approaches to be successful without resorting to extremely large scale virus production. In these cases, whole-genome amplification was carried out using the GenomiPhi DNA amplification kit (Amersham Biosciences/GE Healthcare, Baie d'Urfé, Québec, Canada) prior to construction of the shotgun libraries. This amplification approach is based on an isothermal amplification of the starting material using random primers with a strand displacing DNA polymerase and can generate microgram quantities of DNA from nanogram levels of input template. The initial assessment of libraries constructed using DNA amplified in this manner indicated that the terminal regions (approximately 5 to 7 kb from the ends of the genome) as well as segments of the internal repeat and reiteration regions were severely underrepresented. These effects are known problems or deficiencies with this approach. To overcome this, two sets of linkers were utilized, one for EcoRI and one for NgoMIV-SphI, and enzymes were chosen based on their cleavage patterns in the Dumas sequence and intended to either facilitate recovery of terminal sequences or otherwise split up the underrepresented regions into smaller segments that would potentially be amplified more efficiently. These linkers were then ligated onto blunt-ended nucleocapsid DNA. The resulting DNA was digested with the appropriate enzyme, and the fragments were subjected to conditions favoring intramolecular ligation. These circularized fragments, along with undigested nucleocapsid DNA, were used as templates in the GenomiPhi amplification prior to shearing and shotgun cloning.
Prior to sequencing, plasmid DNA was prepared using Wizard SV96 kits (Promega, Madison, Wis.), and sequencing reactions were carried out using either Big Dye v3.1 (Applied Biosystems, Inc., Foster City, CA) or BigDye containing enhancer A (Invitrogen Canada, Inc., Burlington, Ontario, Canada). Sequencing reactions were run on ABI3100 or ABI3730 DNA/genetic analyzers, and the data were assembled using SeqmanII (DNAStar, Madison, WI) or the Staden Package (49). The sequence data were assembled against the reference sequence VZV-Dumas, and all polymorphisms identified were confirmed as authentic by being present in two or more independent/overlapping clones. In cases where regions were identified as being variable within a strain and/or multiple clones were not available, the sequences were amplified, the PCR products were cloned, and several independent clones were sequenced to arrive at a consensus sequence for that region. Because of the nature of the shotgun approach, it was not always possible to specifically identify sequences originating from the inverted repeats as belonging to either the internal or terminal repeat. As a result, the data for this region were assembled en masse. Sufficient sequence data were obtained which spanned into the unique regions flanking both inverted repeats so that the boundaries of the repeat unit could be identified. The sequence for this region was then transposed and inserted into the sequence data to arrive at the complete sequence.
In addition, PCR primers were also designed to generate amplicons spanning the junction of the genome during its replicative phase (ORF 0 to ORF 62). The terminal sequences were further characterized using a technique commonly employed to obtain cDNA ends (rapid amplification of cDNA ends). Nucleocapsid DNA was blunt ended with T4 and Klenow DNA polymerases, a custom adaptor was ligated on, and PCR was conducted using adaptor-specific and VZV-specific primers targeting either end. In all cases where direct sequencing of the PCR products indicated that multiple sequences were present, these amplicons were cloned and several independent clones were subsequently sequenced. All modifying and restriction enzymes used in this study were purchased from New England Biolabs (Pickering, Ontario, Canada).
Sequence analysis.
In addition to the VZV genome sequences derived in the current study, the analysis included the sequences of the prototype strain VZV-Dumas (10), VZV-Oka, parental and vaccine strains (17), VZV-MSP and VZV-BC (19), and the sequences from the commercially produced VZV vaccine strains VarilRix and VariVax (56). The Virus Bioinformatics Resource at http://www.virology.ca (12) was used extensively in the analysis of the data. The annotated genomes were entered into the Viral Orthologous Clusters module to analyze the gene products, while the aligned genomes were analyzed in the Base-By-Base (BBB) module to identify polymorphic sites and amino acid changes. The typing strategy proposed by Wagenaar et al. (subsequently referred to as the glycoprotein/IE62 method) (57), was used to evaluate the phylogenetic relationship of these strains, and the data for strains described previously (13, 57) were included to provide a background population for comparison. Alignments and phylogenetic analyses were conducted using MEGA v3.1 (25) and ClustalW, employing the Kimura two-parameter model with complete deletion of gaps or missing data. All strains were also examined using the typing strategies of Loparev et al. (subsequently referred to as the ORF 22 method) (30) and Barrett-Muir et al. (subsequently referred to as the scattered SNP method) (4), using the descriptive locations identified to assign genotypes. SimPlot (version 2.5; distributed by the author, Stuart C. Ray, Division of Infectious Diseases, John Hopkins University School of Medicine) in conjunction with PHYLIP (Phylogeny Inference Package, version 3.6; distributed by the author, J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle) was used in the study of potential recombination events.
Nucleotide sequence accession numbers.
The genomic sequences for the newly sequenced isolates described by this work were deposited in GenBank under accession numbers DQ479953 to DQ479963.
RESULTS
Genotyping based on full-genome sequencing.
As part of this project, a total of 11 complete genomes were sequenced. In addition, sequences of 7 previously reported genomes were included in the genomic analyses; the 18 fully sequenced VZV isolates included in this study are listed in Table 1. VZV-Dumas and VZV-pOka are considered to be the European and Japanese reference strains, respectively. The isolates vOka, VarilRix, and VariVax are all vaccine derivatives attenuated from pOka that differ in their manufacturers and production methods (42). Three separate passages of strain VZV-32 were sequenced for this study, passages 5, 22, and 72. All other newly sequenced isolates have a Canadian or American origin. Of note, VZV-03-500 was isolated from a post-bone marrow transplant adult patient with disseminated chicken pox-zoster, presenting as hemorrhagic lesions (not the vesicular-fluid-filled lesions that are typical of normal chicken pox); the patient also had pneumonia. All isolates examined in this study were genotyped according to the previously proposed sequence-based typing schemes, with the results of these analyses presented in Table 2.
TABLE 2.
Differentiation of VZV strains based on currently accepted typing schemes
| Strain | Clade or genotype determined by sequence-based typing schemea:
|
||
|---|---|---|---|
| Faga/Wagenaar (glycoprotein/IE62) clade | Loparev (ORF 22) genotype | Barrett-Muir (scattered SNP) genotype | |
| Dumas | A | E | C |
| MSP | A | E | C |
| BC | A | E | C |
| SD | A | E | C |
| KEL | A | E | C |
| 36 | A | E | C |
| 49 | A | E | C |
| 32 P5 | A | E | C |
| 32 P22 | A | E | C |
| 32 P72 | A | E | C |
| 11 | D | E | B |
| 22 | D | E | B |
| 03-500 | D | E | B |
| 8 | C | M (M2) | J1 |
| pOka | B | J | J1 |
| vOka | B | J | J2 |
| VarilRix | B | J | J2 |
| VariVax | B | J | J2 |
The three strains placed in clade D by glycoprotein/IE62 analysis were classified as genotype B by the scattered SNP scheme but were indistinguishable from the Dumas-like strains according their classification as genotype E by ORF 22 analysis. VZV-8 was unique among the isolates examined in that it was placed in clade C by glycoprotein/IE62 typing and assigned genotype M2 by ORF 22 typing; the scattered SNP typing assigned VZV-8 genotype J1, the same classification assigned to pOka. pOka and the three vaccine strains were all classified the same by the glycoprotein/IE62 and ORF 22 methods but differed in their scattered SNP classification as genotype J1 for pOka and genotype J2 for the vaccine derivatives.
The genotypes that have been described by groups previously that do not have any fully sequenced representatives in this study were genotype M1 from the ORF 22 study (30) and genotype A from the scattered SNP method (4). These both appear to be prevalent in Africa and India and may represent a fifth distinct genotype. For convention's sake, the glycoprotein/IE62 designations for genotyping VZV strains have been retained in describing strains herein (57). These were chosen due to the greater amount of sequence examined per isolate and its ability to distinguish between each of the isolates examined, including those in the same clade. All coordinates used herein refer to the equivalent nucleotide in VZV-Dumas.
Phylogeography and clades.
Based on the hypothesis that VZV has coevolved with humankind, clades representative of different geographic regions should be evident (32). In examining the full-genome alignment, some general observations regarding the clades can be made. Clade A isolates included the prototype VZV-Dumas, with other members of the clade displaying a 99.93 to 99.97% identity to Dumas at the nucleotide level, discounting insertions and deletions, which corresponded to 37 to 43 total site differences (not including VZV-32 P72 with 65). Clade B in the full-genome alignment had only pOka as a wild-type isolate, with a 99.83% identity to Dumas at the nucleotide level consisting of 188 total site differences. Clade C was only represented by VZV-8 in the alignment, which may not be fully representative of the clade as a whole; regardless, VZV-8 had 165 total site differences compared to Dumas, showing a 99.85% identity at the nucleotide level. Clade D strains, all Canadian clinical isolates in the alignment, ranged from 134 to 146 total site differences from Dumas for the three strains, showing a 99.87 to 99.88% identity at the nucleotide level. The similarities suggest that clades A and B were the two most divergent clades studied. A more in-depth characterization of sequence variations between the isolates examined in this full-genome alignment is being prepared as a separate study.
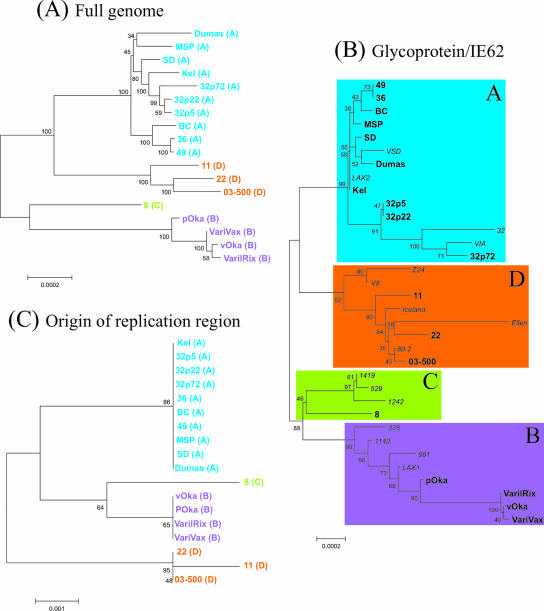
The phylogenetic relationship of all 18 strains based on the full-genome alignment is presented in Fig. 1A. The resulting dendrogram revealed a distinct clustering of the strains as anticipated; however, the placement of VZV-8 in relation to the Japanese isolates was somewhat surprising, as this strain was isolated from a patient in New Brunswick, Canada. The epidemiological information on this isolate was limited to the source being a 73-year-old zoster patient. To determine if this placement was the result of some unique feature found in this isolate, all of the strains were further assessed using the Wagenaar et al. glycoprotein/IE62 scheme (57). As can be seen in Fig. 1B, the clustering of the strains was similar to what was obtained based on the full-genome comparison. Using this typing scheme, the majority of our strains fell within clades A and D, indicative of European/North American origin.
FIG. 1.
Phylogenetic trees of VZV strains based on full-genome sequence (A), aligned sequences of five glycoprotein genes and IE62 (13, 57) (B), and origin of replication region (C). Strains in boldface type are those represented in the full-genome alignment, while strains in italics were taken from previous studies.
Variability in the viral origin of replication.
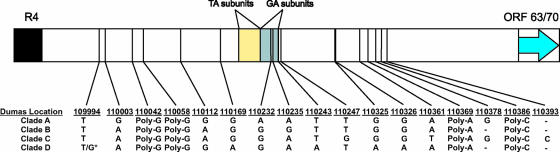
While examining the full-genome alignment, it was noted that the origin-associated region between reiteration R4 and the start codon for ORF 63/70 was variable between the clades, with variability not restricted to the copy numbers of the TA and GA subunits in the ORI. Interestingly, a number of these locations appeared to be clade specific, accurately providing the ability to type strains into distinct clades, as shown in Fig. 1C, and illustrating that the ORI sequences were linked to the ancestral origin of the isolates. Figure 1C displays the resulting dendrogram for this region from all 18 isolates, which is located at IRS coordinates bp 109908 to 110580 and TRS coordinates bp 119317 to 119989.
Only by full-genome sequencing can unpredicted variation be uncovered. At least 17 variable locations have been identified as a result of insertions/deletions and SNPs in the VZV origin-associated region, not including variable copy numbers of the TA and GA subunits in the actual ORI; these variable positions are summarized in Fig. 2. For the newly sequenced isolates, the region was an exact duplicate between the two inverted repeats, but variation existed between the two regions in two sequences, VarilRix and VariVax. It is known that the vaccine preparations are a mixture of distinct genetic subtypes (42, 56). As a result, the sequences reported for the three vaccine strains would likely not fully encompass the genetic variability that would be present in the mixed vaccine batches. Certain portions of the genome have been identified as variable within an isolate, for example, homopolymer stretches and the GA/TA subunits within the ORI, but little information is available on potential sequence differences between the IRS/TRS inverted repeats. The sequencing strategies used in this study did not allow for the distinction of sequences covering the inverted repeats as originating from IRS or TRS.
FIG. 2.
Variable locations within the VZV origin-associated region located between reiteration region R4 and ORF 63/70. As there was noted variability between the copy numbers of TA and GA subunits within the origin of replication between virus sequences within a clade, the differences in copy numbers are not shown. Dumas Location refers to the coordinates of the features in the VZV-Dumas IRS copy of the origin of replication; these locations are duplicated in TRS. The nucleotide at each of these variable locations is listed below the coordinates for the four clades examined. Poly-G, Poly-A, and Poly-C refer to homopolymer stretches that have displayed variability in length both between and within clades. A dash (—) indicates that the clade does not have a base at this location due to an insertion or deletion event. The bases listed for clade C are based on the sequence of VZV-8 only. *, VZV-11 from clade D was the only strain reported to contain a G at location 109994.
SNPs contained within clades.
Earlier analyses of 6 individual VZV genes signaled the value of SNP analysis (13). The full-genome sequencing allowed us to enumerate the number of coding, silent, and intergenic SNPs found to be specific to all members of an individual clade, based on preliminary indications from the full-genome alignment (Table 3). To be included, a polymorphism had to be present in all strains representing an individual clade and not present in any isolates representing other clades. Some putative SNPs from the full-genome alignment were discounted due to their presence or absence from strains not included in the alignment but present in GenBank. The 170 sites identified included duplications due to inverted repeats and were a fraction of the over 700 polymorphic sites identified by the full-genome alignment in conjunction with analysis of other VZV sequences deposited in GenBank. As only one fully sequenced clade C isolate existed, private alleles for C generally referred to locations where VZV-8 differed from all other strains, with the exception being the 6 genes from the three Singapore strains previously described (57). A similar situation existed for clade B, which contained four isolates in the full-genome alignment, but three of these were vaccine strains derived from pOka, the only currently sequenced wild-type clade B strain.
TABLE 3.
Clade-specific SNPs based on the full-genome alignmenta
| Clade | No. of SNPs:
|
|||
|---|---|---|---|---|
| Total | Coding | Silent | Intergenic | |
| A | 19 | 4 | 10 | 5 |
| B | 54 | 21 | 27 | 6 |
| C (VZV-8 only) | 47 | 11 | 29 | 7 |
| D | 50 | 11 | 23 | 16 |
| Total | 170 | 47 | 89 | 34 |
The full list of the variable positions summarized in Table 3 is included in the supplemental material.
SNPs that are specific to all isolates from one particular clade were not the only SNPs with use in genotyping studies. Polymorphic locations contained within isolates from an individual clade were useful in distinguishing between strains within an individual clade as well as distinguishing between certain clades (A/D versus B/C, etc.). In addition to the 19 SNPs found in all clade A strains that are summarized in Table 3, this study has also identified 28 SNPs shared by at least two clade A isolates. These results were interesting when the geographical origins of the strains were considered. The three Canadian clade A isolates sequenced (VZV-36 and VZV-49 from New Brunswick and VZV-BC from British Columbia) have accumulated a number of SNPs, distinguishing them from the American and European clade A isolates examined in this study, in particular, at nucleotides 14345, 17734, 28836, 63448, 99186, 104405, 105264, and 124633. Three strains from the central United States, VZV-32 (Texas), VZV-KEL (Iowa), and VZV-SD (South Dakota), also shared a number of SNPs which distinguished them from other clade A isolates; in particular, at nucleotides 51920, 53523, and 102403. All other shared SNPs in clade A appear to be related to the ancestral origins of the strains, with the exception of the single shared SNP between VZV-MSP and VZV-BC responsible for the D150N mutation in glycoprotein E. The SNPs summarized above did not include the 72 polymorphic sites that were located in only one clade A strain, although these locations may also have had potential use in further distinguishing between clade A isolates.
In addition to the 50 clade D-specific SNPs described in Table 3, 7 shared SNPs restricted to clade D strains have been located, all shared between VZV-22 and VZV-03-500. The 45 polymorphic sites identified in a single clade D isolate are not summarized but may be of eventual use in further classifying clade D isolates. If a greater number of clade D strains were sequenced, there would undoubtedly be more shared SNPs specific to clade D. As the full-genome alignment contained only one strain classified as clade C (VZV-8) and one wild-type clade B strain (VZV-pOka), there was limited information available on potential SNPs that may be localized to these clades. Further sequencing of isolates from these clades should refine the list of SNPs and allow for better differentiation between subgroups within a clade.
The nucleotide identity between strains from clades A and B suggested that these were the two most divergent clades in this study. In the genomic alignment, all polymorphisms shared by clade A and clade B strains were also shared with either clade C or D isolates. No SNPs were located that were shared by clades A and B alone, confirming that these two groups were the most divergent of the clades examined. Interestingly, the 50 clade D-specific SNPs in the full-genome alignment were clustered in three portions of the genome, with 24 located from ORFs 0 to 22, 16 spread between ORFs 35 to 50, and 10 spread between ORFs 62 to 71 in the inverted repeats. This was a relatively constant rate of approximately 1 SNP/1.4 kb in these particular regions. The intervening regions in which no SNPs specific to clade D have been located (ORFs 23 to 34 and 51 to 61) do contain SNPs shared by all clade D strains, but in all instances, these polymorphisms were shared with clade B and, in some cases, with clade C as well.
Evidence for recombination in strain VZV-8.
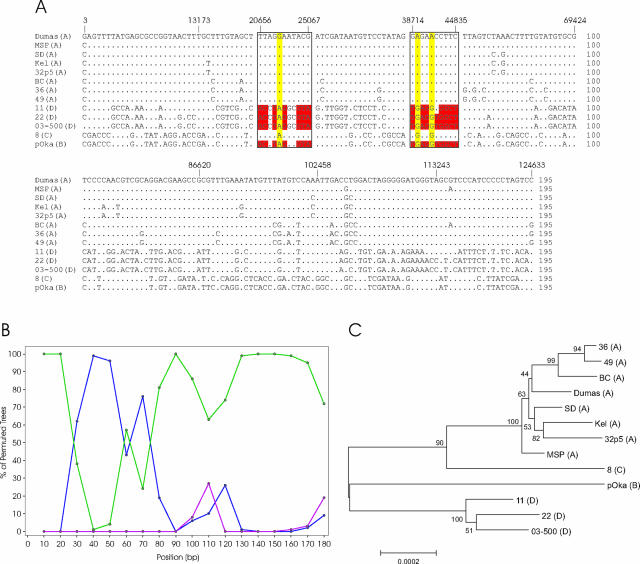
When the dendrogram from the full-genome alignment is examined (Fig. 1A), VZV-8 was found to be a distinct outlier on the branch containing the Japanese isolates. By using the glyocoprotein/IE62 typing scheme, VZV-8 was found to loosely cluster with a group of isolates from Singapore in clade C, which is distinct from clade B harboring the Japanese strains (Fig. 1B). To better understand the factors underlying this divergence, a condensed full-genome alignment in which only the variable sites were represented was created (Fig. 3A). In this alignment, singleton SNPs (polymorphisms occurring in a single isolate) and other variations that were deemed to be uninformative (reiteration regions, homopolymer stretches, and insertions/deletions) were removed.
FIG. 3.
Evidence for recombination in VZV. (A) Alignment of variable sites. The full-genome alignment was condensed to only the variable sites by removing the high-passage isolates derived from a single strain (i.e., 32p22, 32p72, vOka, VarilRix, and VariVax) as well as by removing noninformative variable sites, such as singletons and other strain-specific variations. The Dumas coordinates of the variable sites are indicated above the alignment to provide a relative reference. The boxed regions indicate the segments proposed to be involved in the recombination events and correspond to Dumas coordinates bp 20656 to 25067 for the first segment and bp 38714 to 44835 for the second. Positions in these regions which are conserved between the clade B and D strains are highlighted in red, and the locations in which clade A strains are proposed to have developed mutations after the recombination event are highlighted in yellow. (B) SimPlot analysis of the variable site alignment. Analysis was conducted by grouping the strains based on their phylogenetic designations. Clade A was selected as the query group using a 20-bp window and a 10-bp step value. Similarities to clade D are indicated in green, those to clade C are indicated in blue, and those to clade B are indicated in red. (C) Phylogenetic analysis of the recombination region. The segment of the full-genome alignment between Dumas coordinates bp 13000 to 52000 was assessed using the neighbor-joining algorithm with 500 bootstrap replicates (bootstrap values are indicated).
The polymorphisms responsible for the unique genetic profile of VZV-8 indicated that the majority of the genome showed considerable similarity to the Japanese clade B isolates; however, two regions of the genome, ORFs 14 to 17 and a portion of ORF 22 thru ORF 26, showed greater similarity to the European/North American clade A isolates. This suggested that VZV-8 may be the product of recombination events between members of clade A and clade B. Other than these putative recombination regions, there was only one location in the entire genomic alignment where clade C matched clade A, at bp 88477. This location may be an artifact of previous recombination events, but more likely, it was the result of convergent evolution of SNPs, as it was a silent change in ORF 51 (the DNA origin binding protein).
To test the recombination hypothesis, consensus sequences from clades A and D along with the pOka (clade B) and VZV-8 (clade C) sequences from the modified alignment were compared using the Bootscan function of SimPlot. The software works in conjunction with the phylogenetic analysis suite PHYLIP and plots the results of successive bootstrap analyses on a group of sequences by moving a sliding window across the alignment in incremental steps until the entire alignment has been assessed. SimPlot was developed to study recombination in human immunodeficiency virus (28, 46), and it has been used to study recombination of other viruses (20, 59, 61), including clinical isolates of HSV-1 (39). The SimPlot Bootscan analysis, presented as Fig. 3B, clearly indicated a shift in similarity between the sequences, indicative of a recombination between clades A and C in two regions.
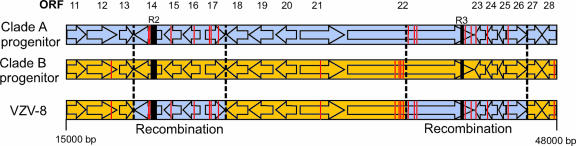
To discount the possibility that editing the alignment had introduced bias into the SimPlot analysis, we performed bootstrap analysis of the unedited alignment covering the region of the putative recombination events (using MEGA3.1). The results, shown in Fig. 3C, also confirm the clustering of VZV-8 with clade A in this region. The region studied, from 13 to 52 kb (ORFs 10 to 29), included a change back to the A/D and B/C homology and three variable reiteration regions (R1, R2, and R3), but the overall influence of the SNPs in this region supported the observations seen with the condensed alignment. The recombination events proposed to have occurred to shape the genetic profile of VZV-8 are presented as Fig. 4.
FIG. 4.
Schematic diagram depicting the recombination events proposed to have shaped the genetic profile of the Canadian clinical isolate VZV-8. Open arrows depict ORFs, and reiteration regions R2 and R3 are depicted by solid black boxes. The superimposed vertical red lines depict locations where VZV-8 shares an SNP with either the clade A or clade B progenitor strain. SNPs which are specific to a single clade, as indicated in Fig. 3A, are not shown.
DISCUSSION
Validity of VZV genotyping analysis.
This full-genome sequence-based typing analysis of VZV is to our knowledge the broadest study of differences between clades/genotypes undertaken in the alphaherpesviruses. Most of the previous sequence-based schemes have focused on a limited number of genes, primarily the glycoprotein genes, IE62, and ORFs 1, 21, 22, 50, and 54; examining previously understudied regions has allowed us to locate a number of clade-specific SNPs that can be used in typing strains. Such sequence-based typing mechanisms are more informative than previous restriction fragment length polymorphism, PCR, single-stranded conformational polymorphism, or heteroduplex-mobility assay-based schemes, although more time-intensive and expensive to carry out. Phylogenetic approaches using a limited number of SNPs across the genome can be misleading, especially when assigning strains subgenotypes within the major clades, i.e., genotypes J1/J2 in the scattered SNP method. Such classifications are sometimes made based on one particular SNP that is shared or differs between strains and may not be indicative of true divergence of the strains. An increased focus on informative SNPs genome-wide should enhance this process.
Each of the previously reported SNP/sequence-oriented typing schemes (4, 13, 30, 57) has its advantages and its drawbacks. The ORF 22 method is unable to distinguish between the European/North American clades A and D, with both classified as genotype E, but is advantageous in ease of use, requiring sequencing of a single 447-bp amplicon to genotype a strain. The scattered SNP method requires sequencing of 4 different ORFs to assign a genotype, which can be somewhat unclear, i.e., VZV-8 and pOka are both classified as genotype J1 but are clearly distinct based on the full-genome alignment, while vOka, VarilRix, and VariVax, clearly related to pOka, are classified as genotype J2. The scattered SNP method is advantageous in that, by studying a large number of geographically separated strains, it is able to identify useful markers for subtyping genotypes A and J in outbreaks. The glycoprotein/IE62 scheme requires the most sequencing of the three, examining 6 genes and nearly 13 kb of the genome. It suffers due to studying a relatively limited number of isolates for comparison between clades, with a focus on laboratory strains. The benefits of this scheme are that, by examining a larger amount of the genome, it is better able to distinguish between closely related isolates within a clade and provides a clearer view of the phylogenetic relationships between strains.
Fifty SNPs specific for clade D have been identified from the full-genome alignment in conjunction with data from previously described strains. It is unclear at this time whether the clustering of clade D SNPs is a result of divergent evolution, recombination between heretofore uncharacterized progenitor strains, or evolutionary artifacts due to selective pressure. Based on our observations, it does not appear as though the intervening regions are less variable on the whole in VZV when the overall polymorphic sites by ORF are examined, though different portions of clade D genomes have likely evolved via different mechanisms, similar to the case in VZV-8.
Variation in the VZV origin of replication.
The variability in the VZV origin of replication was the single most striking observation because of the lack of precedence in HSV sequence analyses. Although others have noted variation in the VZV origin of replication both within and between isolates (17, 19, 50), the extent of the variability observed in the full-genome alignment was unexpected. We have located a number of clade-specific SNPs in the region between R4 and ORFs 63/70 encompassing the ORI which are sufficient to distinguish between isolates from each of the clades studied (Fig. 2). When the phylogenetic tree created using this region (Fig. 3C) is compared to that from the full-genome alignment, it is observed that all strains are classified in a similar manner. With the amount of variation observed between isolates representing the four clades characterized by Wagenaar et al. (57), it is likely that the origin region of any previously uncharacterized genotypes, i.e., African and Indian isolates, would exhibit unique profiles relative to currently classified clades.
An origin-based typing scheme appears to be a useful typing mechanism, but we acknowledge that there are some potential drawbacks to such a method that may limit its ease of use. Aside from the technical issues inherent with amplifying and sequencing a portion of the genome that contains variable homopolymer stretches and a variable number of subunit copies in the actual origin of replication, the region is duplicated in VZV, increasing the potential for variation. At the current time, a limited number of strains have been studied, with the majority of North American isolates classified as clade A. A greater number of clade B and C representative strains would enhance the analysis by refining the number of clade-specific SNPs, along with providing better background data for the construction of phylogenetic trees.
Nevertheless, the cumulative data from this report and earlier genomic analyses described herein have clearly established that there are VZV clades found predominantly in humans of European ancestry (including immigrants to North America) and humans of Asian ancestry. In turn, this observation in more contemporary times extends the hypothesis of McGeoch and coworkers that all mammalian herpesviruses have coevolved with their hosts through geologic times dating back to millions of years ago (31-33). In other words, our data are compatible with the hypothesis that, as humankind migrated out of Africa, not only did humans undergo divergence in Asia and Europe but also their accompanying VZV strains.
Recombination in the alphaherpesviruses.
As the alphaherpesvirus family is so genetically stable with very low rates of nucleotide substitution, recombination may play a crucial role in the evolution of different branches of the phylogenetic tree (32). Interspecific recombinant viruses rarely survive in the natural environment, and the mechanism is inefficient relative to intraspecific recombination (35, 48). However, a natural interspecies recombinant virus between equine herpesvirus 1 (EHV-1) and EHV-4 has recently been reported (40), the first such naturally occurring recombinant described in the alphaherpesviruses. The recombination described was in the ICP4 gene, homologue to IE62. No recombination has been documented in this region of VZV, but it is interesting that one end of the recombination region between EHV-1 and EHV-4 is composed of reiterated 12-bp and 15-bp subunits. This seemingly suggests that reiteration regions can play a role in recombination events between viruses, and the proximity of R2 and R3 to the recombination in VZV-8 may be significant. Interspecific recombinants have been previously generated in vivo between HSV-1 and HSV-2 (60) as well as between bovine herpesvirus 1 (BoHV-1) and BoHV-5 (35).
Intraspecific homologous recombination has been noted both in vitro and in vivo for a number of alphaherpesviruses, including HSV-1 and HSV-2 (23, 38, 55), PRV (16, 21, 22, 24), BoHV-1 (48), and feline herpesvirus 1 (15). In HSV-1, recombination has played an important role in the evolution of genotypes. It has been noted that most full-length genomes consist of a mosaic pattern of fragments from different genotypes, with a number of isolates containing putative recombination points that are located within a gene (39). The BoHV-1 in vivo recombination studies involving coinfection of calves by a natural route of infection have indicated that recombination occurs as frequently in vivo as it does in vitro and that resulting recombinants are persistent and stable after reactivation from latency (48). Putative wild-type intraspecific recombinants have been described for HSV-1 (5, 39), PRV (8), Epstein-Barr virus (36, 58), and human herpesvirus 8 (41), demonstrating the commonality of recombination events of herpesviruses in nature.
Recombination events in the evolutionary history of VZV have likely been underestimated due to the relative genetic stability of worldwide genotypes, and full-genome sequencing discriminating between widely spaced SNPs is the only definitive method of identifying recombinant viruses. The most important prerequisite for recombination to occur is coinfection of a host by the different progenitor strains, with a high degree of genetic similarity between these strains another important factor in homologous recombination (52). The remarkably stable nature of VZV genomes in general suggests that sufficient homology between two strains would not be a hindrance in regard to recombination.
Recombination in VZV is not a new phenomenon. An early study by Dohner et al. (11) noted putative in vitro recombination between viral strains in coinfected cells by mixing two viruses with unique restriction profiles, resulting in hybrid plaque isolates containing neither parental virus in its original form. These hybrids appeared to show differing patterns of recombination, including complete crossover recombination and a partial recombination in which a segment of one parent was inserted into the other. Although the potential recombination regions did not coincide with the regions of VZV-8 associated with recombination, such an event can occur at any point which shows sufficient identity between genomes (52). Additionally, a probable recombinant Brazilian VZV isolate as well as two potential recombinant isolates from the United Kingdom have been described previously (4, 37). The Brazilian genotype A/C recombinant virus matches the profile for genotype C (clade A) throughout the loci studied in ORFs 1 and 21, but at informative sites studied in ORFs 50 and 54, it was found to contain the genotype A (African/Asian clade) markers. It is possible that a single recombination event covering ORFs 50 to 54 has resulted in the mosaic profile observed, but the divergence from its putative progenitor clades may indicate that other factors have played a role in creating its genetic profile.
Recently, the analysis of the complete sequence of herpesvirus papio 2 (54), a simian alphaherpesvirus similar to HSV, has shown evidence of recombination between progenitor viruses. Interestingly, the regions involved in the putative recombination contain genes homologous to those involved in the recombination events described for VZV-8 in this work, consisting of the UL41 to UL44 genes (homologous to VZV ORFs 14 to 17) and a localized portion of the UL36 gene (homologous to VZV ORF 22).
The observations regarding VZV-8 seemingly indicate that there has been at least one recombination event during the evolution of the virus. Such an event is not likely to have been a recent occurrence, as the putative recombination regions have undergone evolution in developing a number of polymorphisms distinguishing them from their progenitor strains. Since VZV-8 shares a number of characteristics with VZV-pOka, it may represent an Asian-like lineage that was involved in recombination with a clade A progenitor that has led to two segments of the genome having a clade A-like profile. Reciprocal (homologous) recombination among precursor viruses during the evolution of the clades is a potential explanation for the observed SNPs. An alternative hypothesis for the SNP clustering could be that certain polymorphisms have arisen independently in a number of different lineages of VZV, but based on the similarity between the different clades and the number of sites involved, this possibility seems unlikely. Further genomic analyses will clarify these issues and likely provide greater insight into the phenomenon of recombination in VZV. If recombination can occur between wild-type isolates, recombination presumably could occur also between wild-type and vaccine strains. Thus, recombination events may need to be considered in strategies to immunize large populations with live attenuated varicella vaccines as well as other herpes vaccines.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the Office of the Chief Scientist, Health Canada. Research by C.G. and C.U. was supported by NIH grant AI22795 and NSERC Strategic Grant STPGP 269665-03, respectively.
We thank the DNA Core Facility, National Microbiology Laboratory, for oligonucleotide synthesis and DNA sequencing, which was invaluable for the completion of this project. We also thank Richard Garceau and Kevin Fonseca for providing strains used in the sequencing and Vasily Tcherepanov with the Virus Bioinformatics Resource (TVBR) at the University of Victoria, Victoria, BC, Canada, for bioinformatics assistance.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. [DOI] [PubMed] [Google Scholar]
- 2.Baker, R. O., and J. D. Hall. 1998. Impaired mismatch extension by a herpes simplex DNA polymerase mutant with an editing nuclease defect. J. Biol. Chem. 273:24075-24082. [DOI] [PubMed] [Google Scholar]
- 3.Barrett-Muir, W., K. Hawrami, J. Clarke, and J. Breuer. 2001. Investigation of varicella-zoster virus variation by heteroduplex mobility assay. Arch. Virol. Suppl. 17:17-25. [DOI] [PubMed] [Google Scholar]
- 4.Barrett-Muir, W., F. T. Scott, P. Aaby, J. John, P. Matondo, Q. L. Chaudhry, M. Siqueira, A. Poulsen, K. Yaminishi, and J. Breuer. 2003. Genetic variation of varicella-zoster virus: evidence for geographical separation of strains. J. Med. Virol. 70:S42-S47. [DOI] [PubMed] [Google Scholar]
- 5.Bowden, R., H. Sakaoka, P. Donnelly, and R. Ward. 2004. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect. Genet. Evol. 4:115-123. [DOI] [PubMed] [Google Scholar]
- 6.Chou, S. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188:388-390. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, L. S., and B. Lomniczi. 1993. High frequency intergenomic recombination of suid herpesvirus 1 (SHV-1, Aujeszky's disease virus). Arch. Virol. 132:37-50. [DOI] [PubMed] [Google Scholar]
- 9.Clark, D. A. 2000. Human herpesvirus 6. Rev. Med. Virol. 10:155-173. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 11.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 12.Esteban, D. J., M. Da Silva, and C. Upton. 2005. New bioinformatics tools for viral genome analyses at Viral Bioinformatics-Canada. Pharmacogenomics. 6:271-280. [DOI] [PubMed] [Google Scholar]
- 13.Faga, B., W. Maury, D. A. Bruckner, and C. Grose. 2001. Identification and mapping of single nucleotide polymorphisms in the varicella-zoster virus genome. Virology 280:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Franti, M., J. T. Aubin, L. Poirel, A. Gautheret-Dejean, D. Candotti, J. M. Huraux, and H. Agut. 1998. Definition and distribution analysis of glycoprotein B gene alleles of human herpesvirus 7. J. Virol. 72:8725-8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, K., K. Maeda, N. Yokoyama, T. Miyazawa, C. Kai, and T. Mikami. 1998. In vitro recombination of feline herpesvirus type 1. Arch. Virol. 143:25-34. [DOI] [PubMed] [Google Scholar]
- 16.Glazenburg, K. L., R. J. Moormann, T. G. Kimman, A. L. Gielkens, and B. P. Peeters. 1994. In vivo recombination of pseudorabies virus strains in mice. Virus Res. 34:115-126. [DOI] [PubMed] [Google Scholar]
- 17.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the Oka varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grose, C., and P. A. Brunel. 1978. Varicella-zoster virus: isolation and propagation in human melanoma cells at 36 and 32 degrees C. Infect. Immun. 19:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grose, C., S. Tyler, G. Peters, J. Hiebert, G. M. Stephens, W. T. Ruyechan, W. Jackson, J. Storlie, and G. A. Tipples. 2004. Complete DNA sequence analyses of the first two varicella-zoster virus glycoprotein E (D150N) mutant viruses found in North America: evolution of genotypes with an accelerated cell spread phenotype. J. Virol. 78:6799-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, M. G., J. R. Smiley, C. Thomas, and L. J. Saif. 2004. Genetic recombination between two genotypes of genogroup III bovine noroviruses (BoNVs) and capsid sequence diversity among BoNVs and Nebraska-like bovine enteric caliciviruses. J. Clin. Microbiol. 42:5214-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, L. M., J. B. Katz, G. A. Erickson, and J. E. Mayfield. 1990. In vivo and in vitro genetic recombination between conventional and gene-deleted vaccine strains of pseudorabies virus. Am. J. Vet. Res. 51:1656-1662. [PubMed] [Google Scholar]
- 22.Henderson, L. M., R. L. Levings, A. J. Davis, and D. R. Sturtz. 1991. Recombination of pseudorabies virus vaccine strains in swine. Am. J. Vet. Res. 52:820-825. [PubMed] [Google Scholar]
- 23.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746-748. [DOI] [PubMed] [Google Scholar]
- 24.Katz, J. B., L. M. Henderson, and G. A. Erickson. 1990. Recombination in vivo of pseudorabies vaccine strains to produce new virus strains. Vaccine 8:286-288. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.LaRussa, P., O. Lungu, I. Hardy, A. Gershon, S. P. Steinberg, and S. Silverstein. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaRussa, P., S. Steinberg, A. Arvin, D. Dwyer, M. Burgess, M. Menegus, K. Rekrut, K. Yamanishi, and A. Gershon. 1998. Polymerase chain reaction and restriction fragment length polymorphism analysis of varicella-zoster virus isolates from the United States and other parts of the world. J. Infect. Dis. 178:S64-S66. [DOI] [PubMed] [Google Scholar]
- 28.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loparev, V. N., T. Argaw, P. R. Krause, M. Takayama, and D. S. Schmid. 2000. Improved identification and differentiation of varicella-zoster virus (VZV) wild-type strains and an attenuated varicella vaccine strain using a VZV open reading frame 62-based PCR. J. Clin. Microbiol. 38:3156-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fickenscher, E. G. Torfason, and D. S. Schmid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., and S. Cook. 1994. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 238:9-22. [DOI] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., and A. J. Davison. 1999. The molecular evolutionary history of the herpesviruses, p. 441-466. In E. Domingo, R. Webster and J. Holland (ed.), Origins and evolution of viruses. Academic Press, London, England.
- 33.McGeoch, D. J., D. Gatherer, and A. Dolan. 2005. On phylogenetic relationships among major lineages of the Gammaherpesvirinae. J. Gen. Virol. 86:307-316. [DOI] [PubMed] [Google Scholar]
- 34.Meng, Y. X., T. J. Spira, G. J. Bhat, C. J. Birch, J. D. Druce, B. R. Edlin, R. Edwards, C. Gunthel, R. Newton, F. R. Stamey, C. Wood, and P. E. Pellett. 1999. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology 261:106-119. [DOI] [PubMed] [Google Scholar]
- 35.Meurens, F., G. M. Keil, B. Muylkens, S. Gogev, F. Schynts, S. Negro, L. Wiggers, and E. Thiry. 2004. Interspecific recombination between two ruminant alphaherpesviruses, bovine herpesviruses 1 and 5. J. Virol. 78:9828-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Midgley, R. S., N. W. Blake, Q. Y. Yao, D. Croom-Carter, S. T. Cheung, S. F. Leung, A. T. Chan, P. J. Johnson, D. Huang, A. B. Rickinson, and S. P. Lee. 2000. Novel intertypic recombinants of epstein-barr virus in the chinese population. J. Virol. 74:1544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 65:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norberg, P., T. Bergstrom, E. Rekabdar, M. Lindh, and J. A. Liljeqvist. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 78:10755-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagamjav, O., T. Sakata, T. Matsumura, T. Yamaguchi, and H. Fukushi. 2005. Natural recombinant between equine herpesviruses 1 and 4 in the ICP4 gene. Microbiol. Immunol. 49:167-179. [DOI] [PubMed] [Google Scholar]
- 41.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlivan, M., A. A. Gershon, S. P. Steinberg, and J. Breuer. 2005. An evaluation of single nucleotide polymorphisms used to differentiate vaccine and wild type strains of varicella-zoster virus. J. Med. Virol. 75:174-180. [DOI] [PubMed] [Google Scholar]
- 43.Quinlivan, M., K. Hawrami, W. Barrett-Muir, P. Aaby, A. Arvin, V. T. Chow, T. J. John, P. Matondo, M. Peiris, A. Poulsen, M. Siqueira, M. Takahashi, Y. Talukder, K. Yamanishi, M. Leedham-Green, F. T. Scott, S. L. Thomas, and J. Breuer. 2002. The molecular epidemiology of varicella-zoster virus: evidence for geographic segregation. J. Infect. Dis. 186:888-894. [DOI] [PubMed] [Google Scholar]
- 44.Safronetz, D., A. Humar, and G. A. Tipples. 2003. Differentiation and quantitation of human herpesviruses 6A, 6B and 7 by real-time PCR. J. Virol. Methods 112:99-105. [DOI] [PubMed] [Google Scholar]
- 45.Sakaoka, H., K. Kurita, T. Gouro, Y. Kumamoto, S. Sawada, M. Ihara, and T. Kawana. 1995. Analysis of genomic polymorphism among herpes simplex virus type 2 isolates from four areas of Japan and three other countries. J. Med. Virol. 45:259-272. [DOI] [PubMed] [Google Scholar]
- 46.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 47.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schynts, F., F. Meurens, B. Detry, A. Vanderplasschen, and E. Thiry. 2003. Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 77:12535-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 50.Stow, N. D., and A. J. Davison. 1986. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J. Gen. Virol. 67:1613-1623. [DOI] [PubMed] [Google Scholar]
- 51.Takada, M., T. Suzutani, I. Yoshida, M. Matoba, and M. Azuma. 1995. Identification of varicella-zoster virus strains by PCR analysis of three repeat elements and a PstI-site-less region. J. Clin. Microbiol. 33:658-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thiry, E., F. Meurens, B. Muylkens, M. McVoy, S. Gogev, J. Thiry, A. Vanderplasschen, A. Epstein, G. Keil, and F. Schynts. 2005. Recombination in alphaherpesviruses. Rev. Med. Virol. 15:89-103. [DOI] [PubMed] [Google Scholar]
- 53.Tipples, G. A., D. Safronetz, and M. Gray. 2003. A real-time PCR assay for the detection of varicella-zoster virus DNA and differentiation of vaccine, wild-type and control strains. J. Virol. Methods 113:113-116. [DOI] [PubMed] [Google Scholar]
- 54.Tyler, S. D., and A. Severini. 2006. The complete genome sequence of herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J. Virol. 80:1214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umene, K. 1985. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 66:2659-2670. [DOI] [PubMed] [Google Scholar]
- 56.Vassilev, V. 2005. Stable and consistent genetic profile of Oka varicella vaccine virus is not linked with appearance of infrequent breakthrough cases postvaccination. J. Clin. Microbiol. 43:5415-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagenaar, T. R., V. T. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 58.Walling, D. M., and N. Raab-Traub. 1994. Epstein-Barr virus intrastrain recombination in oral hairy leukoplakia. J. Virol. 68:7909-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Z., Z. Liu, G. Zeng, S. Wen, Y. Qi, S. Ma, N. V. Naoumov, and J. Hou. 2005. A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J. Gen. Virol. 86:985-990. [DOI] [PubMed] [Google Scholar]
- 60.Yirrell, D. L., C. E. Rogers, W. A. Blyth, and T. J. Hill. 1992. Experimental in vivo generation of intertypic recombinant strains of HSV in the mouse. Arch. Virol. 125:227-238. [DOI] [PubMed] [Google Scholar]
- 61.Zeng, F. Y., C. W. Chan, M. N. Chan, J. D. Chen, K. Y. Chow, C. C. Hon, K. H. Hui, J. Li, V. Y. Li, C. Y. Wang, P. Y. Wang, Y. Guan, B. Zheng, L. L. Poon, K. H. Chan, K. Y. Yuen, J. S. Peiris, and F. C. Leung. 2003. The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39). Exp. Biol. Med. (Maywood) 228:866-873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.