Abstract
Tegument proteins homologous to the essential herpes simplex virus type 1 UL36 gene product (p)UL36 are conserved throughout the Herpesviridae and constitute the largest herpesvirus-encoded proteins. So far, only limited information is available on their functions, which include complex formation with the (p)UL37 homologs via an N-terminal domain and a deubiquitinating activity in the extreme N terminus. For further analysis we constructed deletion mutants lacking 437, 784, 926, 1,046, 1,217, or 1,557 amino acids (aa) from the C terminus. While none of them supported replication of a pseudorabies virus (PrV) UL36 deletion mutant, a mutant polypeptide with an internal deletion from aa 2087 to 2795, which comprises a proline/alanine-rich region, fully complemented the lethal replication defect. Thus, our data indicate that the extreme C terminus of (p)UL36 fulfills an essential role in PrV replication, while a large internal portion of the C-terminal half of the protein is dispensable for replication in cell culture.
Tegument proteins homologous to the UL36 gene product of herpes simplex virus type 1 (HSV-1) are the largest herpesvirus-encoded proteins, ranging in size from 2,241 amino acids (aa) in human cytomegalovirus (5) to 3,534 aa in equine herpesvirus 4 (33). They are conserved throughout the Herpesviridae (25), indicating that they execute important functions for herpesvirus replication. This correlates with the essential character of these proteins, whose deletion, where analyzed, resulted in a complete block in viral replication (6, 8). In stark contrast to these observations, only two functions have so far been assigned to specific regions of the UL36 proteins: interaction with (p)UL37, which is important for secondary envelopment (3, 15, 34), and enzymatic deubiquitinating activity (13, 32), whose importance for viral replication is unclear as yet. The (p)UL37-interacting domain has been localized in pseudorabies virus (PrV) (p)UL36 to an 87-aa region between aa 312 and 398 of the 3,084-aa protein (15). The active enzymatic site is located at a conserved cysteine at position 65 of HSV-1 (p)UL36 (13). The corresponding fragments of the betaherpesvirus murine cytomegalovirus and the gammaherpesvirus Epstein-Barr virus (EBV) also exhibit this activity (32). Thus, both functional regions are located within the N-terminal 400 aa, leaving more than 80% of the proteins without any assigned role. In HSV-1 and PrV (p)UL36 and (p)UL37 are important for virion morphogenesis in the cytoplasm after nuclear egress of DNA-filled capsids (6, 8). In PrV, (p)UL36 is the only tegument protein that has so far been shown to be strictly essential for viral replication (reviewed in reference 26). However, (p)UL36 has to execute functions beyond interacting with (p)UL37. In the absence of PrV (p)UL37, and in the absence of the (p)UL37-interacting domain, viral titers are reduced ca. 100-fold, but productive replication still occurs. In contrast, deletion of (p)UL36 is lethal for PrV (8).
(p)UL36 and (p)UL37 are part of the inner, capsid-proximal portion of the tegument (14, 35); remain associated with the capsid during entry and translocation to the nuclear pore; and are detectable on cytoplasmic capsids before secondary envelopment (10). The presence of the (p)UL36-(p)UL37 proteins on capsids that are located at the nuclear pore suggests that they may interact with nuclear pore proteins to mediate the docking step. Moreover, recent live-cell video microscopy has shown that (p)UL36 and (p)UL37 are required for intracellular transport of capsids during entry and egress, indicating that either or both of the complex partners interact with cellular motor proteins (21, 22). However, no such interactions have been demonstrated as yet, although analysis of temperature-sensitive as well as deletion mutants indicated a role for HSV-1 (p)UL36 in early as well as late steps of viral replication (2, 6, 16).
Construction of C-terminally truncated UL36 proteins.
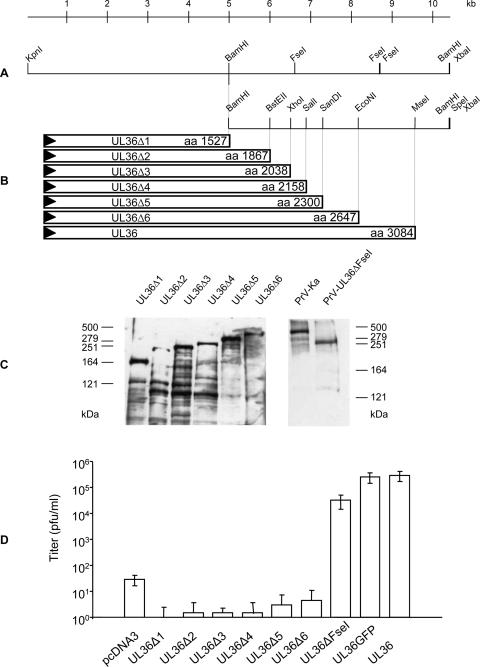
Since in the N-terminal part of (p)UL36 two functional regions had already been identified, we were interested in assaying for functions in the C-terminal part. To this end, the complete PrV UL36 gene from plasmid pUC-UL36 (8) (Fig. 1A) was cloned as a 10.5-kbp KpnI/XbaI fragment into the appropriately cleaved vector pcDNA3 (Invitrogen). Then, C-terminal truncations ending at amino acid position 2647, 2300, 2158, 2038, 1867, or 1527 were engineered using appropriate restriction enzyme sites (Fig. 1A), giving rise to plasmids pcDNA-UL36Δ6 to -UL36Δ1, respectively (Fig. 1B). Since the authentic stop codon of UL36 was also deleted, stop codons provided by the vector were used which are present in all three reading frames, leading to the addition of several non-UL36-specific amino acids. After transfection into rabbit kidney (RK13) cells, expression of truncated proteins was verified by Western blot analysis (Fig. 1C). The apparent molecular masses fitted well with the predicted sizes of the truncated proteins. Expression could also be verified by indirect immunofluorescence using the monospecific anti-UL36-2 serum directed against a glutathione S-transferase-UL36 fusion protein containing aa 624 to 1371 of (p)UL36 (data not shown).
FIG. 1.
Construction and analysis of C-terminally truncated UL36 proteins. (A) Schematic map of pcDNA-UL36 with relevant restriction sites. (B) C-terminal truncations of (p)UL36 were generated using unique restriction enzyme sites as indicated and an SpeI site located in the vector. The amino acid length of the truncated proteins is indicated. (C) Western blot analysis after transfection of RK13 cells with the indicated expression constructs. Purified PrV-Ka virions were tested in parallel for control as were virions of PrV-UL36ΔFseI. On the left and the right the locations of the marker proteins (high-molecular-weight markers; Invitrogen) are indicated in kilodaltons. (D) Progeny virus titers after transfection of the indicated expression constructs and subsequent infection with PrV-ΔUL36GFP. Shown are mean titers of three independent assays. Vertical bars indicate standard deviations.
Functional complementation of PrV-ΔUL36 by the truncated proteins.
To test the truncated UL36 proteins for functional complementation, RK13 cells were transfected either with pcDNA3 as a negative control, pcDNA-UL36 as a positive control, or the respective truncated versions of UL36. Transfections were infected at a multiplicity of infection of 5, 1 day after transfection with phenotypically complemented PrV-ΔUL36GFP, which contains a green fluorescent protein (GFP) marker cassette substituting for the UL36 gene. One day after infection cells and supernatants were harvested and lysed, and the supernatant was titrated on RK13 or RK13-UL36 cells (8). As shown in Fig. 1D, none of the truncated UL36 constructs was able to complement the UL36 defect in PrV-ΔUL36GFP to a significant extent, whereas transfection of the plasmid expressing full-length UL36 led to titers similar to those obtained after infection of RK13-UL36 cells with PrV-ΔUL36GFP. Thus, even the shortest deletion tested (437 aa) abrogated function of the PrV UL36 protein.
Computational sequence analysis of PrV (p)UL36.
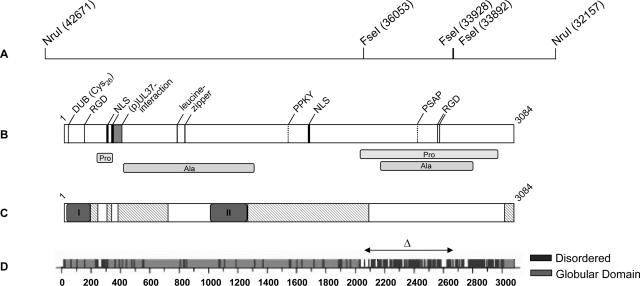
Sequence analysis by PROSITE release 19.24 (12) (http://www.expasy.org/prosite/) and PSORT (27) (http://psort.hgc.jp/) revealed several putative functional amino acid motifs in PrV (p)UL36 (Fig. 2): (i) two leucine zipper motifs at amino acid positions 779 to 800 and 827 to 848; (ii) three RGD motifs at amino acid positions 146 to 148, 2566 to 2568, and 2577 to 2579; and (iii) three nuclear localization signals at amino acid positions 288 to 296, 315 to 321, and 1679 to 1682, as well as (iv) several phosphorylation sites (not shown). Leucine zipper motifs are also predicted for UL36-homologous proteins of members of all three herpesvirus subfamilies except for human herpesvirus 6, human herpesvirus 7, and EBV. RGD motifs could be found only in HSV-2 and varicella-zoster virus (p)UL36 homologs, whereas sequences containing two of the predicted nuclear localization signals [aa 288 to 296 and 1679 to 1682 for PrV (p)UL36] are highly conserved among the alphaherpesvirus subfamily.
FIG. 2.
Computer analysis of PrV (p)UL36 and deletion in (p)UL36ΔFseI. (A) The location of FseI restriction sites used for construction of the internal deletion mutant protein UL36ΔFseI is indicated. Numbers in brackets denote nucleotide positions in the complete genomic sequence of PrV (GenBank accession no. BK001744). (B) Putative amino acid motifs predicted by Prosite release 19.24 (12) (http://www.expasy.org/prosite/) and PSORT II (27) (http://psort.hgc.jp/) are shown (NLS, nuclear localization signal; RGD, Arg-Gly-Asp motif, leucine zipper), and alanine- and proline-rich regions are indicated. Two putative L domain motifs were predicted by ELM (29) (http://elm.eu.org/), PPKY and PSAP. The (p)UL37 interaction domain was identified by yeast two-hybrid screening (15) and is shown as a gray box. The location of the putative active site of the deubiquitinating activity (DUB) is also indicated. (C) Pfam analysis (1) (http://www.sanger.ac.uk/Software/Pfam/) highlighted two highly conserved PfamA domains within homologous UL36 proteins of herpesviruses of all three subfamilies (I = Herpes_teg_N PrV (p)UL36, aa 11 to 178) and of the alphaherpesviruses (II = Herpes_UL36 PrV (p)UL36, aa 1000 to 1251). Besides, there are also matches to PfamB (hatched boxes) that likely indicate a true relationship. (D) Globplot 2.1 (20) (http://globplot.embl.de/) prediction for PrV UL36 protein. Globular (light gray) and disordered (dark gray) regions are displayed (graphic modified from the website). The region removed from (p)UL36ΔFseI (aa 2087 to 2795) by deleting 2.1 kb of the PrV UL36 gene is indicated by the double-headed arrow.
Moreover, there are two large stretches of proline- and alanine-rich sequences (alanine rich, aa 397 to 1293 and 2167 to 2801; proline rich, aa 226 to 299 and 2026 to 2970) within PrV (p)UL36 (Fig. 2B). A putative late domain motif which had been described in retroviral Gag proteins (PT/SAP) was predicted by ELM (29) (http://elm.eu.org/), PSAP at amino acid positions 2419 to 2422, and an additional late domain motif, PPXY (7), could also be found in the PrV (p)UL36 sequence (PPKY, aa 1528 to 1531) (Fig. 2B). Secondary structure prediction using the software GlobPlot 2.1 (20) (http://globplot.embl.de/) revealed a large disordered region in the carboxy-terminal part of PrV (p)UL36 ranging from aa 2025 to 2976 (Fig. 2D). This region presumably contains random coils due to the presence of numerous proline residues (see above) which break α-helical structures.
Sequence alignment of homologous UL36 proteins from all herpesvirus subfamilies showed several regions with highly conserved amino acids mainly within the N-terminal part defined as PfamA (1) domains Herpes_teg_N (I, Fig. 2C), showing high sequence homology for members of all three herpesvirus subfamilies, and Herpes_UL36 (II, Fig. 2C) being conserved within the alphaherpesviruses only. Moreover, there are also matches to PfamB (hatched boxes) which include the C-terminal 62 aa that are highly conserved within the alphaherpesviruses.
Identification of a large nonessential internal region in PrV (p)UL36.
To analyze whether the inability to obtain functional C-terminally truncated (p)UL36 is exclusively associated with the loss of the highly conserved C terminus, and whether the internal unstructured region predicted by GlobPlot may be dispensable for (p)UL36 function, we deleted a 2.1-kb fragment encoding amino acids 2087 to 2795 of (p)UL36 using FseI restriction sites (Fig. 2A). To restore the correct reading frame of the C-terminal 289 aa, an EcoRI linker was inserted which was subsequently cut and blunt ended by Klenow polymerase, resulting in insertion of four additional nonauthentic amino acids. Expression of the truncated (p)UL36ΔFseI was verified by Western blot analysis and immunofluorescence (data not shown). Furthermore, an in-frame UL36-GFP fusion protein was constructed by addition of GFP to the C terminus of PrV UL36, whose correct expression could be verified after transfection microscopically by the observation of green-fluorescing cells and by indirect immunofluorescence using UL36-specific antisera.
In transient-transfection/infection assays, (p)UL36ΔFseI complemented the PrV-ΔUL36 mutant to significant levels, resulting in only approximately 10-fold-lower titers in viral progeny compared to full-length (p)UL36 protein or the (p)UL36-GFP fusion protein (Fig. 1D).
Isolation and characterization of PrV-UL36ΔFseI.
Since (p)UL36ΔFseI complemented the UL36 deletion mutant, marker rescue was performed to transfer the mutation into the viral genome of mutant PrV-ΔUL36F (8). To this end, DNA of PrV-ΔUL36F was cotransfected with pcDNA-UL36ΔFseI into RK13 cells. Infectious progeny was plated onto RK13 cells, and one plaque isolate was further characterized by restriction enzyme analysis and Southern blotting (data not shown).
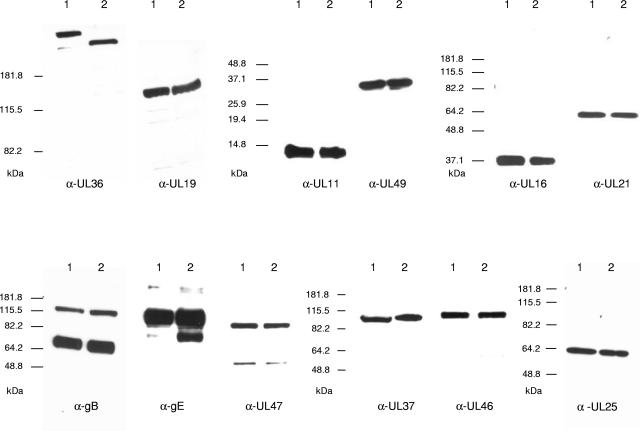
Western blot analysis of PrV-UL36ΔFseI using the anti-UL36-2 serum showed that the truncated protein was incorporated into purified virions (Fig. 3). The size of the detected protein (∼260 kDa) was as expected, and the deletion of the internal 709 aa had no significant effect on the incorporation of other viral components including the interaction partner (p)UL37 as well as other tegument proteins [(p)UL11, (p)UL49, (p)UL16, (p)UL21, (p)UL47, and (p)UL46], capsid or capsid-associated proteins [(p)UL19 or (p)UL25], and envelope glycoproteins [gB and gE] (Fig. 3).
FIG. 3.
Western blot analysis of purified PrV-Ka and PrV-UL36ΔFseI virions. Purified PrV-Ka (lanes 1) and PrV-UL36ΔFseI (lanes 2) virions were separated on sodium dodecyl sulfate-7.5%, -10%, or -15% polyacrylamide gels. Parallel blots were probed with the indicated antisera against PrV tegument proteins (UL36, UL37, UL11, UL16, UL21, UL46, UL47, and UL49), capsid or capsid-associated proteins (UL19 and UL25), or envelope glycoproteins (gB and gE). Locations of molecular mass marker proteins are indicated on the left of each panel.
In one-step growth analysis on RK13 cells, PrV-UL36ΔFseI replicated with only slightly delayed kinetics compared to wild-type PrV-Ka, reaching the same final viral titers (data not shown). Moreover, plaque sizes (88% ± 2% compared to PrV-Ka set as 100%) and penetration kinetics were only marginally affected. In ultrastructural analyses by electron microscopy all stages of herpesvirus maturation were observed in RK13 cells infected with PrV-UL36ΔFseI including unimpeded nuclear egress and secondary envelopment in the cytoplasm (data not shown). Thus, deletion of 709 amino acids from the 3,084-aa PrV (p)UL36 did not significantly impair function of this protein during early and late stages of viral infection.
In summary, we show that deletion of the C terminus of PrV (p)UL36 abrogates (p)UL36 function. The C-terminal 62 aa are highly conserved between (p)UL36 homologs of the alphaherpesviruses and, thus, apparently serve a hitherto-unknown function which is critical for herpesvirus replication. Interestingly, a fusion protein containing GFP linked to the C terminus of PrV (p)UL36 is functional, which indicates that the role of this C-terminal region is not influenced by the downstream addition of the 30-kDa GFP protein.
In contrast, deletion of 709 aa from amino acid positions 2087 to 2795 of the 3,084-aa PrV (p)UL36 proved to retain function of the protein. This large deletion covers a region which is predicted to contain a mostly unstructured randomly coiled domain whose sequence is not conserved in different UL36 homologs but is rich in proline and alanine residues. Corresponding proline-rich regions could also be detected in the UL36 proteins of alphaherpesviruses HSV-1, HSV-2, equine herpesviruses 1 and 4, bovine herpesvirus 1, and Marek's disease virus as well as in the gammaherpesviruses EBV and human herpesvirus 8 but not in varicella-zoster virus, human cytomegalovirus, or human herpesviruses 6 and 7. Thus, our results show that ca. one-quarter of PrV (p)UL36 can be removed without interfering with its essential function. Besides yielding a truncated protein which is significantly better to handle and analyze than the full-length (p)UL36, this also indicates that in the UL36 proteins of herpesviruses large nonessential regions are retained for as-yet-unknown reasons. Apparently, this internal deletion does not impair virion morphogenesis and structure as demonstrated by electron microscopy.
Since the conserved (p)UL37 interaction (3, 15, 34) and deubiquitination (13, 32) domains reside within the N-terminal 400 aa of (p)UL36, the internal deletion was not expected to interfere with these functions. Apparently, it also did not interfere with the role of (p)UL36 and (p)UL37 in intracytoplasmic movement of capsids, since no significant difference in either entry or egress from that of wild-type PrV-Ka could be observed. This indicates that whatever the putative domains for interaction with nuclear pore, cellular motor, or other viral proteins are, they are located outside the deleted region.
Sequence analysis of PrV (p)UL36 revealed putative motifs like leucine zippers which are often found in dimerizing DNA binding proteins (19), RGD motifs able to interact with integrins of the extracellular matrix (30), and nuclear localization signals responsible for targeting proteins to the nuclear pore complex (9). Surprisingly, also two putative late domain sequences (PSAP and PPKY), which hitherto have been described only for RNA viruses, in particular in retroviral Gag proteins, and have been shown to be necessary for efficient budding (7, 23), are also present in PrV (p)UL36. PT/SAP motifs can also be found in HSV-1 and HSV-2 (p)UL36 (C. Crump, personal communication). In addition, another alternative viral L domain sequence motif described as essential for the function of the Gag protein of equine infectious anemia virus with the consensus sequence YXXL (28) was found in PrV, HSV-1, and HSV-2 (p)UL36, located mainly within the N-terminal parts of the proteins. Since removal of two of the three putative RGD motifs (aa 2566 to 2568 and aa 2577 to 2579) as well as the PT/SAP motif PSAP (aa 2419 to 2422) exerts no significant effect on viral replication, PrV (p)UL36 apparently can fulfill its essential function in the absence of these motifs.
So far it is unclear why such a large nonessential region is retained in a protein which exhibits gigantic proportions anyway. Presently, we cannot exclude the possibility that this region plays a role during infection in vivo. Furthermore, using two FseI restriction sites for deletion, a stretch of about 200 amino acids of the described proline-rich region still remains in the PrV UL36ΔFseI protein. A number of proline recognition domains have been described. For example, SH3 (Src homology 3) domains, which are present in many eukaryotic proteins, are involved in signal transduction or mediate protein-protein interactions of cytoskeletal proteins (11, 17, 24, 31). Hence for the remaining C-terminal stretch of nearly 290 amino acids motifs essential for interactions with cellular proteins may still be present. More extensive deletions will demonstrate whether they are indeed functional.
It is also conceivable that deletion of this region has a more subtle influence on viral replication, e.g., modulating the composition of the capsid-associated inner tegument. However, detailed quantitative Western blot and mass spectrometric analysis did not reveal any differences in protein composition between PrV-Ka and PrV-UL36ΔFseI virions (data not shown).
Redundancy in molecular interaction has been demonstrated for numerous tegument proteins, resulting in minor phenotypes after deletion of single tegument components, which, however, can have serious consequences when additional mutations are introduced (4, 18). Thus, there may also be redundancy in the functional role that this nonessential region plays.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/8-1).
We thank Egbert Mundt for help in preparation of the antiserum; Petra Meyer, Kathrin Müller, and Diana Werner for expert technical assistance; and Mandy Jörn for photographic help.
REFERENCES
- 1.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W. D., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, and J. A. Martignetti. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 6.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Görlich, D. 1997. Nuclear protein import. Curr. Opin. Cell Biol. 9:412-419. [DOI] [PubMed] [Google Scholar]
- 10.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, W., and V. Helms. 2005. Dynamical binding of proline-rich peptides to their recognition domains. Biochim. Biophys. Acta Proteins Proteomics 1754:232-238. [DOI] [PubMed] [Google Scholar]
- 12.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. A. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 14.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, C. A., D. Anderson, M. F. Moran, C. Ellis, and T. Pawson. 1991. Sh2 and Sh3 domains—elements that control interactions of cytoplasmic signalling proteins. Science 252:668-674. [DOI] [PubMed] [Google Scholar]
- 18.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper—a hypothetical structure common to a new class of DNA-binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 20.Linding, R., R. B. Russell, V. Neduva, and T. J. Gibson. 2003. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31:3701-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luxton, G. W. G., J. I.-H. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luxton, G. W. G., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer, B. J., and M. J. Eck. 1995. SH3 domains: minding your p's and q's. Curr. Biol. 5:364-367. [DOI] [PubMed] [Google Scholar]
- 25.McGeoch, D. J., F. J. Rixon, and A. J. Davison. 2006. Topics in herpesvirus genomics and evolution. Virus Res. 117:90-104. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 27.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 28.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puntervoll, P., R. Linding, C. Gemund, S. Chabanis-Davidson, M. Mattingsdal, S. Cameron, D. M. A. Martin, G. Ausiello, B. Brannetti, A. Costantini, F. Ferre, V. Maselli, A. Via, G. Cesareni, F. Diella, G. Superti-Furga, L. Wyrwicz, C. Ramu, C. McGuigan, R. Gudavalli, I. Letunic, P. Bork, L. Rychlewski, B. Kuster, M. Helmer-Citterich, W. N. Hunter, R. Aasland, and T. J. Gibson. 2003. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruoslahti, E., and M. D. Pierschbacher. 1986. Arg-Gly-Asp: a versatile cell recognition signal. Cell 44:517-518. [DOI] [PubMed] [Google Scholar]
- 31.Schlessinger, J. 1994. SH2/SH3 signaling proteins. Curr. Opin. Genet. Dev. 4:25-30. [DOI] [PubMed] [Google Scholar]
- 32.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 79:1197-1203. [DOI] [PubMed] [Google Scholar]
- 34.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]