Abstract
Epstein-Barr virus (EBV) is a herpesvirus that infects cells by fusing its lipid envelope with the target cell membrane. The fusion process requires the actions of viral glycoproteins gH, gL, and gB for entry into epithelial cells and additionally requires gp42 for entry into B cells. To further study the roles of these membrane-associated glycoproteins, purified soluble forms of gp42, gH, and gL were expressed that lack the membrane-spanning regions. The soluble gH/gL protein complex binds to soluble gp42 with high affinity, forming a stable heterotrimer with 1:1:1 stoichiometry, and this complex is not formed by an N-terminally truncated variant of gp42. The effects of adding soluble gp42, gH/gL, and gH/gL/gp42 were examined with a virus-free cell-cell fusion assay. The results demonstrate that, in contrast to gp42, membrane fusion does not proceed with secreted gH/gL. The addition of soluble gH/gL does not inhibit or enhance B-cell or epithelial cell fusion when membrane-bound gH/gL, gB, and gp42 are present. However, the soluble gH/gL/gp42 complex does activate membrane fusion with B cells, similarly to soluble gp42, but it does not inhibit fusion with epithelial cells, as observed for gp42 alone. A gp42 peptide, derived from an N-terminal segment involved in gH/gL interactions, binds to soluble gH/gL and inhibits EBV-mediated epithelial cell fusion, mimicking gp42. These observations reveal distinct functional requirements for gH/gL and gp42 complexes in EBV-mediated membrane fusion.
Epstein-Barr virus (EBV) is an extremely prevalent herpesvirus among human populations worldwide, with an estimated 95% of adults infected. The virus is transmitted through saliva, and it can infect epithelial cells, as well as B cells, which provide the host cell latency reservoir (27). EBV-infected individuals carry the virus life long within B cells, and reactivation of the virus can occur intermittently, allowing virus infection of other hosts (1). Although primary infection during childhood is reasonably benign, the first exposure in adolescence or adulthood results in infectious mononucleosis 30 to 50% of the time and recovery typically takes about 1 month (14).
EBV infection has also been associated with several human tumors. EBV has been established to play a role in the etiology of nasopharyngeal carcinoma and endemic Burkitt's lymphoma, with nearly 100% association (34, 38). It is also implicated in Hodgkin's disease, gastric carcinoma, and other cancers (35). Immunodeficient hosts, such as individuals with AIDS or patients undergoing immunosuppressive treatments associated with organ transplantation or cancer therapy, can develop illnesses that are strongly associated with EBV, including oral hairy leukoplakia and lymphoproliferative diseases such as B-cell lymphoma of the central nervous system (36). Thus, understanding and inhibiting the mechanism of EBV entry into cells may have important implications for developing medical therapeutics for a wide range of EBV-related illnesses.
EBV is an enveloped virus that must fuse its own lipid membrane with that of the host cell membrane for infection to occur. Initially, EBV entry into B cells involves binding of the viral gp350/220 protein to cell surface receptor CD21 (also known as complement receptor type 2), resulting in attachment of the virus (4, 21). This interaction enhances infection efficiency but it is not required for membrane penetration and entry (11, 31). In vitro experiments reveal that the minimal requirement for viral fusion with B cells includes EBV envelope glycoproteins gH, gL, gB, and gp42 (7). For infection of B cells, gp42 is known to specifically bind the host cell major histocompatibility complex (MHC) class II proteins to trigger viral-cell membrane fusion (6, 8, 10, 16, 32). Only those MHC class II receptors having a glutamic acid at beta chain residue 46, which includes all HLA-DR and -DP alleles but only some HLA-DQ alleles, can bind gp42 and thereby activate membrane fusion (6, 30). In contrast, for membrane fusion and infection of epithelial cells, gp42 is not required but instead appears to inhibit this process. The EBV gH, gL, and gB proteins are necessary and sufficient for efficient membrane fusion in this case (18).
The mechanistic actions of gH and gL in virus entry are not understood, although the proteins are conserved and serve an essential function among almost all herpesviruses (29). EBV gH/gL exists as a noncovalently associated complex. EBV lacking gH is unable to attach to epithelial cells, suggesting the existence of a specific epithelial cell receptor for gH (19, 22). In addition, soluble gH/gL has been shown to bind to epithelial cells, but the identity of this putative receptor remains elusive (3). EBV gL can be expressed independently of gH, but in order for EBV gH to fold properly and traffic to the cell surface, gL must also be present (17). Both the EBV gL protein and the related varicella-zoster virus gL protein function effectively in mediating the folding and expression of EBV gH protein (15).
Although a crystal structure of the gp42-MHC class II complex has been determined (20), the mechanism of EBV-mediated membrane fusion remains unclear. In particular, it remains to be established which viral protein(s) may actively and efficiently drive membrane merger (20). Since several proteins are involved in membrane fusion for EBV and other herpesviruses, the process may be different from the better-understood class 1 and class 2 viral fusion protein mechanisms, in which trimeric fusion proteins assemble into hairpin-like conformations that bring the viral and cellular membranes together (12). By analogy to the class 1 and class 2 mechanisms, it is likely that EBV-mediated membrane fusion will involve the insertion of one of the membrane-anchored viral proteins (gH, gB, or gp42) into the target cell membrane, with an intermediate structure physically bridging the intermembrane gap before protein refolding, reorganization, or complex formation drives the membrane fusion process.
Since multiple viral membrane-associated proteins are required for the EBV fusion process, any one of them could potentially act in the membrane-merging step by itself, by forming heterologous complexes with each other, or even by engaging cellular receptors. It has previously been demonstrated that soluble gp42-Fc dimers can stimulate gp42-null virus entry into B cells when added to supernatants in an infection assay (37), indicating that the transmembrane anchoring of gp42 is not required for membrane merger. However, for virus entry into epithelial cells, gp42-Fc protein is actually inhibitory (37). Since full-length gp42 and gp42-Fc both associate with gH/gL in immunoprecipitation assays, it seems likely that both effects of gp42-Fc are mediated by its interactions with gH/gL. On the basis of these and other observations, it has been hypothesized that gp42 acts as a tropism switch that controls virus infection of these two different cell types (2). Viruses that predominantly carry gH/gL/gp42 complexes would infect B cells more efficiently, while viruses depleted of gp42 and containing a prevalence of gH/gL complexes would be more likely to infect epithelial cells (37).
In order to better understand the roles of EBV gH, gL, and gp42 in the virus entry mechanism, we have expressed and purified soluble forms of these essential proteins. The secreted proteins assemble into very stable complexes with an apparent stoichiometry of 1:1:1. Deletion of 58 N-terminal residues of gp42 disrupts this interaction of the purified proteins (37). In order to probe the roles of gH/gL and gp42 in membrane fusion, the effects of the soluble proteins on quantitative cell-cell fusion assays were studied. By transfecting various combinations of the expression plasmids for the full-length viral glycoproteins, the ability of the soluble gH/gL, gp42, and gH/gL/gp42 proteins to activate or inhibit membrane fusion could be tested. These experiments demonstrated that soluble, monomeric gp42 acts similarly in the cell-cell fusion assay to the dimeric gp42-Fc fusion protein used in virus infection assays. The gp42-mediated activation of membrane fusion with B cells and the inhibition of membrane fusion with epithelial cells both occurred in the 1- to 10-nM concentration range, suggesting that gp42 acted in both cases through its high-affinity interaction with gH/gL. While soluble gH/gL had no effects on membrane fusion with either cell type over a similar concentration range, the gH/gL/gp42 complex exhibited a unique functional profile. The addition of soluble gH/gL/gp42 to cells expressing membrane-bound gH/gL and gB activated membrane fusion with B cells similarly to soluble gp42. However, in contrast to soluble gp42, the gH/gL/gp42 complex did not significantly inhibit fusion with epithelial cells over a comparable concentration range. Finally, a peptide derived from the N terminus of gp42, comprising a region implicated in binding to gH/gL, was synthesized and was observed to bind to gH/gL but not to gH/gL/gp42 complexes. The gp42-derived peptide showed specific inhibition of membrane fusion with epithelial cells, albeit with significantly reduced affinity compared to intact gp42. These results provide novel insights into the roles of gp42, gH, and gL in EBV entry into B cells and epithelial cells.
MATERIALS AND METHODS
Cells and antibodies.
High Five insect cells (BTI-TN-5B1-4; Invitrogen) were grown in shaker flasks in Excell-405 medium (JRH Biosciences). SF+ insect cells (Protein Sciences) were grown in shaker flasks in HyQ medium (HyClone). Sf9 insect cells (Invitrogen) were grown in shaker flasks in Sf900 medium (Gibco) and used for baculovirus production in 150-cm2 cell culture flasks (Corning) in TNM-FH medium (BD Biosciences).
Mammalian cells were grown in 75-cm2 or 150-cm2 cell culture flasks (Corning) in medium always supplemented with 5% or 10% FetalPlex animal serum complex (Gemini Bio-Products) and 1% penicillin-streptomycin (BioWhittaker). Mammalian epithelial cells were human embryonic kidney 293 cells that were expressing simian virus 40 large T antigen (American Type Culture Collection, Manassas, VA) and modified to stably express T7 RNA polymerase under selection of 100 μg/ml zeocin in Dulbecco modified Eagle medium (BioWhittaker) with 10% FetalPlex, which is known as line 14 as described previously (24). Mammalian B cells were Daudi B lymphocytes that are EBV positive and express HLA class II and CD21 (American Type Culture Collection) and modified to stably express T7 RNA polymerase under selection of G418 (700 μg/ml) in RPMI 1640 medium (BioWhittaker) (28). Chinese hamster ovary cells (CHO-K1) were kindly provided by Nanette Susmarski and were grown in Ham's F-12 medium (BioWhittaker). Trypsin-EDTA (BioWhittaker) or EDTA (1 mM EDTA in phosphate-buffered saline [PBS]) was used to detach adherent cells.
The polyclonal HL-800 antibody, which recognizes gH and gL, was obtained as described elsewhere (24). Monoclonal antibody E1D1, which recognizes the gH/gL complex, was a gift from L. Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport) (33). The Northwestern Monoclonal Antibody Facility generated mouse monoclonal antibodies 3H3, 1G11, and 3E9 against soluble gp42.
Constructs of soluble gH, gL, and gp42.
Soluble forms of the EBV gp42, gH, and gL proteins were made with the baculovirus expression vector system. DNA encoding the ectodomain residues but excluding the N-terminal signal sequence of each protein was cloned into the pBACgus-3 vector following the insect cell gp64 signal sequence to direct protein secretion. For these constructs, the following residues were used: gp42 residues 33 to 223; gH residues 18 to 679; and gL residues 22 to 137 (residue numbers are for the full-length precursor proteins). The gp42 construct had included the vector-derived N-terminal six-His and S tags. Constructs for gL were made both with and without the N-terminal six-His and S tags. Constructs for gH were made both with and without a C-terminal six-His tag. The gp42 and gL tags had an enterokinase cleavage site adjacent to the EBV protein sequence. The constructs for gH and gL were subsequently cloned into the same pBACgus4x-1 expression vector, which can accommodate up to four different genes for simultaneous expression from a single recombinant baculovirus. The p10 and polyhedrin promoters separately controlled expression of gH and gL, respectively. Baculovirus stocks were grown and amplified in Sf9 insect cells as monolayer in 150-cm2 T flasks (Corning) with serum-containing TNM-FH medium for 7 to 10 days at each amplification generation.
Protein expression.
Expression of gp42 was performed as previously described (20). Briefly, High Five insect cells in shaker flasks were infected with gp42-encoding baculovirus and cell supernatant was harvested 48 to 72 h after infection. Expression of gH and gL was performed in SF+ insect cells grown in shaker flasks. Baculovirus was added, and cell supernatants were harvested at 48 to 72 h postinfection. Coexpression of gH, gL, and gp42 was performed by simultaneous infection with baculoviruses for gp42 and gH/gL in High Five cells. Recombinant baculovirus stock volumes were experimentally optimized to maximize the amount of gH/gL/gp42 complex and minimize the amount of uncomplexed protein after purification. Cell supernatants were harvested 48 to 72 h after infection. After centrifugation, all supernatants were sterile filtered and stored at 4°C with 0.02% sodium azide added.
Protein purification.
Expressed six-His-tagged gp42 protein was purified from insect cell supernatants by cobalt metal affinity resin purification. Talon SuperFlow resin (BD Biosciences) was used in batch method according to the manufacturer's protocol. Elution buffer consisted of 50 mM sodium phosphate (pH 7.0), 300 mM NaCl, and 150 mM imidazole. After elution, the protein was exchanged into TBS (25 mM Tris [pH 7.4], 150 mM NaCl, 0.02% sodium azide). The protein could be stored at or below a concentration of 0.1 mg/ml for several weeks at 4°C. Amicon Ultra-4 (Millipore) centrifugation tubes with a 10-kDa molecular mass cutoff were used to concentrate the protein. Gel filtration with Superdex 200 or Superdex 75 HR 10/30 analytical columns (Amersham Biosciences) provided a final purification step.
Expressed gH and gL proteins were isolated from insect cell supernatants by immunoaffinity purification when expressed in the absence of gp42. To generate the affinity purification matrix, 10 mg of purified mouse monoclonal antibody E1D1 was coupled through oxidized carbohydrates to 5 ml of UltraLink Hydrazide resin (Pierce) as instructed by the manufacturer. Proteins were purified from liter volumes of supernatant that were passed through the column by siphoning gravity flow. After washing with PBS, gH/gL was eluted with 0.1 M glycine-HCl (pH 2.5) and neutralized to pH 7 by dripping into one-fifth volume of 10× basic PBS (14.7 mM K2HPO4, 81 mM Na2HPO4, 1.37 M NaCl, 26.8 mM KCl, pH 9). Typical yields of gH/gL protein were 0.5 to 1 mg/liter supernatant. Immunoaffinity column elutions were concentrated with Amicon Ultra apparatuses with a 10-kDa molecular mass cutoff, and buffer was exchanged for PBS through repeated concentration and dilution. The proteins were stored with 0.02% azide at 4°C. With PBS as the running buffer, gel filtration chromatography with a Superdex 200 sizing column was used to further purify the soluble gH/gL.
Coexpressed gH, gL, and gp42 as a soluble three-part complex were purified from insect cell supernatants by Talon SuperFlow resin (BD Biosciences) by the same protocol as gp42 purification and by E1D1 immunoaffinity purification with the same protocol as gH/gL purification (see above).
Peptides.
Synthetic peptide gp42-36-65, corresponding to gp42 residues 36 to 65 (RVAAAAITWVPKPNVEVWPVDPPPPVNFNK), was obtained commercially (EZBioLab) at the high-performance liquid chromatography-purified grade (95% purity). Lyophilized peptide was used without further purification and was reconstituted in 25 mM Tris, pH 7.4 (with or without 150 mM NaCl). Concentration of peptide was measured by absorbance at 280 nm with the theoretical absorbance 0.1% value of 3.4, and this concentration was within 5% of the observed peptide peak area on gel filtration (see below). A control 19-mer peptide (YKTKYLINSARLLETSMVD), which was also obtained commercially (Genemed Synthesis, Inc.), was also high-performance liquid chromatography purified (95% purity). This synthesized peptide was dissolved in deionized water prior to use.
Gel filtration chromatography.
An AKTA Explorer was used to carry out gel filtration chromatography to separate proteins by size. A Superdex 200 column was used in the final purification of gH/gL and gH/gL/gp42, as well as to follow the formation of the heterotrimeric gH/gL/gp42 complex. Protein molecular mass sizing standards (Sigma) were used to calibrate the column as instructed by the manufacturer. A peptide HR 10/30 column (Amersham Biosciences) was used with gp42-36-65 peptide to study its binding to gH/gL. The running buffer was TBS or PBS supplemented with 0.02% azide. Absorbance was monitored at 280 nm. Fractions were collected at 0.5- or 1-ml intervals with a flow rate of 0.5 to 0.7 ml/min.
PAGE.
Both sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and native PAGE were used to demonstrate the purity and identity of the soluble proteins. SDS-PAGE was performed with 12% acrylamide resolving gels typically run at 160 V for 1 h. Native PAGE was carried out with 4 to 20% acrylamide gradient gels (Bio-Rad) that were run for at least 3 h at 100 V. If not used for Western blotting (see below), gels were stained in Coomassie blue dye for at least 1 h and then destained in methanol-acetic acid to visualize protein bands.
Western blotting.
After PAGE, proteins were transferred to nitrocellulose membrane at 80 V for 45 min. Membranes were blocked in 10% nonfat milk in TBS-T (25 mM Tris, 250 mM NaCl [pH 7.4], 0.1% Tween 20) and then incubated at 1:10,000 dilutions of first the primary antibody, followed by secondary antibody coupled to horseradish peroxidase (HRP) for at least 1 h each. Blots were developed by enhanced chemiluminescence detection (Amersham Biosciences) with a 1-min film exposure time. S-tagged proteins were detected in a single step with S protein-HRP (Novagen). His-tagged proteins were identified with anti-His tag rabbit polyclonal antibody (Rockland), followed by anti-rabbit immunoglobulin G (IgG)-HRP (R&D Systems). Soluble gp42 was identified by anti-gp42 mouse monoclonal antibody 3H3, followed by anti-mouse IgG-HRP (R&D Systems). Soluble gH/gL proteins were identified by anti-gH/gL rabbit polyclonal antibody HL-800 (1:2,000 dilution), followed by anti-rabbit IgG-HRP.
Transfection.
CHO-K1 cells were transfected in Opti-MEM I medium (Gibco) by a uniform protocol with Lipofectamine 2000 (Invitrogen). Cells were plated in a six-well format, and after 24 h each well received 5 μl Lipofectamine 2000 and various combinations of expression vectors with the following amounts: 0.5 μg for gH, 0.5 μg for gL, 0.5 μg for gB, 0.8 μg for luciferase, 0.5 μg for green fluorescent protein, and 1.5 μg for the pCAGGS vector control (7, 23). In the case of gp42-transfected CHO cells, green fluorescent protein and pCAGGS plasmids were replaced with 2 μg gp42 vector. The transfection was confirmed by observation of illuminated cells containing green fluorescent protein, as well as by cell enzyme-linked immunosorbent assay (CELISA) detection of gH, gL, and gp42 (see below).
Fusion assay.
CHO-K1 cells were transiently transfected as described above. After 12 h posttransfection, the cells were detached with EDTA and 2.5 × 105 cells/well in 0.5 ml were transferred to a 24-well format for the addition of peptide and/or soluble proteins (in TBS without azide) and subsequently overlaid with 0.5 ml target cells, either Daudi B cells or 293-T epithelial cells. Equal numbers of CHO cells plated and Daudi or 293-T cells overlaid were used in each experiment, and the total volume was 1 ml per well. Both of the target cell types, Daudi and 293-T, used in the fusion assay stably expressed T7 RNA polymerase and were under selection by G418 and zeocin, respectively (24). After a 24-h overlay, cells were washed with PBS and lysed with 100 μl passive lysis buffer (Promega) per well. The luciferase activity was quantified by transferring triplicate 20-μl aliquots of lysed cells to a 96-well opaque plate with clear well bottoms (Wallac), and luminescence was measured on a Perkin-Elmer Victor plate reader immediately after adding 100 μl of luciferase assay reagents (Promega).
CELISA.
The CELISA was performed with CHO cells transfected with the glycoproteins of interest, as described above. Transfected CHO cells were plated in a 96-well format. After 24 h incubation, the medium was removed, cells were washed, and primary antibody was added. Mouse monoclonal antibody E1D1 was used for detection of gH/gL, and mouse monoclonal antibody 3H3 was used for detection of gp42. Cells were fixed and blocked with bovine serum albumin (Sigma). Biotinylated anti-mouse IgG was added as the secondary antibody, followed by streptavidin-HRP. 3,3′,5,5′-Tetramethylbenzidine substrate (Sigma) was added, and quantitative colorimetric measurement was performed at 10-min intervals at 370 nm with a Victor plate reader, achieving maximum signal over background measurement by 30 min. Negative control wells were used that either lacked transfected glycoproteins or separately omitted addition of the primary antibody.
RESULTS
Production and purification of soluble EBV gH and gL proteins.
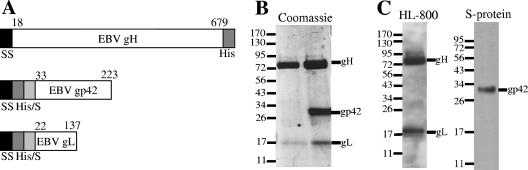
Since EBV gH and gL are complex, disulfide-containing glycoproteins, we pursued the expression of the secreted proteins in insect cells with the baculovirus expression system. In addition, the expression of EBV gH with recombinant baculovirus has been reported previously (26). Infection of cells with recombinant baculovirus for full-length gH alone did not allow proper transport of the protein to the cell surface. However, the coexpression of full-length EBV gH and gL proteins in insect cells did result in expression at the cell surface, and deletion of 27 C-terminal gH residues allowed secretion of gH (25). Following these results, we designed secreted, soluble EBV gH constructs that removed the C-terminal transmembrane domain and cytoplasmic tail residues. Signal sequences of gH, gL, and gp42 were replaced with baculovirus gp64 signal sequence to enhance secretion (Fig. 1A). The regions omitted from the sequences corresponded to the hydrophobic stretches, predicted by hydropathy plots (5) (data not shown).
FIG. 1.
Soluble gH/gL and gH/gL/gp42. (A) Schematic of soluble protein constructs for EBV gp42, EBV gH, and EBV gL. SS is the signal sequence from gp64. His/S represents six-His and S tags. (B) SDS-PAGE showing gel filtration-purified samples of soluble gH/gL purified by E1D1 affinity column (left side) and soluble gH/gL/gp42 purified by Talon resin (right side). The gL protein has no N-terminal tags. (C) SDS-PAGE and Western blotting of the single gel filtration peak containing the purified soluble gH/gL/gp42 complex. Anti-gH/gL antibody HL-800 detects gH and gL (left side), while the tag-specific S protein detects gp42 (right side). The values on the left are molecular sizes in kilodaltons.
Insect cells are well suited to the task of expressing soluble EBV glycoproteins in liters of cells in shaker suspension (20, 25, 26), and the pBACgus4x-1 vector allowed simultaneous expression of both glycoproteins from a single recombinant baculovirus. The soluble gp42 and gH/gL proteins were optimally expressed 48 to 72 h after infection, based on test infections following protein expression by Western blotting of supernatants (data not shown). Initial metal chelate affinity and anti-His tag antibody purification experiments suggested that these approaches were not optimal for the purification of the gH/gL proteins from infected-cell supernatants. For efficient purification of gH/gL, an immunoaffinity column was constructed with milligrams of the anti-gH/gL E1D1 antibody coupled through carbohydrate moieties. The E1D1 column proved very effective at purifying gH/gL from infected-cell supernatants. Concentrated samples of immunoaffinity-purified gH/gL were further purified by Superdex 200 gel filtration chromatography. Final purification was complete, as demonstrated by SDS-PAGE (Fig. 1B, left lane). Observed molecular masses on SDS-PAGE corresponded well to the combination of theoretical amino acid sequences (soluble gH is 77 kDa, soluble gp42 is 26.5 kDa, and soluble gL is 17.6 kDa) and putative N-linked glycosylation (five sites for gH, four sites for gp42, and three sites for gL). Since the molecular mass of gL is much smaller than that of gH, the Coomassie staining would be expected to be less for gL compared to gH, since Coomassie staining is dependent on the overall number of amino acids.
Bipartite soluble gH/gL forms a complex with a 1:1 component ratio.
A single predominant peak containing both the gH and gL proteins was observed by gel filtration chromatography when loading E1D1-purified and concentrated samples (Fig. 2A). Comparison to molecular mass sizing standard proteins revealed that the gH/gL complex migrates with an apparent molecular mass of ∼130 kDa, corresponding to a heterodimeric complex with 1:1 stoichiometry. Samples of gel filtration-purified protein analyzed by SDS-PAGE and Western blotting confirmed the presence of both gH and gL subunits at their appropriate molecular masses and also containing His tags for some of the constructs studied. Furthermore, native gel electrophoresis revealed a single band by Coomassie staining (Fig. 2C) that was also identified as the gH/gL complex by Western blotting with both tag-specific antibodies and gH/gL-specific HL-800 antibody (data not shown).
FIG. 2.
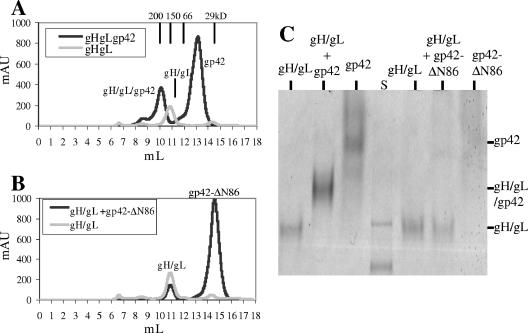
Demonstration of the soluble gH/gL/gp42 complex. (A) Gel filtration traces (overlay) showing the soluble gH/gL/gp42 complex in a 1:1:1 ratio as determined by size. (B) Gel filtration traces (overlay) showing soluble gH/gL unable to bind soluble gp42-ΔN86. For panels A and B, peak positions for sizing standard proteins are 200 kDa at 9.99 ml, 150 kDa at 10.87 ml, 66 kDa at 11.98 ml, and 29 kDa at 14.5 ml. (C) Native PAGE showing the soluble gH/gL/gp42 complex and lack of gH/gL binding to soluble gp42-ΔN86. S is the prestained standard proteins used to guide the gel running time.
Soluble gH/gL binds soluble gp42 with high affinity and forms a complex with a 1:1:1 component ratio.
The formation of a complex containing soluble gH/gL/gp42 proteins could be readily observed by gel filtration chromatography by adding excess purified gp42 to gH/gL (Fig. 2A). The new peak migrates with a retention volume corresponding to an apparent molecular mass of ∼180 kDa, based on the migration of a set of size standards. This molecular mass corresponds to the formation of a 1:1:1 complex of the three proteins, based on the individual mobilities and molecular masses of the gH/gL and gp42 proteins. The gH/gL/gp42 complex was observed to be very stable since reinjections of the isolated 180-kDa peak onto the gel filtration column produced only the expected complex peak again, with no observable dissociation of the complex (data not shown). As a negative control, an N-terminally truncated mutant form of gp42, starting at residue 86 (gp42-ΔN86), was also used in this binding assay. It has previously been shown that truncation of the N-terminal 58 residues of gp42 significantly decreases its interaction with gH/gL, and loss of the N-terminal 90 residues completely abolishes interaction with gH/gL and its ability to function in membrane fusion (37). We therefore anticipated that the purified gp42-ΔN86 protein would not be able to form stable complexes with soluble gH/gL, and indeed the addition of excess gp42-ΔN86 protein to purified gH/gL did not lead to any detectable complex formation (Fig. 2B).
Since the gH/gL/gp42 heterotrimer appeared to be very stable, we also attempted to detect complex formation with a native PAGE assay. Samples of gH/gL and gp42 alone in separate lanes migrated as single bands with distinct mobilities in the native gel. Smearing of gp42 with minimal migration was likely observed because its isoelectric point is predicted to be similar to the running-buffer pH. After mixing of the two components, the appearance of a new band that corresponded to an intermediate migration distance between the gH/gL and gp42 bands could be readily observed (Fig. 2C). Western blotting of this new protein band with anti-gp42 antibody 3H3 and anti-gH/gL antibody HL-800 demonstrated the presence of all three proteins, consistent with the formation of the heterotrimeric complex (data not shown). As a control, samples of gH/gL were mixed with gp42-ΔN86 and these proteins showed no interaction, maintaining the migration for each component independently.
Following the demonstration of the formation of the stable gH/gL/gp42 complex, the coexpression of the proteins in insect cells was explored. Simultaneously infecting Hi5 insect cells with the baculoviruses for gp42 and for gH/gL directly allowed quantitative production of the three-part complex (Fig. 1B, right lane). This complex could be readily purified with either Talon resin or E1D1 immunoaffinity columns and migrated at the appropriate molecular mass of 180 kDa during gel filtration chromatography (data not shown). The single peak on gel filtration contained both gH/gL and gp42, as demonstrated by SDS-PAGE and Western blotting with HL-800 and S protein, respectively (Fig. 1C).
Soluble gp42 enhances B-cell fusion and inhibits epithelial cell fusion in a dose-dependent manner.
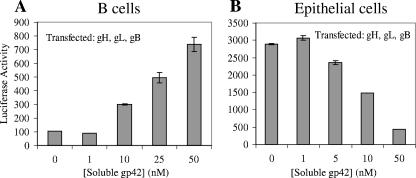
A previously described virus-free cell-cell fusion assay was used to test the functional effects of adding soluble gp42, gH/gL, and gH/gL/gp42 on the membrane fusion process (7). CHO-K1 cells were transiently transfected with combinations of EBV glycoproteins, and after 12 h the purified, soluble proteins were added along with the target cells. After 24 h, luciferase production was measured to determine the quantity of cell fusion. As shown in Fig. 3A, when gH, gL, and gB were transfected into CHO-K1 cells, the addition of increasing concentrations of soluble gp42 caused increased fusion with B cells. This result is consistent with a previous observation that dimeric gp42-Fc protein can activate gp42-null virus infection of B cells (37). Thus, soluble, monomeric gp42 functioned similarly to gp42-Fc and to transfected full-length gp42 to mediate membrane fusion with B cells. Membrane-bound gp42 and Fc stabilization of gp42 dimers are not required for the activation of B-cell fusion (37).
FIG. 3.
Dose-dependent effects of soluble gp42 on membrane fusion. Fusion assay graphs showing concentration-dependent effects of added soluble gp42 on B-cell fusion (A) and epithelial cell fusion (B); CHO cells were transfected with gH, gL, and gB. The data shown are representative examples of at least three independent experiments, and error bars indicate standard deviations.
The cell-cell fusion assay also allowed a quantitative investigation of the effects of added glycoproteins. When 2.5 × 105 effector cells were overlaid with 2.5 × 105 target cells in a 1-ml volume, a threshold for gp42 activation of membrane fusion was observed, where a final concentration of the soluble protein of 1 to 10 nM provided a fusion signal above the background (Fig. 3A). At a final concentration of 50 nM soluble gp42, the resulting luciferase activity was comparable to the levels observed in parallel experiments in which full-length gp42 was expressed by cotransfection with the other glycoprotein expression vectors (data not shown). Soluble gp42-ΔN86 did not trigger fusion with B cells when added to CHO cells transfected with gH, gL, and gB (data not shown). Furthermore, increased levels of fusion (up to ∼150%) were typically observed when soluble gp42 was added to cells already expressing transfected gH, gL, gB, and gp42 in the B-cell fusion assay. Other combinations of transfected glycoproteins (gB alone, gH/gL alone, and gB and gp42) did not demonstrate significant fusion even when soluble gp42 was added (data not shown).
In contrast to this ability of exogenously added gp42 to activate fusion with B cells, the addition of soluble gp42 to CHO cells transfected with gH, gL, and gB inhibited epithelial cell fusion in a dose-dependent manner (Fig. 3B). Inhibition arises in the range of 1 to 10 nM, and higher concentrations of soluble gp42 more significantly inhibited epithelial cell fusion. At the highest concentration tested of 50 nM soluble gp42, epithelial cell fusion is almost completely abrogated, showing a decrease to ∼15% of the uninhibited reaction. CHO cells that were transfected with gH, gL, gB, and gp42 also exhibited a relative decrease (∼50%) in epithelial cell fusion that was comparable to the values observed when ∼10 nM soluble gp42 was added to CHO cells transfected with gH, gL, and gB (data not shown). Regardless of whether gp42 was cotransfected with gH, gL, and gB or not, addition of the higher concentrations of soluble gp42 leads to greater inhibition of fusion with epithelial cells. Soluble gp42-ΔN86 added to an epithelial cell fusion with transfected gH, gL, and gB does not inhibit fusion (data not shown).
Soluble gH/gL cannot mediate fusion and has no discernible effect on membrane fusion with either B or epithelial cells.
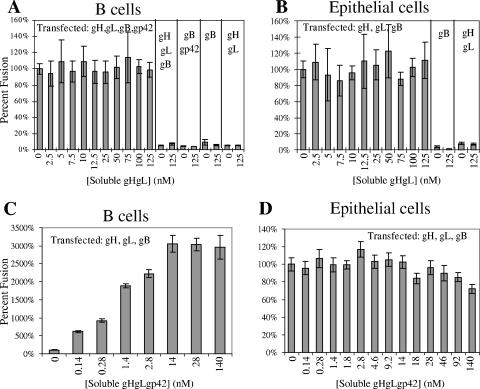
The addition of soluble gH/gL appeared to have no effect on fusion when tested at concentrations of up to 125 nM, either with CHO cells transfected with gH, gL, gB, and gp42 and overlaid with B cells (Fig. 4A) or with CHO cells transfected with gH, gL, and gB and overlaid with epithelial cells (Fig. 4B). Even though insect cell-derived soluble gH/gL bound to epithelial cell surfaces, as determined by fluorescence-activated cell sorter (FACS) experiments with HL-800 antibody (data not shown), and the purified gH/gL complex bound soluble gp42 (Fig. 2A and C), the fusion activity of B cells and epithelial cells remained unchanged by the presence of soluble gH/gL. Unlike soluble gp42, soluble gH/gL did not influence fusion with B or epithelial cells with any combination of transfected gH, gL, gB, and gp42. Importantly, soluble gH/gL at concentrations of up to 125 nM could not trigger membrane fusion when cells were transfected with gB only or with both gB and gp42. Thus, the C-terminal transmembrane region of gH, lacking in the soluble gH construct, appeared essential to the process of membrane fusion with both B cells and epithelial cells. Soluble gH/gL at concentrations of up to 125 nM did not appear to compete with transfected gH/gL and inhibit membrane fusion or alternatively cooperate with membrane-bound gH and enhance membrane fusion, with either B or epithelial cells.
FIG. 4.
Effects of soluble gH/gL and soluble gH/gL/gp42 on membrane fusion. Fusion assay graphs showing concentration-dependent effects of added soluble gH/gL on B-cell fusion (A) and epithelial cell fusion (B) and of added soluble gH/gL/gp42 on B-cell fusion (C) and epithelial cell fusion (D). The data shown are the average of at least three independent experiments, and error bars represent standard deviations. CHO cells were transfected with gH, gL, and gB (B, C, D); transfected gp42 was additionally present in the B-cell fusion with soluble gH/gL (A).
Soluble gH/gL/gp42 complexes act similarly to soluble gp42 to enhance B-cell fusion but show significantly weaker inhibition of epithelial cell fusion.
In the membrane fusion assay with B cells, soluble gH/gL/gp42 functioned very similarly to soluble gp42 in its ability to stimulate membrane fusion in the absence of transfected gp42 (Fig. 4C) and may show a greater effect at lower concentrations. Soluble gH/gL/gp42 also acted similarly to soluble gp42 in enhancing fusion when full-length gp42 was cotransfected with the other EBV glycoproteins (data not shown). It has been shown that N-terminal deletions of gp42 disrupt gH/gL binding, but not MHC binding, and the resulting mutant gp42 proteins are nonfunctional in fusion, suggesting that the simultaneous binding of the same gp42 protein to both MHC and gH/gL is important for fusion activation. In the cell-cell fusion assays, soluble gH/gL was not able to substitute for membrane-bound gH/gL (see above), indicating a requirement for membrane anchoring. Similarly, soluble gH/gL/gp42 complexes do not activate the fusion of gB-expressing cells or gB- and gp42-expressing cells with B cells (data not shown). These data further demonstrate the requirement for membrane-anchored gH/gL in membrane fusion and that preformed gH/gL/gp42 complexes can in some way provide the gp42 for fusion activation. This effect could potentially arise by transfer of the gp42 from the soluble gH/gL complex to the membrane-bound proteins or potentially by the participation of the soluble gH/gL/gp42 complex in activating the membrane-bound proteins. In this regard, it is potentially interesting that the gH/gL/gp42 complex may be a more potent activator of fusion.
In fusion assays with epithelial cells, soluble gH/gL/gp42 behaves quite differently from soluble gp42 and did not inhibit epithelial cell fusion as soluble gp42 does (Fig. 4D). However, a small decrease in epithelial cell fusion is observed at very high concentrations of soluble gH/gL/gp42. This difference in activity is likely due to the fact that gp42 is tightly bound by soluble gH/gL and cannot efficiently saturate the gp42-binding sites of membrane-bound gH/gL to inhibit fusion. This would be consistent with the observation that soluble gp42 can inhibit epithelial cell fusion over a concentration range (nanomolar) similar to that required for activation of B-cell fusion (Fig. 3) and that the binding affinity between gp42 and gH/gL is also within this range. The addition of soluble gH/gL/gp42 to cells transfected with gB only also does not result in fusion with epithelial cells (data not shown). The fusion assay data obtained by adding the soluble proteins to the B-cell and epithelial cell fusion assays are summarized in Table 1.
TABLE 1.
Summary of fusion assay data obtained with B and epithelial cellsa
| Transfected glycoproteins(s) | Native fusion | Soluble gp42 | Soluble gp42-ΔN86 | Soluble gH/gL | Soluble gH/gL/gp42 | Peptide gp42-36-65 |
|---|---|---|---|---|---|---|
| B-cell fusionb | ||||||
| gH, gL, gB, gp42 | High | ↑ | — | — | ↑ | ? |
| gH, gL, gB | None | ↑ | — | — | ↑ | — |
| gH, gL | None | — | NDd | — | — | ND |
| gB, gp42 | None | — | ND | — | — | ND |
| gB | None | — | ND | — | — | ND |
| Epithelial cell fusionc | ||||||
| gH, gL, gB | High | ↓ | — | — | —e | ↓ |
| gH, gL | None | — | ND | — | — | ND |
| gB | None | — | ND | — | — | ND |
Native fusion levels measured as quantified luciferase are listed as above the background level (high) or at the background level (none). The overall fusion trend of adding increasing amounts of soluble proteins or peptide to the fusion assay is indicated as increased fusion (↑), decreased fusion (↓), or no effect on fusion (—). ?, overall trend of fusion change is unclear (see text).
Fusion of Daudi B cells and transfected CHO cells.
Fusion of 293T epithelial cells and CHO cells.
ND, not done.
At a high concentration, a small decrease in fusion was observed.
A peptide corresponding to gp42 residues 36 to 65 binds to soluble gH/gL and is blocked by soluble gp42 binding.
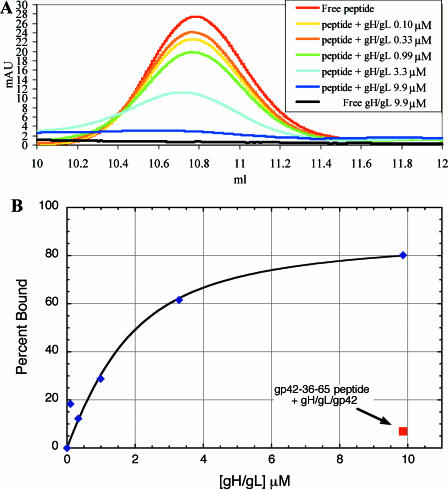
The N-terminal region of gp42 between residues 40 and 58 has been implicated as the binding site for gH/gL (37). In the gp42-HLA-DR1 crystal structure, the N-terminal residues (33 to 85) were disordered, suggesting that these may be in an extended, flexible conformation in the absence of gH/gL binding (20). To investigate this further with the soluble gH/gL and gp42 proteins, a peptide from gp42 residues 36 to 65 was tested for the ability to bind to soluble gH/gL. Gel filtration chromatography was used to monitor the presence of free peptide in binding experiments. In each 200-μl injection, the concentration of peptide was held constant at 7.55 μM while the concentration of soluble gH/gL was varied (0.099 μM to 9.86 μM). The observed free-peptide peak at 10.77 ml in the gel filtration trace decreased systematically as increasing concentrations of soluble gH/gL were added (Fig. 5A) and was almost completely absent when gH/gL was higher in concentration than peptide. The plot of the percentage of peptide bound versus the concentration of gH/gL added yields a hyperbolic curve with ∼50% binding of peptide occurring in the 1- to 3-μM range. When the tripartite gH/gL/gp42 complex is used in analogous binding experiments, very little peptide binding is observed (Fig. 5B), indicating that gp42 competes effectively for peptide binding to gH/gL. The peptide binding affinity appears to be significantly weaker than that of gp42, since gp42 activation of B-cell membrane fusion and inhibition of epithelial cell fusion occurs in the nanomolar concentration range (Fig. 3). In fact, the peptide peak maximum shifts slightly to a larger size (smaller retention volume of <0.1 ml), possibly because of the character of equilibrium binding.
FIG. 5.
Peptide from gp42 residues 36 to 65 binds to soluble gH/gL and competes with soluble gp42 for binding to gH/gL. (A) Gel filtration traces (overlay) on a peptide column monitored at 280 nm showing the free gp42-36-65 peptide peak (10.77 ml) as gH/gL was increasingly titrated from 0.099 μM to 9.86 μM. (B) Percent peptide binding derived from area-under-the-curve analysis of gel filtration experiments; control data (red square) show peptide unable to efficiently bind gH/gL/gp42 at 9.86 μM (6.9% binds). In all experiments, the gp42-36-65 peptide concentration was constant at 7.55 μM.
The gp42-36-65 peptide inhibits epithelial cell fusion in a dose-dependent manner.
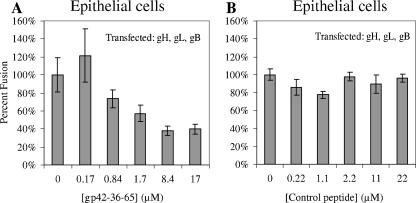
Since the gp42 peptide appears to bind to gH/gL at an overlapping site with soluble gp42, albeit with apparently lower affinity, it was of interest to test its activity in cell-cell fusion assays. Peptide was added to the fusion assay mixture immediately prior to adding any soluble proteins and before overlaying the target cells. Dose-dependent inhibition of fusion was observed when the gp42-derived peptide was used in the epithelial cell fusion assay (Fig. 6A), and no corresponding trend was observed with a control peptide (Fig. 6B). Thus, the peptide appears to act in the same way as soluble gp42, resulting in the inhibition of epithelial cell fusion. However, the peptide does exhibit significantly weaker activity, compared to gp42, consistent with the lower binding affinity estimated from the gel filtration binding experiments (Fig. 5). To reach the same effect of gp42 at a nanomolar concentration, approximately 1,000-fold more peptide (micromolar concentration) is needed. Nevertheless, the fusion assay with epithelial cells reveals the peptide's dose-dependent trend of inhibition. A strong effect of added peptide on B-cell fusion was not observed, most likely because of the higher affinity of the intact gp42 protein for gH/gL. At micromolar concentrations of the peptide, even low nanomolar concentrations of soluble gp42 would be expected to compete effectively for binding to gH/gL, overcoming potential peptide inhibition of B-cell fusion (data not shown). Lower concentrations of soluble gp42 are below the activity range for stimulating B-cell fusion, so it was difficult to observe the effect of peptide in relation to gp42 on B-cell fusion.
FIG. 6.
Effect of peptide from gp42 residues 36 to 65 on membrane fusion. Epithelial cell fusion assay graphs showing that incubation with the gp42-36-65 peptide can inhibit fusion in a dose-dependent manner (A) and a control peptide does not inhibition fusion (B). The data shown are representative examples of at least three independent experiments, and error bars represent standard deviations. The wild-type fusion was normalized to 100%, and CHO cells were transfected with gH, gL, and gB.
DISCUSSION
EBV gH/gL was efficiently purified by immunoaffinity chromatography, and gH/gL/gp42 was efficiently purified by immobilized metal affinity and immunoaffinity chromatography.
The E1D1 antibody was produced and purified in several milligram quantities and coupled via carbohydrates to UltraLink Hydrazide resin (Pierce) to create an E1D1 affinity column. Since IgG is glycosylated primarily on the Fc portion, this coupling strategy should help orient the antibody and lead to a greater percentage of antibody remaining active. The E1D1 immunoaffinity purification approach also precluded the binding and purification of any misfolded gH and/or gL protein, such as disulfide-linked oligomers of gL that are produced when soluble gL is expressed in the absence of gH (data not shown). Purified gH/gL was identified by its expected molecular mass, as demonstrated by SDS-PAGE and gel filtration chromatography, as well as by Western blotting with tag-specific and protein-specific (HL-800) antibodies (Fig. 1C).
The Talon affinity resin (BD Biosciences) was also used to purify the gp42-containing complex. The completeness of purification was quite high, with nearly all of the gp42 extracted from the cell supernatants. In contrast, gH/gL expressed without gp42 was inefficiently purified by Talon resin, which suggests that the six-His tags present on gH and gL were somewhat inaccessible to resin binding or perhaps structurally sequestered by the protein conformation. When gp42 is coexpressed with gH/gL, the tripartite complex is efficiently purified by Talon resin, presumably through the gp42 six-His tag. The gH/gL/gp42 complex can also be isolated by E1D1 immunoaffinity purification with the same protocol as purification of gH/gL. In all of the supernatant samples tested, gp42 was present in excess and all E1D1-purified protein was found by gel filtration to be in the three-part complex. The properties of Talon-purified and E1D1-purified soluble gH/gL/gp42 remained identical, suggesting that both methods were acceptable purification approaches. All three proteins are present in the single gel filtration-purified peak, as demonstrated by SDS-PAGE with Western blotting by tag-specific S protein for gp42 and protein-specific anti-gH/gL antibody HL-800 (Fig. 1C).
Several lines of evidence suggest that the baculovirus-expressed gH/gL and gH/gL/gp42 protein complexes were obtained in a properly folded and functional form.
First, the monoclonal E1D1 antibody recognized the gH/gL and gH/gL complexes, which is thought to require a conformation-specific interaction with the heterodimer (17). Not only did E1D1 provide an efficient way to purify the gH/gL and gH/gL/gp42 protein complexes, but the purification method also established that the captured complex was in its native conformational epitope for E1D1 binding. The E1D1 antibody also detected purified soluble gH/gL binding to CHO cells transfected with gp42 in CELISA experiments, suggesting that low-pH elution does not cause a permanent change in gH/gL for E1D1 binding (data not shown). Second, after elution from the E1D1 affinity column, the soluble gH/gL and gH/gL/gp42 complexes were each observed as a single peak by gel filtration and a single band by native PAGE (Fig. 2A and C). The eluted proteins behaved well in solution and can be concentrated to 10 mg/ml without difficulty, further arguing that there is no major disruption of the protein structure, which is very often linked to the formation of aggregates and reduced solubility. This indicates that short exposure to a low pH can release gH/gL and gH/gL/gp42 from the E1D1 antibody but the proteins remain a stable complex when rapidly returned to a neutral pH.
Third, the soluble gH/gL complex binds gp42 to form a tripartite species that is stable on gel filtration as a single peak and native PAGE as a single band (Fig. 2A and C). This indicates that purified gH/gL maintains the proper binding site for the interaction with gp42. Furthermore, there is no nonfunctional fraction of the low-pH-eluted gH/gL protein, suggesting that the recovered material is homogeneous, at least in terms of gp42 binding activity. Importantly, we have shown that the complexes that are formed by low-pH-eluted gH/gL are indistinguishable from complexes that are formed by the coexpression of gH/gL and gp42 in insect cells and that are subsequently purified by cobalt affinity chromatography by ligand substitution elution with imidazole at pH 7. The observation that affinity-purified gH/gL can quantitatively be converted into gH/gL/gp42 complexes that are similar to those produced by coexpression of the proteins with neutral-pH elution argues against a major disruption of the protein structure due to low-pH elution. Finally, FACS analysis with the anti-gH/gL HL-800 antibody revealed that the soluble gH/gL complex binds to AGS epithelial cell surfaces, presumably through direct binding to a putative gH receptor (data not shown). In addition, FACS experiments demonstrated soluble gH/gL/gp42 binding to Daudi B cells, presumably through gp42 directly binding to MHC class II receptor, and all three proteins were detected by probing with anti-His antibody, polyclonal anti-gp42 antibody PB1114, and anti-gH/gL antibody HL-800 (data not shown).
The tripartite gH/gL/gp42 complex has a component protein ratio of 1:1:1.
Gel filtration was used to investigate the predominant oligomeric species of gH/gL/gp42. Comparison with molecular mass standards demonstrated that the size of the tripartite complex is ∼180 kDa, consistent with a 1:1:1 ratio of the glycoproteins. This finding was observed consistently with TBS (pH 7.4) as the running buffer and with sodium acetate buffer at pH 5.0. The 1:1:1 complex was first identified by gel filtration performed by mixing separately purified soluble gp42 (in excess) with soluble gH/gL (Fig. 2A). Complete disappearance of the gH/gL peak and appearance of a new peak corresponding to the size of soluble gH/gL/gp42 were observed. Analysis of the new peak fractions by SDS-PAGE demonstrated the presence of gH/gL/gp42 by Coomassie staining and Western blotting. Reinjection of the gH/gL/gp42 peak revealed the appearance of only the same single peak again. Gel filtration also demonstrated the 1:1:1 complex when testing purified gH/gL/gp42 coexpressed together by simultaneous infection of insect cells. Native gel electrophoresis was also used to demonstrate that the soluble gH/gL/gp42 complex is stable and migrates as a single band with a mobility that is intermediate between those of soluble gp42 and soluble gH/gL (Fig. 2C).
Soluble gH/gL has no effect on membrane fusion, indicating an essential role for the gH transmembrane domain.
An essential role of gH in membrane fusion has been demonstrated by in vitro experiments showing that EBV gH, gL, and gB are the minimal combination required for optimal cell-cell fusion with epithelial cells, and gp42 is additionally required for B-cell fusion (18). Interestingly, the addition of soluble gH/gL does not effect B-cell fusion or epithelial cell fusion (Fig. 4A and B). Unlike soluble gp42, the fusion assay data demonstrates that soluble gH/gL cannot act as a trigger for fusion when only transfected membrane-bound gB or gp42 and gB are present (where gH/gL or only gH is absent). Although the purified soluble gH/gL complexes are in their native conformation with binding epitopes for E1D1 recognition, gp42 binding, and putative gH/gL receptor binding, the soluble gH/gL complexes are not “fully functional” in activating membrane fusion compared to the intact membrane-bound protein. Therefore, these data suggest that the membrane fusion mechanism requires the transmembrane domain of gH. This is consistent with evidence from virions engineered to be gH null, which are not capable of infecting target cells (9). The data also agree with studies of herpes simplex virus type 1 gH, in which the transmembrane domain and cytoplasmic tail were replaced with a glycosylphosphatidylinositol anchor and the resulting gH protein did not function in fusion (13). Data presented herein strongly suggest that the EBV gH is centrally involved in the physical merging of cellular membrane and viral envelope, or alternatively, the gH transmembrane domain and/or cytoplasmic tail play some other critical role during viral fusion.
Soluble gH/gL also did not inhibit membrane fusion with epithelial cells, which might have been expected on the basis of previous studies suggesting the presence of a gH receptor on these cells. It may be that at the highest concentrations of gH/gL used in this study, the putative epithelial cell receptors were not saturated with soluble gH/gL, either because of their number or because the affinity of the receptor-gH/gL interaction is relatively weak. Perhaps if higher concentrations of soluble gH/gL were tested, a receptor saturation phenomenon could be observed with concomitant inhibition of membrane fusion. The soluble gH/gL protein may also prove useful in identifying the putative gH/gL receptor.
The soluble gH/gL/gp42 heterotrimeric complex enhances fusion with B cells but not with epithelial cells.
Soluble gp42 enhances B-cell membrane fusion and inhibits epithelial cell membrane fusion (Fig. 3A and B). The soluble gH/gL/gp42 complex is able to enhance B-cell fusion (Fig. 4C) and has minimal effects on epithelial cell fusion until much higher concentrations (Fig. 4D). The difference in the function of the three-part protein complex strongly suggests that the mechanism of fusion inhibition by soluble gp42 is binding of membrane-bound gH/gL. Since the soluble heterotrimer has already sequestered gp42, it is unable to inhibit epithelial cell fusion until a considerably higher concentration of soluble protein is present, presumably allowing some fraction of the gp42 to dissociate from the three-part complex. This is also consistent with the in vitro peptide binding studies that show that a small fraction of peptide is able to bind the three-part complex, presumably when a small amount of soluble gp42 dissociates (Fig. 5B). These observations are also consistent with the idea that gp42 acts as a tropism switch for EBV governed by the amount of gp42 present in the virion envelope (2). It is possible that the binding of gp42 to gH/gL may directly interfere with gH/gL interactions with an epithelial cell receptor, blocking the activation of membrane fusion.
A gp42 peptide is an inhibitor of membrane fusion.
Soluble gp42 inhibits fusion with epithelial cells in a dose-dependent manner at concentrations in the low nanomolar range. Paralleling this inhibitory activity, we have demonstrated that the gp42-36-65 peptide inhibits epithelial cell fusion in the low micromolar concentration range in a dose-dependent manner (Fig. 6A). A control peptide does not have any effect on fusion of B cells or epithelial cells. Moreover, the gp42 peptide binds to purified gH/gL and this binding is blocked by the presence of gp42 (Fig. 5). The peptide studies indicate that this domain of gp42 provides part, but not all, of the binding interactions with gH/gL. Thus, the gp42-36-65 peptide likely inactivates the epithelial cell fusion process by acting similarly to soluble gp42 through an interaction with gH/gL. The peptide inhibition can be further investigated by making sequence and length variants to further enhance its binding affinity. The inhibitory effect may be due to a steric block of gH/gL binding to a receptor on epithelial cells or induction of a conformational change in gH/gL that makes gH/gL unable to engage its receptor. The gp42-36-65 peptide provides an interesting peptide-based inhibitor for the EBV entry process. If the peptide affinity could be increased sufficiently, it should compete for gp42 binding to gH/gL and might have the potential to provide a single inhibitor that would block viral entry into both epithelial and B cells. The possibility of developing a peptide therapeutic for EBV adds to the nascent field of viral entry inhibitors. This gp42-derived peptide and others like it may also provide new tools for studying herpesvirus entry.
Acknowledgments
We thank Lindsey Hutt-Fletcher for generously donating the E1D1 antibody cell line and both the Northwestern Monoclonal Antibody Facility and the National Cell Culture Center for producing milligram quantities of the E1D1 and 3H3 antibodies. We thank Hisae Matsuura for providing gp42-ΔN86 and Marisa McShane for FACS analysis. We appreciate the help and support provided by the members of the Jardetzky and Longnecker laboratories.
T.S.J. is a Scholar and R.L. is a Stohlman Scholar of the Leukemia and Lymphoma Society of America. This research was supported by Public Health Service grants CA62234 (R.L.), CA73507 (R.L.), and CA93444 (R.L. and T.S.J.) from the National Cancer Institute. This work was supported in part by a predoctoral fellowship from Northwestern's Biotechnology Training Program from NIH (A.N.K.) and by a predoctoral fellowship from the American Heart Association, Midwest Affiliate (J.O.).
REFERENCES
- 1.Amon, W., and P. J. Farrell. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149-156. [DOI] [PubMed] [Google Scholar]
- 2.Borza, C. M., and L. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 3.Borza, C. M., A. J. Morgan, S. M. Turk, and L. M. Hutt-Fletcher. 2004. Use of gHgL for attachment of Epstein-Barr virus to epithelial cells compromises infection. J. Virol. 78:5007-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fingeroth, J. D., M. E. Diamond, D. R. Sage, J. Hayman, and J. L. Yates. 1999. CD21-dependent infection of an epithelial cell line, 293, by Epstein-Barr virus. J. Virol. 73:2115-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, N.J.
- 6.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 8.Haan, K. M., and R. Longnecker. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 97:9252-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haddad, R. S., and L. M. Hutt-Fletcher. 1989. Depletion of glycoprotein gp85 from virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J. Virol. 63:4998-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutt-Fletcher, L. M., and C. M. Lake. 2001. Two Epstein-Barr virus glycoprotein complexes. Curr. Top. Microbiol. Immunol. 258:51-64. [DOI] [PubMed] [Google Scholar]
- 11.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jardetzky, T. S., and R. A. Lamb. 2004. Virology: a class act. Nature 427:307-308. [DOI] [PubMed] [Google Scholar]
- 13.Jones, N. A., and R. J. Geraghty. 2004. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology 324:213-228. [DOI] [PubMed] [Google Scholar]
- 14.Katz, B. Z. 2003. Principles and practice of pediatric infectious diseases, 2nd ed. Churchill Livingstone, New York, N.Y.
- 15.Li, Q., C. Buranathai, C. Grose, and L. M. Hutt-Fletcher. 1997. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J. Virol. 71:1667-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane, M. P., and R. Longnecker. 2004. Cell-surface expression of a mutated Epstein-Barr virus glycoprotein B allows fusion independent of other viral proteins. Proc. Natl. Acad. Sci. USA 101:17474-17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 21.Nemerow, G. R., R. A. Houghten, M. D. Moore, and N. R. Cooper. 1989. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56:369-377. [DOI] [PubMed] [Google Scholar]
- 22.Oda, T., S. Imai, S. Chiba, and K. Takada. 2000. Epstein-Barr virus lacking glycoprotein gp85 cannot infect B cells and epithelial cells. Virology 276:52-58. [DOI] [PubMed] [Google Scholar]
- 23.Okuma, K. M., S. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 24.Omerovic, J., L. Lev, and R. Longnecker. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J. Virol. 79:12408-12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulford, D., P. Lowrey, and A. J. Morgan. 1995. Co-expression of the Epstein-Barr virus BXLF2 and BKRF2 genes with a recombinant baculovirus produces gp85 on the cell surface with antigenic similarity to the native protein. J. Gen. Virol. 76(Pt. 12):3145-3152. [DOI] [PubMed] [Google Scholar]
- 26.Pulford, D., P. Lowrey, and A. J. Morgan. 1994. Expression of the Epstein-Barr virus envelope fusion glycoprotein gp85 gene by a recombinant baculovirus. J. Gen. Virol. 75(Pt. 11):3241-3248. [DOI] [PubMed] [Google Scholar]
- 27.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Silva, A. L., J. Omerovic, T. S. Jardetzky, and R. Longnecker. 2004. Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion. J. Virol. 78:5946-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Speck, P., K. M. Haan, and R. Longnecker. 2000. Epstein-Barr virus entry into cells. Virology 277:1-5. [DOI] [PubMed] [Google Scholar]
- 31.Speck, P., and R. Longnecker. 1999. Epstein-Barr virus (EBV) infection visualized by EGFP expression demonstrates dependence on known mediators of EBV entry. Arch. Virol. 144:1123-1137. [DOI] [PubMed] [Google Scholar]
- 32.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Mullberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strnad, B. C., T. Schuster, R. Klein, R. F. Hopkins III, T. Whitmer, R. H. Neubauer, and H. Rabin. 1982. Production and characterization of monoclonal antibodies against the Epstein-Barr virus membrane antigen. J. Virol. 41:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takada, K. 2001. Role of Epstein-Barr virus in Burkitt's lymphoma. Curr. Top. Microbiol. Immunol. 258:141-151. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, M. P., and R. Kurzrock. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803-821. [DOI] [PubMed] [Google Scholar]
- 36.Volpi, A. 2004. Epstein-Barr virus and human herpesvirus type 8 infections of the central nervous system. Herpes 11(Suppl. 2):120A-127A. [PubMed] [Google Scholar]
- 37.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, W., and J. Sham. 2005. Nasopharyngeal carcinoma. Lancet 365:2041-2054. [DOI] [PubMed] [Google Scholar]