Abstract
Human herpesvirus 8 interleukin-6 (vIL-6) displays 25% amino acid identity with human IL-6 (hIL-6) and shares an overall four-helix-bundle structure and gp130-mediated STAT/mitogen-activated protein kinase signaling with its cellular counterpart. However, vIL-6 is distinct in that it can signal through gp130 alone, in the absence of the nonsignaling gp80 α-subunit of the IL-6 receptor. To investigate the structural requirements for gp80 independence of vIL-6, a series of expression vectors encoding vIL-6/hIL-6 chimeric and site-mutated IL-6 proteins was generated. The replacement of hIL-6 residues with three vIL-6-specific tryptophans implicated in gp80 independence from crystallographic studies or the A and C helices containing these residues did not confer gp80 independence to hIL-6. The N- and C-terminal regions of vIL-6 could be substituted with hIL-6 sequences with the retention of gp80-independent signaling, but substitutions of other regions of vIL-6 (helix A, A/B loop, helix B, helix C, and proximal half of helix D) with equivalent sequences of hIL-6 abolished gp80 independence. Interestingly, the B helix of vIL-6 was absolutely required for gp80 independence, despite the fact that this region contains no receptor-binding residues. Point mutational analysis of helix C, which contains residues involved in physical and functional interactions with gp130 domains 2 and 3 (cytokine-binding homology region), identified a variant, VI120EE, that was able to signal and dimerize gp130 only in the presence of gp80. gp80 was also found to stabilize gp130:g130 dimers induced by a distal D helix variant of vIL-6 that was nonetheless able to signal independently of gp80. Together, our data reveal the crucial importance of overall vIL-6 structure and conformation for gp80-independent signaling and provide functional and physical evidence of the stabilization of vIL-6-induced gp130 signaling complexes by gp80.
Human herpesvirus 8 (HHV-8) is associated with the human malignancies Kaposi's sarcoma, primary effusion lymphoma (PEL), and multicentric Castleman's disease (7, 8, 21, 25). In each of these, HHV-8 encoded viral interleukin-6 (vIL-6), like its human counterpart (hIL-6), is believed to play a role via its proproliferative, proangiogenic, and proinflammatory activities (1, 3, 6, 13, 14). Therefore, understanding receptor recognition and the functional properties of vIL-6, especially those that might differ from hIL-6, is important for elucidating the contribution of the viral cytokine to HHV-8 neoplasia, in addition to virus biology, and devising potentially therapeutic strategies to abrogate vIL-6 activities specifically.
There has been considerable research effort directed towards determining the expression of vIL-6 during virus replication and in Kaposi's sarcoma, PEL, and multicentric Castleman's disease tissues. These studies have revealed that vIL-6 is primarily expressed as a lytic gene, being greatly induced upon lytic reactivation in PEL cell lines (20, 22). However, the expression of vIL-6 appears to be distinct from other lytic genes and vIL-6 protein and transcripts can be detected in the absence of other lytic gene expression (10, 23). Indeed, vIL-6 can be induced specifically by alpha interferon in PEL cells and protect these cells from alpha interferon-mediated cell cycle arrest and apoptosis (9). These data indicate that vIL-6 might function during latency as well as during lytic replication and that it may be involved in viral pathogenesis, even in the absence of full productive replication, and could therefore mediate autocrine, in addition to paracrine, functions during HHV-8-induced malignancy.
Several published studies have focused on identifying the structural determinants of vIL-6 receptor recognition and function. The elucidation of the crystal structure of vIL-6 bound to the three proximal cytokine-interacting domains of gp130 was a major achievement that both confirmed predictions about the binding interfaces and the involved residues of ligand and receptor and revealed novel aspects of vIL-6-gp130 recognition (11). Of note, these structural studies implicated three vIL-6-specific “site II” tryptophan residues as key elements in interactions with domain 2 (D2) and D3 (cytokine-binding homology region [CHR]) of gp130 and it was suggested that these residues may account for the gp80 independence of vIL-6. Also, the CD loop of gp130 domain 2 (proximal domain of CHR) was found to interact with vIL-6, in addition to the EF loop of this domain and BC loop of domain 3 (distal domain of CHR), which was previously suspected of contributing ligand-binding residues. Mutational analysis of gp130 and vIL-6 coupled with vIL-6-gp130 interaction, vIL-6-induced gp130 dimerization, and signal transduction studies identified several of the same gp130 residues as contributing to site II interactions to mediate functional complexing and confirmed the central importance of W167 (numbering from the first methionine of the vIL-6 open reading frame [ORF]) at the tip of the D helix for “site III” interactions with gp130 domain 1 (immunoglobulin [Ig] homology region) (15, 16, 26). Domain 2 EF loop variants of gp130 unable to support vIL-6 signaling through gp130 alone were helpful in establishing that gp80 could indeed complex functionally with vIL-6 and gp130, as gp80 was able to rescue these otherwise nonfunctional variants (15, 16, 26). The reciprocal situation was reported by utilizing a vIL-6 site III variant (W167G) that could not signal through gp130 alone but could mediate appreciable signal transduction if gp80 was coexpressed (5). The physical association of vIL-6 directly with gp80 and/or with gp80 in association with gp130 has been determined by enzyme-linked immunosorbent assay, coprecipitation, and cosedimentation procedures (5, 16), and neutralizing antibodies to gp80 have been shown to inhibit vIL-6 signaling when applied either alone or along with gp130 antibodies (6, 22). While there are published reports arguing against the involvement of gp80 in vIL-6 signaling (2, 19), it now seems evident that vIL-6 does indeed have two modes of signaling, one involving gp80 and the other not. However, the significance of these with respect to signal transduction and biological effects is unresolved.
Here we report on structure-function studies of vIL-6 and hIL-6 aimed at determining the structural requirements for gp80-independent signaling by vIL-6. We exploited the analogous secondary and tertiary structures of the two cytokines to generate a panel of vIL-6/hIL-6 chimeras and point-mutated vIL-6 and hIL-6 proteins for use in various functional experiments to address this question. Our results reveal that the overall structure of vIL-6, as opposed to specific residues, is critically important for gp80-independent signaling and that gp80 can stabilize gp130 dimerization induced by vIL-6.
MATERIALS AND METHODS
Cell culture, transfections, and cell lines.
HEK293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For transfections, cells were passaged 12 to 24 h prior to transfection to produce monolayers that were 60% confluent and these were transfected by the calcium phosphate-DNA coprecipitation procedure with HEPES-buffered saline. Cells were harvested 48 h posttransfection for determinations of chloramphenicol acetyltransferase (CAT) activity, STAT3 activation, gp130 phosphorylation, induced receptor complexing, or protein expression.
Plasmids.
pSG5-based eukaryotic expression plasmids for vIL-6 and hIL-6 have been described previously (16, 26). Expression plasmids for gp80 (IL-6R) and gp130, containing the receptor coding sequences cloned between the XbaI (gp80) or SacI and BamHI (gp130) sites of the pEF-BOS vector (18), were provided by M. Narazaki and T. Kishimoto. The pα2MCAT reporter plasmid contains rat α2-macroglobulin promoter sequences cloned upstream of the CAT gene (24) and was provided by T. Schaefer. A pSG5-based expression vector, pSG5.S-CBD, containing chitin-binding domain (CBD) coding sequences was generated by cloning a 227-bp BamHI fragment from the pTYB4 vector (New England Biolabs, Beverly, Mass.) between the BglII and BamHI sites of a modified version of pSG5, pSG5.S, containing additional 5′ cloning sites. ORFs encoding vIL-6, hIL-6, or chimeric or point-mutated IL-6 proteins were PCR amplified to exclude stop codons and cloned in frame upstream of the CBD coding sequences in pSG5.S-CBD, between SmaI (5′) and BamHI (3′) sites, to generate IL-6-CBD fusions.
Construction of IL-6 chimeras and point-mutated proteins.
An overlap extension PCR strategy (12) was used to generate coding sequences specifying vIL-6/hIL-6 hybrid (chimeric) proteins and IL-6 proteins containing specific amino acid substitutions. Partially overlapping mutagenic oligonucleotide primers were used for PCR amplification of N- and C-terminal sequences on either side of the vIL-6/hIL-6 junction or substituted codon(s), followed by gel isolation and annealing of the denatured PCR products and PCR amplification of the entire IL-6 ORF with ORF 5′ and 3′ cloning primers. Full-length PCR products were then restriction digested (SmaI and BglII) and ligated into a cloning site-modified pSG5 vector, pSG5.S (for eukaryotic expression).
Western blotting and coprecipitations.
Cytokine expression and ligand-induced receptor complexing were investigated by Western blotting of cell extracts or protein A-agarose-precipitated material (see below) using antiserum to vIL-6 (generated in this laboratory [26]) or commercially available polyclonal or monoclonal antibodies to hIL-6 (R&D Systems, Minneapolis, Minn.; catalog no. AB-206-NA), gp80 (Santa Cruz Biotechnology, Santa Cruz, Calif.; catalog no.sc-661), gp130 (BD Biosciences, Rockville, Md.; catalog no. 555755), and CBD (New England Biolabs, Beverly, Mass.; catalog no. E8034S) to identify native or epitope-tagged ligand and receptor proteins. Total and tyrosine-phosphorylated (active) STAT3 were detected by Western blotting of cell extract using appropriate antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif., catalog. no. sc-482, and Cell Signaling Technology, Danvers, Mass.; catalog no. 9131). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) size fractionation of cell extracts or protein A-agarose precipitated proteins and electrophoretic transfer to nitrocellulose membranes were carried out using standard procedures. Membranes were blocked in phosphate-buffered saline (PBS)-T (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, 0.1% Tween 20) containing 5% nonfat milk prior to the addition of primary antibody (0.1 to 1.0 μg/ml) and incubation at 4°C overnight. After washing in PBS-T, horseradish peroxidase-conjugated anti-mouse (for CBD, gp130, and gp80) or anti-rabbit (for p-STAT3 and STAT3) IgG secondary antibody diluted 1:5,000 in PBS-T containing 5% nonfat milk was used to detect filter-bound primary antibody. Horseradish peroxidase on PBS-T-washed filters was visualized by chemiluminescence assay. For gp130-gp130 and gp130-gp80 complexing assays, gp130-Fc was precipitated from cell lysates with protein A-agarose following incubation overnight at 4°C and sedimented material was washed three times with either PBS or Tris-EDTA (TE) containing 0.1% NP-40 prior to heating in sodium dodecyl sulfate-PAGE load buffer and gel electrophoresis. Coprecipitated proteins were identified by Western blotting using appropriate antibodies to the native ligands and receptors or CBD epitope tag. Protein cross-linking of IL-6-induced gp130-containing complexes was performed by treatment of cells with 0.1 mM dithiobis(succinimidylpropionate) (DSP) cross-linker (Pierce, Rockford, IL; catalog no. 22585) in PBS at room temperature for 20 min. The reaction was stopped by the addition of Tris-HCl, pH 7.5, to a final concentration of 100 mM and incubation at room temperature for 15 min prior to cell lysis in PBS containing 1% NP-40 and proteinase inhibitors. Cross-linking was reversed by treatment with 5% β-mercaptoethanol, present in the protein gel load buffer. For non-cross-linking experiments, cell lysates were made by incubating cells at 4°C in either TE (10 mM Tris-HCl, pH 7.5, 5 mM EDTA) or PBS containing 0.1% NP-40 and proteinase inhibitor cocktail.
RESULTS
vIL-6-specific site II residues are not sufficient for gp80 independence.
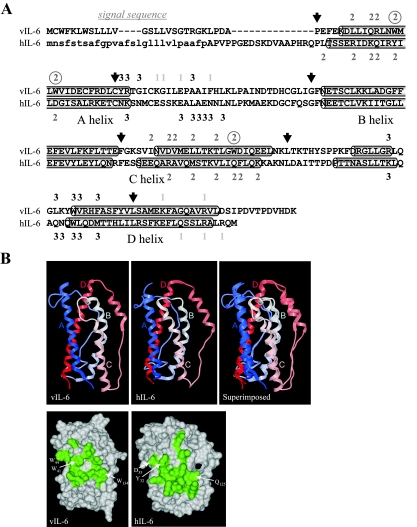
The elegant work of Chow and colleagues (11) solved the crystal structure of vIL-6 bound to the D1-D3 region of gp130. This study identified three vIL-6-specific tryptophan residues, W41, W44, and W134, as being involved in hydrophobic interactions with gp130 CHR and as potential mediators of gp80-independent interactions with the signal transducer (Fig. 1). Therefore, we mutated the vIL-6 ORF to change these tryptophans to alanines and introduced tryptophan residues into hIL-6 in place of the colinear tyrosine-32, aspartate-35, and glutamine-125 residues. Signal transduction by these vIL-6 and hIL-6 variants was compared to that by vIL-6 and hIL-6 in gp80-independent and gp80/gp130 transient transfection signaling assays employing a STAT/AP-1-responsive CAT reporter, pα2MCAT (24, 26). The appropriate receptor subunit(s) and relevant ligand were overexpressed in HEK293T cells, which were also cotransfected with the reporter construction. The results of representative experiments are shown in Fig. 2. We found that alanine substitution of W41 or all three of the vIL-6 site II tryptophans led to around 30 or 50% reductions, respectively, in gp80-independent reporter induction, suggesting that these residues play a role in vIL-6-gp130 complexing but are not required for gp80 independence of vIL-6. The introduction of the three tryptophans into hIL-6 led to a slight but reproducible increase in CAT activity, consistent with the notion that these residues contribute positively to site II interactions but are insufficient to support substantial gp80-independent signaling. The data suggested that other vIL-6-unique residues are necessary. To address this issue, we also investigated in our reporter assays whether replacing hIL-6 helices A and C with those of vIL-6 could confer gp80-independent signaling. These helices contain all known site II (gp130 CHR-interacting) residues. No gp80-independent signaling was detected for the helix A and C substitution variant, but this was fully functional in cells overexpressing gp80 along with gp130. These data suggest at least two possibilities: first, that increased site II interactions in vIL-6 relative to that in hIL-6 do not account for gp80 independence (perhaps vIL-6 site III interactions also are important), and second, that the particular tertiary conformation of vIL-6 is critically important for signaling through gp130 alone.
FIG. 1.
Primary and three-dimensional structures of viral and human IL-6. (A) Alignment of the amino acid sequences specified by the human and HHV-8 IL-6 ORFs. Indicated are the α-helices, residues predicted (vIL-6 site I) or demonstrated to interact with gp80 (“1”) and gp130 (“2” and “3”), and vIL-6-specific site II tryptophan residues (circled) predicted to contribute significantly to site II interactions with gp130 CHR (domains 2 and 3) (4, 5, 11). Site III residues of IL-6 interact with gp130 Ig-like domain 1, and site I residues with CHR residues of gp80. Also shown (arrows) are the junctions of splices between vIL-6 and hIL-6 sequences for the generation of IL-6 chimeric proteins (listed in Fig. 3A). The cleaved signal sequence of hIL-6 is indicated in lowercase. (B) Three-dimensional structures of vIL-6 (5) and hIL-6 (11) as illustrated by ribbon and “skin” models, orientated with helices A and C in the front plane to show the site II interface. The ribbon diagram on the right shows the vIL-6 and hIL-6 structures superimposed to emphasize the highly conserved secondary and tertiary structures of the two cytokines. The skin models below are orientated equivalently to the ribbon diagrams and show site II residues (shaded) of vIL-6 and hIL-6, including the three vIL-6-specific tryptophan residues and colinear residues of hIL-6 (labeled). Numbering of hIL-6 residues is from the first amino acid of the mature, cleaved protein.
FIG. 2.
Functional analyses of hIL-6 proteins with introduced vIL-6-specific site II tryptophan residues and site II-containing vIL-6 helices A and C and vIL-6 proteins with the tryptophans altered to alanine residues. (A) The altered vIL-6 and hIL-6 proteins that were generated are indicated. Mutated residues correspond to W41, W44, and W134 of vIL-6 and Y32, D35, and Q125 of hIL-6. Each point-mutated or chimeric ORF was cloned into pSG5 to allow the expression of the encoded proteins in transfected eukaryotic cells. (B) Data from reporter-based assays utilizing the STAT-responsive reporter pα2MCAT (24) and expression vectors for gp80 and gp130. These vectors were cotransfected into HEK293T cells with each of the IL-6 expression constructions listed in panel A (1 to 4; v, vIL-6; h, hIL-6) or pSG5 (p, negative control) to determine the signaling activities of the IL-6 proteins via gp130 or gp130+gp80, as described previously (26). The mutation of the site II tryptophans in vIL-6 reduced but did not eliminate gp80-independent signaling by the viral cytokine, and substitution of the A and C helices of hIL-6 with those of vIL-6 had no effect on gp80 dependence of hIL-6. Introduction of the three site II tryptophans into hIL-6 allowed modest gp80-independent signaling by hIL-6. Transfections were carried out in triplicate. Error bars indicate standard deviations.
Chimeric proteins reveal nonequivalence of vIL-6 and hIL-6 domains.
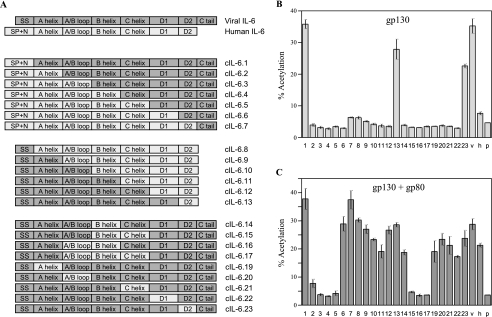
To try to identify regions of vIL-6 that are dispensable or necessary for gp80-independent signaling, we created a series of vIL-6/hIL-6 chimeric proteins for use in our reporter assays. The junctions of the vIL-6 and hIL-6 regions that were swapped and spliced are indicated in Fig. 1. The constructions, shown in Fig. 3A, were each cloned into pSG5 for expression in transfected cells. Successive N- and C-terminal replacements of vIL-6 sequences with those of hIL-6 determined that while sequences N terminal to helix A and C terminal to the proximal half of helix D did not affect gp80 independence, further encroachment into vIL-6 abolished gp80-independent signaling and also gp80+gp130 signaling in the case of the hIL-6(N)/vIL-6(C) variants c2 to c5 (which were expressed appropriately [data not shown]) (Fig. 3B). Thus, the proximal part of helix D and adjacent CD loop region (containing site III residues) and helix A (containing site II residues) are not directly equivalent between vIL-6 and hIL-6. Internal substitutions of vIL-6 helix A (site II), AB loop (site III with site I), B helix (nonbinding), C helix (site II), and CD loop with proximal helix D (site III) with colinear hIL-6 sequences resulted in gp80-dependent chimeras (variants c14 and c19 to c22) (Fig. 3). The cointroduction of hIL-6 B helix with the AB loop and/or helix C into vIL-6 resulted in proteins (variants c15 to c17) that were unable to signal either with or without gp80. Each of the internal domains, with the exception of helix B, contains either site II or site III residues that interact with gp130-CHR (D2 and D3) or gp130-Ig (D1), respectively. Together, our data demonstrate that most of the vIL-6 molecule is required for gp80-independent signal transduction, including helix B that does not bind gp80 or gp30, suggesting that the overall structure of vIL-6 is crucially important for this activity.
FIG. 3.
Generation and analyses of vIL-6/hIL-6 chimeras to identify regions involved in gp80-independent signaling by vIL-6. (A) Diagrammatic illustration of vIL-6/hIL-6 chimeric ORFs generated and cloned into pSG5 for eukaryotic expression. SS, signal sequence; SP+N, signal peptide plus N terminus. (B) Functional analyses of chimeric proteins relative to native vIL-6 (v) and hIL-6 (h) by STAT-CAT reporter (pα2M-CAT) assays to detect signaling through overexpressed gp130. Numbers correspond to designations given to each of the chimeric constructions in panel A. Cultures transfected with pSG5 (p) were used as negative controls. Error bars indicate standard deviations. (C) An experiment similar to that described for panel B was carried out to assay for signaling activities of each of the chimeric proteins through overexpressed gp130+gp80. Triplicate transfections were carried out for each chimera in each experiment. Error bars indicate standard deviations.
vIL-6 helix B and gp80 independence.
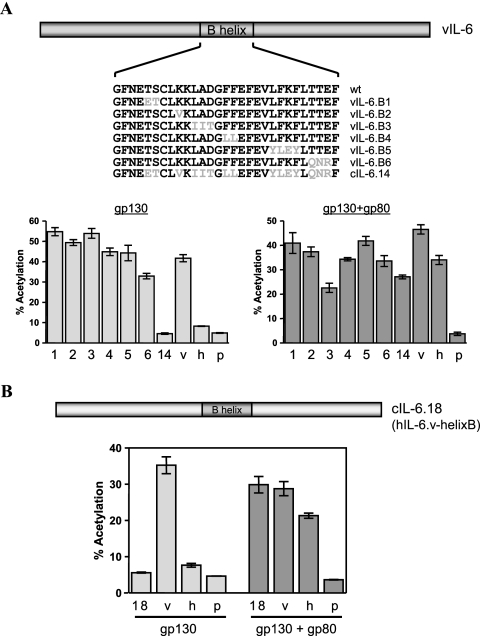
The finding that helix B from hIL-6 could not substitute functionally for that of vIL-6 with respect to gp80-independent signal transduction suggested that the precise conformation of vIL-6 was critical. To further investigate the importance of helix B and to try to identify key vIL-6-specific residues of helix B required for gp80 independence, we generated a series of two- to four-amino-acid substitutions in this region (Fig. 4A). The substitutions corresponded to colinear amino acids in hIL-6. These constructions were tested in reporter assays as before, with cotransfection of gp130 or gp130+gp80 expression vectors. The results of these experiments are shown in Fig. 4A. None of the grouped substitutions had a substantial effect on gp80-independent signaling by vIL-6 or on gp80+gp130 signaling. The retention of gp80 independence by all of the helix B variants suggests that the overall structure of the entire B helix in wild-type vIL-6 is important for gp80 independence.
FIG. 4.
Contribution of vIL-6 helix B to gp80-independent signaling. (A) Generation and analysis of B helix substitution variants of vIL-6. Single or grouped codon substitutions were introduced into vIL-6 to match the equivalent colinear codons in hIL-6. These altered ORFs were then cloned into pSG5. Reporter assays were carried out to determine the effects of the introduced B helix changes on gp80-independent signaling by vIL-6. Assays for signaling in gp80+gp130-overexpressing cells were performed in parallel. None of the introduced paired substitutions significantly altered gp80-independent signaling by vIL-6. (B) Coding sequences for the B helix of hIL-6 were replaced by those of vIL-6, and the resulting encoded protein was tested for its ability to signal via gp130 alone or via gp130+gp80. The B helix of vIL-6 could not confer gp80 independence to hIL-6. All transfections were performed in triplicate. Error bars indicate standard deviations. v, vIL-6; h, hIL-6; p, pSG5.
We next generated an hIL-6 molecule containing vIL-6 helix B in place of the native sequences to see if gp80 independence could be conferred to hIL-6 through vIL-6 helix B-induced conformational changes (Fig. 4B). Replacing helix B of hIL-6 with vIL-6 helix B did not confer gp80-independent signaling ability, although signal transduction through gp80+gp130 was increased somewhat to levels matching those of vIL-6. Our data suggest that the conformational effect of vIL-6 helix B in the context of vIL-6 is necessary for gp80 independence of the viral cytokine but that such conformational influences are insufficient to confer gp80 independence to hIL-6.
Mutational analysis of helix C.
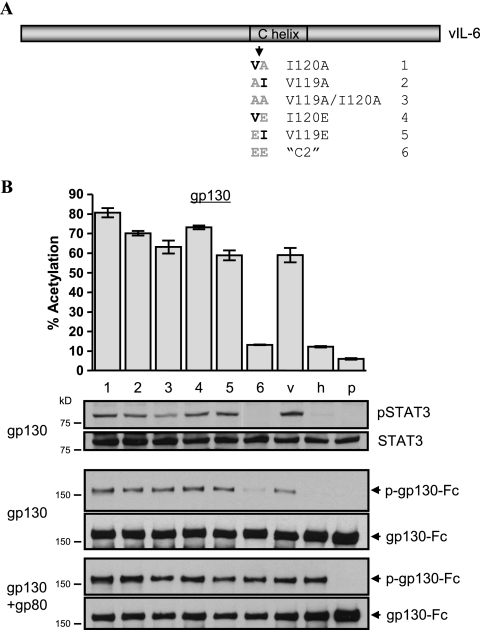
We next turned our attention to helix C, a major contributor of gp130 CHR-interacting (site II) residues. Nonconserved residues in the helix C region of vIL-6 were changed in a pairwise fashion to those found in hIL-6, via generation of the appropriate codon mutations within the vIL-6 ORF (Fig. 5A). The positions of these residues within the three-dimensional structure of vIL-6 and interactions of site II residues with gp130-CHR are shown in Fig. 5B. The panel of C helix variants was tested for signaling in gp130 and gp130+gp80 reporter assays (Fig. 5C). Also tested were the STAT3-inducing activities of these proteins in transfected cells in the context of gp80-independent signaling through overexpressed gp130 or via overexpressed gp80+gp130 (Fig. 5D). Analogous transfections employing a gp130-Fc expression vector were undertaken to measure ligand-induced gp130 phosphorylation (Fig. 5E). These functional analyses revealed that variant C2 (VI120EE) was unique in its dependence upon gp80 for signal transduction.
FIG. 5.
Mutational analysis of the C helix of vIL-6. (A) Pairwise substitution mutagenesis of the C helix of vIL-6. Substitutions introduced into the vIL-6 ORF match colinear hIL-6 codons. (B) Positions within the three-dimensional structure of vIL-6 of C helix site II residues involved in interactions with gp130-CHR. (C) gp130 and gp130+gp80 reporter-based signaling assays to determine the effects of the introduced amino acid substitutions on gp80-independent signaling by vIL-6. All but the “C2” variant (VI120EE) were able to signal at around wild-type levels through overexpressed gp130 in the absence of overexpressed gp80. Error bars indicate standard deviations. (D) Parallel Western blot-based assays to identify directly vIL-6-induced STAT3 activation in cells overexpressing gp130 or gp130+gp80. Cells were cotransfected with receptor and vIL-6 expression constructions and harvested 48 h posttransfection. Tyrosine-phosphorylated (active) STAT3 (“pSTAT3”) was detected by immunoprobing using a phosphospecific antibody; total STAT3 was detected on stripped blots using a phosphorylation-independent antibody to STAT3. (E) Western analysis for g130 phosphorylation in cells cotransfected with ligand and receptor expression constructions. Here, a gp130-Fc expression vector was used to allow direct protein A-agarose precipitation of gp130 from cell extracts. A phosphotyrosine (PY) antibody was used to detect phosphorylated gp130, and total precipitated gp130-Fc was detected on stripped blots using an appropriate antibody to gp130.
Further mutations of “C2” residues V119 and I120 were made (Fig. 6A) to determine the importance of each of these residues to gp80-independent signaling and whether the loss of such signaling in the EE-substituted C2 variant was due to the introduction of these charged residues specifically or due to changing of the native VI residues. Also, I120 has been implicated in site II interactions with gp130 (5) and so we wanted to investigate the effects of substitutions at this position alone. Reporter-based signaling assays and Western analyses for induced STAT3 and gp130 phosphorylation were undertaken using the panel of C2-position-altered proteins. The results obtained demonstrated that each variant (other than VI120EE) was able to signal independently of gp80 (Fig. 6B). Therefore, V119 and I120 are not essential for gp80-independent dimerization of gp130 but rather the introduction of the glutamate residues affects local structure and/or vIL-6-gp130 interactions, resulting in a loss of gp80 independence.
FIG. 6.
Signaling by C2-position point variants of vIL-6. (A) Further mutations at positions 119 and 120 were generated as indicated. (B) These variants were tested in pα2MCAT reporter and phospho-STAT3 Western blot assays for their abilities to signal via gp130 independently of gp80. Reporter assay transfections were performed in triplicate. Error bars indicate standard deviations. v, vIL-6; h, hIL-6; p, pSG5.
gp80 stabilization of vIL-6-induced gp130 dimerization.
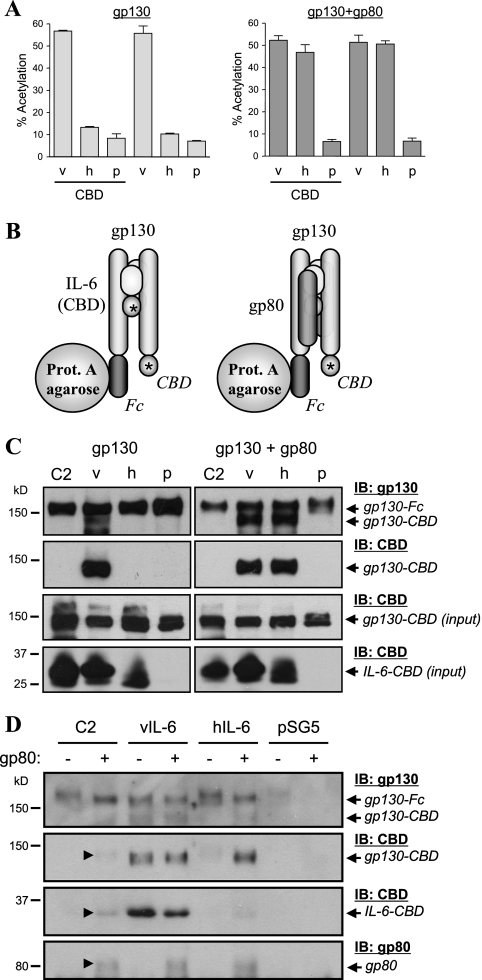
Having identified the VI120EE (C2) variant as dependent on gp80, we next wanted to examine its ability, relative to that of wild-type vIL-6, to induce gp130 dimerization in the absence and presence of gp80. For coprecipitation assays, wild-type and mutated vIL-6 and hIL-6 were expressed as CBD fusion proteins to facilitate analysis and comparison of their expression. The CBD fusion did not alter the signaling activities of vIL-6 or hIL-6, as detected by reporter and STAT3 activation assays (Fig. 7A). Assays for gp130 dimerization involved coprecipitation from transfected cell lysates of CBD-tagged gp130 with gp130-Fc in response to cytokine expression (Fig. 7B). Receptor and ligand components were introduced into HEK293T cells by cotransfection of the appropriate expression vectors, thereby replicating the conditions used for the reporter, STAT3 activation, and gp130 phosphorylation assays. Coprecipitated material was washed repeatedly with PBS (containing 137 mM NaCl) prior to gel electrophoresis and Western blotting (see Materials and Methods). The data from these experiments are shown in Fig. 7C. vIL-6.C2, as expected, could not induce gp130 dimerization (coprecipitation of gp130-CBD with gp130-Fc) in the absence of gp80 but, unexpectedly, did not do so in the presence of gp80 either. In contrast, vIL-6 induced gp130 dimerization in a gp80-independent manner and hIL-6 did so in the presence of gp80, thereby validating the assay. Similar coprecipitation experiments using low- or no-salt buffers for the lysis and washing steps also failed to detect vIL-6.C2-induced gp130 dimerization in the presence of gp80 (data not shown). The lack of gp80-induced dimerization by vIL-6.C2 suggested that this vIL-6 variant had reduced affinity of binding to gp130, leading to the disruption of complexes during the coprecipitation procedure, despite the clear functional evidence of gp80-dependent signaling complex formation in intact cells (Fig. 5 and 6). To detect vIL-6.C2-induced gp130-gp80 complexes and gp130 dimerization directly, we utilized DSP to cross-link signaling receptor complexes in vIL-6.C2- and gp130-Fc-expressing (transfected) HEK293T cells, either in the presence or absence of cooverexpressed gp80. Cross-linking was reversed with 5% β-mercaptoethanol (in gel load buffer) prior to gel fractionation for Western blotting. As before, vIL-6 and hIL-6 were used as positive controls, along with transfected empty vector pSG5-CBD (negative control). The results (Fig. 7D) revealed that vIL-6.C2, like hIL-6, could induce gp130-gp130 and gp130-gp80 complexing in a gp80-dependent manner.
FIG. 7.
Dimerization of gp130 by vIL-6.C2. (A) Reporter (pα2MCAT) assays were undertaken, as outlined for previous experiments, to confirm the functional integrities of vIL-6-CBD (v) and hIL-6-CBD (h) fusion proteins used in the gp130 dimerization assays. Empty expression plasmids (p) pSG5-CBD and pSG5 were used as negative controls for the CBD fusion and native proteins, respectively. Values shown are averages from duplicate transfections, with bars indicating the range. (B) Diagrammatic representation of coprecipitation experiments performed to analyze gp130 dimerization induced by vIL-6.C2. HEK293T cells were cotransfected with expression vectors for gp130-Fc, gp130-CBD, and IL-6-CBD (vIL-6, vIL-6.C2, or hIL-6), or pSG5-CBD (negative control), with or without gp80 expression vector. Protein (prot.) A-agarose was used to precipitate gp130-Fc from cell lysates, and this and coprecipitated gp130-CBD (indicative of gp130 dimerization) were identified by Western blotting using antibodies specific for gp130 or CBD. (C) Assays for gp130 dimerization induced by vIL-6.C2 relative to wild-type vIL-6 and hIL-6. Stable, gp130-Fc-containing complexes were precipitated directly from cell lysates, and PBS was used to wash protein A-agarose-bound complexes prior to gel loading. (D) Cells were treated with protein cross-linker (DSP, 0.1 mM) prior to cell lysis and protein A-agarose precipitation of gp130-Fc-containing complexes. Cross-linking was reversed in 5% β-mercaptoethanol before gel loading. +, presence of; −, absence of; IB, immunoblot.
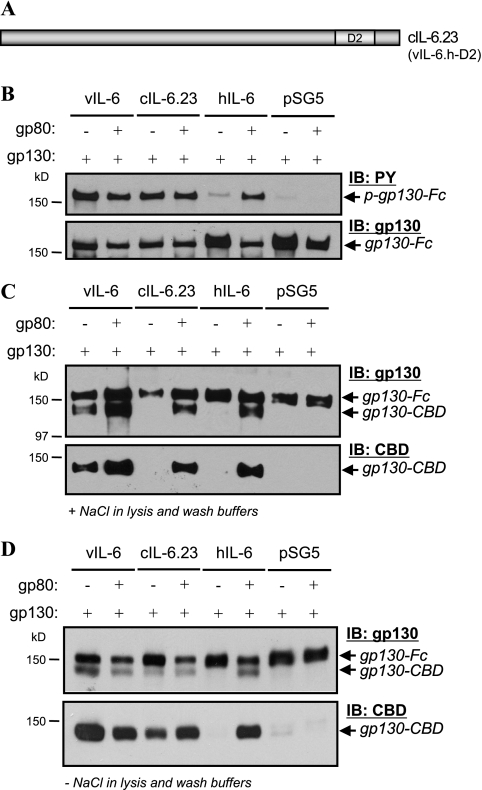
Further coprecipitation analyses were carried out with the variant cIL-6.23, in which the distal half of helix D was replaced with colinear sequences of hIL-6 (Fig. 8A). The distal region of the D helix of vIL-6, like that of hIL-6, is predicted to specify site I (gp80-interacting) residues (5). Chimeric protein cIL-6 was one of only three chimeric IL-6 proteins to retain gp80 independence in our reporter assays (Fig. 3), and we wanted to further investigate its functional and gp130 dimerization properties. Again we employed gp130-Fc-based coprecipitation assays to examine gp130 dimerization. Parallel gp130 phosphorylation assays were performed to verify gp80-independent signaling by cIL-6.23, and the expected result was obtained (Fig. 8B). However, cIL-6.23, in contrast to vIL-6, was unable to induce gp80-independent gp130-gp130 complexing that was stable enough to be detectable by regular coprecipitation (in PBS, containing 137 mM NaCl) from cell lysates (Fig. 8C). Coexpression of gp80 did allow the detection of these complexes, as for hIL-6. When these experiments were repeated using salt-free lysis and washing buffers (see Materials and Methods), gp80-independent gp130 dimerization induced by vIL-6.23 could now be detected (Fig. 8D). As for the results obtained with vIL-6.C2, these data demonstrate that vIL-6.23-induced signaling complexes are less stable than those induced by wild-type vIL-6 and that gp80 can stabilize vIL-6-induced gp130-gp130 complexing.
FIG. 8.
Analysis of gp130 dimerization and phosphorylation induced by cIL-6.23 (vIL-6.h-D2) in the absence (+) and presence (−) of gp80. (A) Diagrammatic representation of cIL-6.23, in which the distal half of helix D of vIL-6 is replaced by that of hIL-6. (B) Verification of gp80-independent signaling by cIL-6.23 using gp130 phosphorylation assay. gp130-Fc was precipitated from HEK293T cells cotransfected with gp130-Fc vector with or without gp80 vector and pSG5-based expression plasmids for vIL-6, vIL-6.23, or hIL-6; empty vector, pSG5, was used as a negative control. Precipitated gp130-Fc was subjected to PAGE and blotted onto membrane, and phosphotyrosine (PY) residues were detected by using anti-PY antibody. (C) gp130-Fc-based coprecipitation assays for gp130 dimerization, showing gp80-dependent coprecipitation of coexpressed gp130-CBD from lysates of cells expressing vIL-6.23. Wild-type vIL-6 and hIL-6 were used as positive controls for gp80-independent and gp80-dependent gp130 dimerization, respectively, and empty pSG5 vector provided the negative control. Cells were lysed in PBS containing 0.1% NP-40, and precipitates were washed with PBS prior to gel loading. (D) Repeat of the experiment described for panel C except that salt-free TE buffer was used to lyse cells and wash protein A-bound complexes prior to gel loading. IB, immunoblot.
DISCUSSION
Much progress has been made in understanding the molecular interactions that are involved in signal transduction by vIL-6 through the gp130 signal transducer, from mutational studies of ligand and receptor to the solution of the structure of vIL-6 complexed with the cytokine-binding region of gp130 (5, 11, 15, 16, 26). However, still unresolved is the actual basis of the unique gp80 independence of vIL-6; the nonsignaling receptor subunit is required for signaling by cellular IL-6 proteins. The data that we have presented here demonstrate that the three vIL-6-specific site II tryptophans (W41, W44, and W134), predicted from X-ray crystallographic studies to be important for gp80 independence (11), are not required for gp80-independent signaling by vIL-6, nor do they confer gp80 independence to hIL-6. However, their mutation to alanines did lead to a 50% reduction in gp80-independent signaling (Fig. 2) and we have preliminary data (D. Chen and J. Nicholas, unpublished) that indicate that the W3-to-A3 variant induces gp130 dimers that are less stable than those induced by wild-type vIL-6. This is similar to the situation for the gp80-independent D helix variant cIL-6.23 (Fig. 3). This variant, altered in residues predicted to interact with gp80, and a gp80-dependent vIL-6 protein (cIL-6.14) in which the receptor-noninteracting B helix was substituted for hIL-6 helix B are assumed to have lost the ability to dimerize gp130 (in the absence of gp80) via indirect effects on vIL-6 conformation, because neither region contains gp130-interacting residues. Thus, a key determinant of gp80-independent signaling through gp130 appears to be the particular tertiary structure of vIL-6, a conclusion supported by the finding that most vIL-6/hIL-6 chimeric proteins were unable to support gp80-independent signaling but retained function in the presence of gp80 (Fig. 3, Table 1).
TABLE 1.
Properties of vIL-6 variants
| Variant(s)a | Change(s) | gp80-independent CAT/STAT/gp130-PYg | gp130 Dimerization −gp80/+gp80 (CPS/NS, XL)h |
|---|---|---|---|
| vIL-6(A3) | Site II Ws to Asb | Y/-/- | - |
| hIL-6(W3) | Site II Ws introducedb | N/-/- | - |
| cIL-6.1 | v/hSP+Nc | Y/-/- | - |
| cIL-6.13 | v(ΔC-tail)/hC′D helixc | Y/-/- | - |
| cIL-6.14 | v/hB-helixc | N/-/- | - |
| cIL-6.19 | v/hA-helixc | N/-/- | - |
| cIL-6.20 | v/hAB-loopc | N/-/- | - |
| cIL-6.21 | v/hC-helixc | N/-/- | - |
| cIL-6.22 | v/hN′D helixc | N/-/- | - |
| cIL-6.23 | v/hC′D helixc | Y/Y/Y | N/Y (CPS), Y/Y (CPNS) |
| Other cIL-6s | Multiple domain substitutionsd | N/-/- | - |
| vIL-6s B1-6 | Multiple B helix substitutionsd | Y/-/- | - |
| hIL-6.vHel-B | vIL-6 B helix in hIL-6d | N/-/- | - |
| vIL-6.C1 | GK117ESe | Y/Y/Y | - |
| vIL-6.C2 | VI120EEe | N/N/N | N/N (CPS), N/N (CPNS)i, N/Y (XL) |
| vIL-6.C3 | NV122QAe | Y/Y/Y | - |
| vIL-6.C4 | DV124RAe | Y/Y/Y | - |
| vIL-6.C5 | ME126VQe | Y/Y/Y | - |
| vIL-6.C6 | LL128MSe | Y/Y/Y | - |
| vIL-6.C7 | TLG133VLIe | Y/Y/Y | - |
| vIL-6.C8 | WD135QFe | Y/Y/Y | - |
| vIL-6.C9 | IQE138LQKe | Y/Y/Y | - |
| vIL-6.C10 | EL140KAe | Y/Y/Y | - |
| vIL-6.C11 | NK142KNe | Y/Y/Y | - |
| vIL-6.C2-1 | I120Af | Y/Y/Y | Y/Y (CPS)i |
| vIL-6.C2-2 | V119Af | Y/Y/Y | Y/Y (CPS)i |
| vIL-6.C2-3 | VI120AAf | Y/Y/Y | Y/Y (CPS)i |
| vIL-6.C2-4 | I120Ef | Y/Y/Y | Y/Y (CPS)i |
| vIL-6.C2-5 | V119Ef | Y/Y/Y | Y/Y (CPS)i |
All variants listed were found to be functional in the presence of gp80.
See Fig. 2.
See Fig. 3.
See Fig. 4.
See Fig. 5.
See Fig. 6.
PY; phosphotyrosine; Y, yes; N, no; -, not determined.
Coprecipitation (CP) in presence (superscript “S”) and absence (superscript “NS”) of salt (NaCl) in the lysis and wash buffers; XL, cross-linked prior to coprecipitation.
Data not shown.
It is notable that other than the “C2” mutation (VI120EE) within the site II residue-containing helix C, none of the mutations introduced into the C helix led to loss of gp80-independent signaling by vIL-6 (Table 1). As only one of the C2-position residues, I120, has been implicated in gp130 interaction and as this is not a major part of the site II interface with gp130 CHR (11), it is unlikely that loss of gp80 independence of vIL-6.C2 is due to direct effects on vIL-6-gp130 interaction via site II. Indeed, the mutation of the C2 position VI residues to alanines had no detectable effect on gp80-independent signaling (Fig. 6) or gp130 dimerization (data not shown). Therefore, as with the B helix and distal D helix variants cIL-6.14 and cIL-6.23 discussed above, it appears that conformational changes induced by the introduction of glutamate residues at positions 119 and 120 account for the loss of gp80 independence. Attempts to model the predicted structural changes induced by these substitutions and those in cIL-6.14 and cIL-6.23 failed to reveal significant conformational changes (data not shown), which is perhaps not altogether surprising considering the limitations of protein modeling software. The fact that vIL-6.C2 is fully functional in the presence of gp80 indicates that gp80 can stabilize vIL-6-gp130 interactions via hexameric (vIL-62-gp1302-gp802) complexing and/or induce appropriate vIL-6 conformation to allow ligand-induced gp130 dimerization. The rescue of vIL-6.C2-induced gp130-gp130 complexing and signaling by gp80, like the stabilization of cIL-6.23-induced gp130 dimerization by gp80, provides clear evidence of gp80 involvement in and augmentation of vIL-6 signaling.
At present, we can only speculate about the biological significance, with respect to virus biology and viral pathogenesis, of gp80-independent versus gp80/gp130 signaling by vIL-6. Data from this laboratory (F. Hu and J. Nicholas, unpublished) have revealed differences in the relative levels and durations of STAT1 and STAT3 activation and support of IL-6-dependent cell growth by vIL-6 as a function of gp80. This suggests that signaling via the two modalities, gp80-independent tetrameric and gp80-dependent hexameric complexes, could lead to different biological outcomes. However, to determine whether this is the case will involve detailed investigations of signal transduction by vIL-6 and selected vIL-6 variants (such as vIL-6.C2 and vIL-6.23) to different downstream targets or signaling intermediates (e.g., STAT1, STAT3, AP-1, SHP2, and phosphoinositide 3-kinase) under carefully controlled conditions. Biological relevance could be addressed, for example, by investigating the effects of signaling by these proteins on cell proliferation, cell survival under stress conditions, and HHV-8 replication during reactivation. The prediction would be that the particular balance of signaling achieved via gp80-containing and -devoid signaling complexes by vIL-6 would favor HHV-8 replication in the particular cell types in which the virus replicates productively. Latency could also be promoted via proliferative or survival effects if vIL-6 can be expressed appreciably in the absence of lytic replication, as may be the case (9, 10, 23). In this regard, it is relevant that vIL-6 is secreted inefficiently, is largely intracellular, and can signal intracellularly (17) and therefore could be biologically active, even if its gene is transcribed and transcripts translated at low levels.
In summary, the structure-function studies presented here have established that the overall structure of vIL-6, in addition to particular gp130-interacting residues (11), is crucially important for gp80-independent signaling by vIL-6 and that gp80 can stabilize vIL-6-gp130 complexes. Studies are underway to determine the biological relevance of gp80-independent versus gp80/gp130 signaling to virus replication and cytokine-induced pathogenesis.
Acknowledgments
We thank Gordon Sandford and Fang Hu for advice about various experimental procedures and other members of the Nicholas laboratory, Young Bong Choi and Liping Feng, for general support.
This work was supported by NIH grant CA76445 from the National Cancer Institute.
REFERENCES
- 1.Aoki, Y., E. S. Jaffe, Y. Chang, K. Jones, J. Teruya-Feldstein, P. S. Moore, and G. Tosato. 1999. Angiogenesis and hematopoiesis induced by Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6. Blood 93:4034-4043. [PubMed] [Google Scholar]
- 2.Aoki, Y., M. Narazaki, T. Kishimoto, and G. Tosato. 2001. Receptor engagement by viral interleukin-6 encoded by Kaposi's sarcoma-associated herpesvirus. Blood 98:3042-3049. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, Y., R. Yarchoan, K. Wyvill, S. Okamoto, R. F. Little, and G. Tosato. 2001. Detection of viral interleukin-6 in Kaposi's sarcoma-associated herpesvirus-linked disorders. Blood 97:2173-2176. [DOI] [PubMed] [Google Scholar]
- 4.Boulanger, M. J., D. C. Chow, E. E., Brevnova, and K. C. Garcia. 2003. Hexameric structure and assembly of the interleukin-6/IL-6α-receptor/gp130 complex. Science 300:2101-2104. [DOI] [PubMed] [Google Scholar]
- 5.Boulanger, M. J., D. C. Chow, E. Brevnova, M. Martick, G. Sandford, J. Nicholas, and K. C. Garcia. 2004. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J. Mol. Biol. 335:641-654. [DOI] [PubMed] [Google Scholar]
- 6.Burger, R., F. Neipel, B. Fleckenstien, R. Savino, G. Ciliberto, J. R. Kalden, and M. Gramatzki. 1998. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood 91:1858-1863. [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of hepesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, M., J. Osborne, G. Bestetti, and Y. Chang. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, C.-J., L. J. Poole, P. S. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J.-C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow, D., X. He, A. L. Snow, S. Rose-John, and K. C. Garcia. 2001. Structure of an extracellular gp130 cytokine receptor complex. Science 291:2150-2155. [DOI] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Jones, K. D., Y. Aoki, Y. Chang, P. S. Moore, R. Yarchoan, and G. Tosato. 1999. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi's sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood 94:2871-2879. [PubMed] [Google Scholar]
- 14.Klouche, M., N. Brockmeyer, C. Knabbe, and S. Rose-John. 2002. Human herpesvirus 8-derived viral IL-6 induces PTX3 expression in Kaposi's sarcoma cells. AIDS 16:F9-F18. [DOI] [PubMed] [Google Scholar]
- 15.Li, H., and J. Nicholas. 2002. Identification of amino acid residues of gp130 signal transducer and gp80 α receptor subunit that are involved in ligand binding and signaling by human herpesvirus 8-encoded interleukin-6. J. Virol. 76:5627-5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, H., H. Wang, and J. Nicholas. 2001. Detection of direct binding of human herpesvirus 8-encoded interleukin-6 (vIL-6) to both gp130 and IL-6 receptor (IL-6R) and identification of amino acid residues of vIL-6 important for IL-6R-dependent and -independent signaling. J. Virol. 75:3325-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meads, M. B., and P. G. Medveczky. 2004. Kaposi's sarcoma-associated herpesvirus-encoded viral interleukin-6 is secreted and modified differently than human interleukin-6: evidence for a unique autocrine signaling mechanism. J. Biol. Chem. 279:51793-51803. [DOI] [PubMed] [Google Scholar]
- 18.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molden, J., Y. Chang, Y. You, P. S. Moore, and M. A. Goldsmith. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625-19631. [DOI] [PubMed] [Google Scholar]
- 20.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 21.Nador, R. G., E. Cesarman, A. Chadburn, D. B. Dawson, M. Q. Ansari, J. Sald, and D. M. Knowles. 1996. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood 88:645-656. [PubMed] [Google Scholar]
- 22.Nicholas, J., V. R. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H.-G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 23.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer, T. S., L. K. Sanders, and D. Nathans. 1995. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl. Acad. Sci. USA 92:9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatum, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences inmulticentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 26.Wan, X., H. Wang, and J. Nicholas. 1999. Human herpesvirus 8 interleukin-6 (vIL-6) signals through gp130 but has structural and receptor-binding properties distinct from those of human IL-6. J. Virol. 73:8268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]