Abstract
To study the regulation of herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) expression and processing in the absence of other cis and trans viral functions, a transgenic mouse containing the region encompassing the LAT promoter (LAP1) and the LAT 5′ exon through the 2.0-kb intron was created. LAT expression was detectable by reverse transcriptase PCR (RT-PCR) in a number of tissues, including the dorsal root ganglia (DRG), trigeminal ganglia (TG), brain, skin, liver, and kidney. However, when the accumulation of the 2.0-kb LAT intron was analyzed at the cellular level by in situ hybridization, little or no detectable accumulation was observed in the brain, spinal cord, kidney, or foot, although the 2.0-kb LAT intron was detected at high levels (over 90% of neurons) in the DRG and TG. Northern blot analysis detected the stable 2.0-kb LAT intron only in the sensory ganglia. When relative amounts of the spliced and unspliced LAT within the brain, liver, kidney, spinal cord, TG, and DRG were analyzed by real-time RT-PCR, splicing of the 2.0-kb LAT intron was significantly more efficient in the sensory ganglia than in other tissues. Finally, infection of both transgenic mice and nontransgenic littermates with HSV-1 revealed no differences in lytic replication, establishment of latency, or reactivation, suggesting that expression of the LAT transgene in trans has no significant effect on those functions. Taken together, these data indicate that the regulation of expression and processing of LAT RNA within the mouse is highly cell-type specific and occurs in the absence of other viral cis- and trans-acting factors.
Herpes simplex virus type 1 (HSV-1) latency has long been characterized by the accumulation of a single abundant RNA, the latency-associated transcript (LAT). From the 8.3- to 8.5-kb polyadenylated primary transcript, a very stable 2.0-kb intron, which can be readily detected in the nuclei of latently infected sensory neurons, is spliced (6, 21, 29, 32). Further processing of the 2.0-kb intron can yield a 1.5-kb product in a subpopulation of neurons (20, 31). In sensory ganglia, LAT expression seems to be tightly regulated; during the lytic infection, most neurons express either the LAT or lytic genes, not both (21). In addition, there is a sharp drop in the amount of LAT during reactivation (3, 30), with a decrease in transcriptional permissiveness of the LAT promoter occurring as early as 30 min postexplant (1). These observations, combined with genetic data describing LAT mutants as being defective in reactivation (2, 10, 13, 16, 24), have led to the model showing that the LAT RNA may play a role in transcriptional silencing of lytic genes during latency (2, 7, 15, 36). Additionally, a cis element encompassing the promoter and enhancer (rcr) may regulate LAT expression to allow the establishment of latency and the occurrence of reactivation (1, 14). Two central questions that have been difficult to address in vivo, however, are (i) how dependent the regulation of LAT transcription on other viral elements is and (ii) whether functions of the LAT rcr, or LAT transcription itself, can be substituted in trans to influence latency and reactivation or whether those functions act in the context of the HSV-1 genome (in cis).
Transgenic mice are a useful tool to isolate the effects of individual viral proteins on viral pathogenesis in the initial absence of other viral functions (25). Two HSV transgenic mouse lines containing portions of the LAT have been previously described. The first transgenic line contained the 2.0-kb HSV-1 intron under the control of a constitutive promoter (cytomegalovirus [CMV]) and was used to examine the effect of expression of the 2.0-kb LAT intron on acute and latent HSV-1 infections in vivo (18). In this study, high levels of expression were seen in most tissues, with the highest being in the heart and skeletal muscles, consistent with the known expression profile of the CMV promoter. Following infection with wild-type HSV-1, no significant difference in establishment of latency between the transgenic and nontransgenic mice was observed. In contrast, transgenic mice that were infected with an HSV-1 strain KOS-based LAT mutant reactivated this mutant more efficiently than nontransgenic mice. This suggested that the 2.0-kb LAT intron was able to rescue reactivation defects of LAT mutants in trans. A second transgenic mouse, containing the first 2.5 kb of the HSV-2 LAT (5′ exon and 2.2-kb intron) under control of its native promoter, was constructed (34). In this study, large amounts of the HSV-2 2.2-kb intron were detected in many tissues; however, no differences in viral replication, establishment or reactivation were observed as a consequence of expressing this portion of the LAT in trans. Significantly, and in contrast to the case with the HSV-1 LAT transgenic mouse, the reactivation phenotype of an HSV-2 LAT mutant was not able to be rescued in the HSV-2 transgenic mouse. It is not clear whether this discrepancy is due to inherent differences between HSV-1 and HSV-2, the fact that different portions of the LAT were used in these two transgenic mice, or that the HSV-1 transgenic mouse used the CMV promoter, which may confer a different expression profile. Neither of these studies examined quantitative differences in LAT expression or RNA processing in different tissues of the mouse in the context of the native LAT promoter. In addition, the previous HSV-1 transgenic mouse did not include the region of the LAT (rcr) that has been shown to be critical for induced reactivation. Therefore, the goal of the present study was to construct an HSV-1 transgenic mouse containing the native LAT promoter and first 2.5 kb of the primary transcript in order to (i) assess quantitative differences in LAT expression in different tissues and (ii) determine whether expression of the HSV-1 LAT in trans has an effect on viral replication during the acute infection, establishment of latency, or the ability to reactivate.
In order to study HSV-1 LAT transcription in vivo in the absence of cis- and trans-acting factors, we generated a transgenic mouse containing a 3,549-bp fragment from HSV-1 strain 17syn+ encompassing the native HSV-1 LAT promoter through the 2.0-kb LAT intron splice acceptor site. Northern blotting, quantitative reverse transcriptase PCR (RT-PCR), and in situ hybridization (ISH) revealed that although LAT RNA is detectable in a variety of tissues, accumulation and efficient splicing of the 2.0-kb LAT intron is limited to nervous tissue and primarily sensory ganglia. Notably, abundant nuclear LAT intron accumulation was detectable in approximately 90% of the primary sensory neurons, suggesting that all sensory neurons are capable of activating the LAT promoter and processing the LAT intron. This is significantly more than the one third of neurons containing latent HSV DNA that express LAT during a normal HSV infection (8, 22), and this result suggests that while most neurons are capable of expressing LAT, LAT expression is repressed during a natural infection in the majority of latently infected neurons. No LAT was detected in nonneuronal cells within the sensory ganglia.
We also found, upon infection of transgenic mice with wild-type HSV-1, no differences in viral yields during acute infection, establishment of latency, or reactivation from latency versus nontransgenic mice. This suggests that in order to exert its regulatory influence on latency and reactivation, LAT must be present within the context of the HSV-1 genome and does not act in trans.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain 17syn+ was propagated and its titers were determined on rabbit skin cells grown in Eagle's minimal essential medium (Invitrogen-Life Technologies, Carlsbad, CA) supplemented with 5% calf serum (Life Technologies), 292 μg of l-glutamine/ml, and antibiotics (250 U of penicillin/ml and 250 μg of streptomycin/ml).
Plasmids.
Plasmid pLAT/LAT was used to construct the transgenic mouse and contains the DraI-AatII fragment of the HSV-1 strain 17syn+ LAT region (nucleotides [nt] 116,516 to 121,549) inserted into the SmaI site of pBluescript SK(+) (Stratagene). The simian virus 40 poly(A) tail sequence was inserted into the pBluescript XbaI site, located immediately downstream of the LAT sequence. Plasmid pAatII (containing the 4.1-kb AatII fragment of nt 4,822 to 9,271 from HSV-1 strain 17+) and the Xist pB1/B10 plasmid (a generous gift of Jeannie Lee) (28) were used as standards for PCR analyses. Plasmids pATD17 (nt 118,863 to 119,343) and pATD19 (nt 119,628 to 119,975) were used to generate hybridization probes for the LAT 5′ exon and 2.0-kb intron, respectively.
Generation of the LAT transgenic mouse.
The LAT transgene contained in plasmid pLAT/LAT was microinjected into fertilized oocytes obtained from C3H/HeJ mice, which were then implanted into surrogate pseudopregnant females as described previously (11, 26). Founder mice were identified by both Southern blot and PCR analyses.
Analysis of transgenic mice by conventional PCR was performed on DNA prepared from mouse tails. Reaction mixtures contained 600 ng each of forward and reverse primers (Table 1), 20 μl Hot Master PCR mix (Brinkman Eppendorf), and 200 ng tail DNA in a 50-μl final reaction volume. PCRs were performed in an Ericomp thermal cycler (San Diego, CA) with 3 min each of 94°C, 55°C, and 72°C (1 cycle) and 1 min each of 94°C, 55°C, and 72°C (30 cycles), with the exception of primer AG31, which used an annealing temperature of 68°C.
TABLE 1.
Conventional PCR primer sequences
| PCR producta | Primer | Sequence (5′ to 3′) | Genome location (nt) |
|---|---|---|---|
| 1 | Promar 1 | GCA CGA TCC CGA CAA CAA TAA CAA C | 118,248-118,272 |
| Promar 2 | ACT TCC ACT TCC CGT CCT TCC ATC C | 118,329-118,353 | |
| 2 | DB60 | CGG CGA CAT CCT CCC CCT AAG C | 118,890-118,911 |
| DB61 | GAC AGA CGA ACG AAA CAT TCC G | 119,017-119,038 | |
| 3 | M int 1 | GAC ACG CAT TGG CTG GTG TAG TGG G | 120,797-120,821 |
| M int 2 | ACG AGG GAA AAC AAT AAG GGA CGC C | 120,874-120,896 | |
| 4 | M2 up | AGA CCC GCT GGT GTG TGG TG | 120,750-120,769 |
| M2 down | GAT GCC CCC CGA GTA CCC GA | 121,046-121,065 | |
| 5 | AG 29 | CGG GTA CTC GGG GGG CA | 121,047-121,063 |
| AG 30 | CTC GGG GGT CTC TAG CGT GG | 121,255-121,274 | |
| 6 | Splice Acc. F | CCT CTT GTC TCC CTC CCA GG | 121,400-121,419 |
| AG 31 | CGC CTC TTC CTC CTC TGC CT | 121,517-121,536 |
Designation of the PCR product that results from the indicated pair of primers. Locations of these products with respect to the LAT transgene are shown in Fig. 1C.
Founder mice were subsequently intercrossed with C57BL/6 mice to expand the lineages. A total of 10 such backcrosses were performed.
RNA extraction and analysis.
Transgenic mice were euthanized by halothane overdose and cervical dislocation. Cortex, rostral and caudal hypothalamus, cerebellum, spinal cord, olfactory bulb, TG, DRG, spleen, liver, kidney, skin, foot, heart, intestine, eye, and lung tissues were dissected and homogenized in 1.2 ml TRIzol reagent (Gibco). Following the addition of 0.2 volumes of chloroform, samples were centrifuged for phase separation. RNA was precipitated from the aqueous phase with 0.7 volumes of isopropanol, followed by DNase treatment using DNA-free (Ambion) according to the manufacturer's directions.
cDNA was prepared from tissue RNA by using Moloney murine leukemia virus reverse transcriptase and random hexamer priming. For each tissue sample, 500 ng of total RNA was added to 4 μl 5 × RT buffer (Invitrogen), 10 pmol random hexamers, 12.5 μM each of dATP, dTTP, dGTP, and dCTP, 200 units of Moloney murine leukemia virus reverse transcriptase (Invitrogen), and 20 units of RNasin (Promega) in a final volume of 20 μl. Reaction mixtures were incubated at 37°C for 1 h followed by 10-min heat inactivation at 100°C.
Quantitative real-time PCR.
Real-time PCRs were run in triplicate on an ABI Prism 7700 thermal cycler (Applied Biosystems) and contained cDNA or tail DNA, TaqMan universal PCR mix (Applied Biosystems), and target-specific TaqMan dye-labeled primer/probe (Assays by Design; Applied Biosystems). Primer and probe sequences are shown in Table 2. Standard curves were generated from serial dilutions (10 to 10,000 molecules) of the LAT transgenic plasmid (pLAT/LAT), plasmid pAatII, the Xist pB1/B10 plasmid, and HSV-1 genomic DNA. Cytoplasmic 18S rRNA (a generous gift from S. Moyer) was used as a positive control, and a range of 1,000 to 10,000,000 molecules was spiked in to generate standard curves. PCR was performed under the following conditions: 50°C for 2 min (1 cycle), 95°C for 10 min (1 cycle), and 95°C for 15 s followed by 60°C for 1 min (40 cycles).
TABLE 2.
Real-time PCR primer and probe sequences
| Targeta | Sequence (5′ to 3′) |
|---|---|
| 5′ LAT exon (a and a′) | GGC TCC ATC GCC TTT CCT (forward) |
| AAG GGA GGG AGG AGG GTA CTG (reverse) | |
| TCT CGC TTC TCC CC (probe) | |
| Spliced LAT(b and b′) | CAA CAA AGA CGC CGC GTT T (forward) |
| CCG CTT CCG CCT CCT C (reverse) | |
| CCG TCG GTG CCC TGG (probe) | |
| 2.0-kb LAT intron(c and c′) | CGC CCC AGA GGC TAA GG (forward) |
| GGG CTG GTG TGC TGT AAC A (reverse) | |
| CCA CGC CAC TCG CG (probe) | |
| HSV-1 polymerase | AGA GGG ACA TCC AGG ACT TTG T (forward) |
| CAG GCG CTT GTT GGT GTA C (reverse) | |
| ACC GCC GAA CTG AGC A (probe) | |
| Murine Xist | GCT CTT AAA CTG AGT GGG TGT TCA (forward) |
| GTA TCA CGC AGA AGC CAT AAT GG (reverse) | |
| ACG CGG GCT CTC CA (probe) |
Notations in parentheses correspond to positions in Fig. 4A.
Northern blot analysis.
Briefly, 5 μg of RNA was heated to 65°C for 15 min in the presence of 15.5 μl denaturing loading buffer (10 parts formamide, 3.5 parts 37.5% formaldehyde, and 2 parts 5× MOPS [morpholinepropanesulfonic acid]). Samples were loaded with RNA dye (50% glycerol, 1 mM EDTA, 0.25% bromophenol blue, 300 μg/μl ethidium bromide) onto 1% agarose-formaldehyde-MOPS denaturing gels. RNA was transferred to a ZetaProbe GT (Bio-Rad) nylon membrane according to the manufacturer's instructions. Hybridization was performed with random hexamer-primed (random labeling kit; Roche) [32P]dCTP-labeled pATD19 probe (see below) to detect expression of the 2.0-kb LAT intron. The washed and dried membrane was exposed to a phosphor screen for 5 h, and the screen was then scanned by a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Radiolabeled ISH.
DRG, kidney, foot, brain, and spinal cord tissues were harvested from transgenic mice, nontransgenic littermates, and infected nontransgenic mice. Tissues were fixed overnight in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS). Tissues were paraffin embedded and sectioned, and hybridization was performed as described previously (9, 33) with 35S-dCTP-labeled probes (1.5 × 105 cpm/site) derived from pATD17 (nt 118,863 to 119,343) and pATD19 (nt 119,628 to 119,975) plasmids. Slides were counterstained with Giemsa stain.
FISH.
For fluorescent in situ hybridization (FISH) analysis, mice were anesthetized and perfused transcardially with PBS followed by 4% PFA fixative in 0.1 M PBS. Tissue sections were postfixed with 4% PFA in 0.1 M PBS for 10 min and then sequentially washed in 2 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), diethyl pyrocarbonate-treated water, and 0.1 M triethanolamine hydrochloride (TEA). Tissue sections were then treated with 0.25% acetic anhydride in 0.1 M TEA (3 min), washed in 2 × SSC, and incubated with prehybridization buffer (50% formamide, 40 μg/ml salmon sperm DNA, 5 × SSC) at 45°C for 2 h. The LAT-specific riboprobe made from pATD19 was heated to 80°C for 10 min, chilled on ice, and then diluted to 1 μg/ml in preheated (55°C) hybridization buffer (50% formamide, 1 × Denhardt's solution, 10 mM EDTA, 10% dextran sulfate, 0.5 mg/ml yeast tRNA, 0.5 mg/ml salmon sperm DNA, 3 × SSC). Prehybridized tissue was incubated with the probe at 55°C overnight. After hybridization, the tissue was washed with 2 × SSC (10 min) and treated with 20 μg/ml of RNase A in 2× SSC at 37°C for 30 min, followed by serial washing with graduated dilutions of SSC at 55°C. Tissue was then equilibrated with 0.1 M PBS and incubated with anti-digoxigenin-fluorescein Fab fragments (Roche) diluted in 0.1 M PBS with 1 × blocking solution (Roche) for 30 min. Stained tissue sections were then washed with 0.1 M PBS, and coverslips were applied with Vectashield mounting medium (Vector Labs).
Statistical analyses.
Data were analyzed by analysis of variance and paired t tests using InStat version 3.05 for Macintosh (GraphPad Software, Inc.).
Footpad infection of mice.
For HSV-1 infections, HSV-1 LAT transgenic mice and nontransgenic littermates at least 6 weeks of age and of at least the F6 generation were used. Mice were anesthetized with halothane and subcutaneously injected with 0.05 ml of a 10% saline solution in each of the rear footpads. Four hours after saline treatment, mice were anesthetized intramuscularly with 0.010 to 0.020 ml of a ketamine cocktail (2.5 to 3.75 mg acepromazine/kg of body weight, 7.5 to 11.5 mg/kg xylazine, 30 to 45 mg/kg ketamine). Both rear footpads were abraded with an emery board to remove the keratinized layer of skin tissue. By use of a pipette tip, a total of 1 × 106 PFU (for acute infections) or 500 PFU (for latent analyses) of virus per mouse in a 50-μl volume was applied to footpads and allowed to adsorb for one hour, while mice remained under anesthesia. Mice were sacrificed by halothane overdose followed by cervical dislocation at acute times (days 1 to 4) or at latent times (at least 28 days postinfection).
Explant cocultivation of latently infected DRG.
Latently infected transgenic and nontransgenic mice were euthanized as described above and DRG dissected. Intact DRG were cultured in minimal essential medium with supplements on a rabbit skin cell monolayer at 37°C with 5% CO2 to detect reactivating virus as previously described (4). Cells were monitored daily for 14 days for the presence of a cytopathic effect (CPE).
RESULTS
The LAT 3549 transgenic mouse contains a single copy of the LAT 5′ exon through the 2.0-kb intron under the control of the native LAT promoter.
A LAT transgenic mouse line (LAT 3549) containing a 3,549-bp region encompassing the LAT promoter and first 2.5 kb of the primary LAT (Fig. 1) was generated as described in Materials and Methods. This transgenic line was initially characterized to determine the approximate number of copies of the LAT transgene in each cell, as well as to ensure that the transgene was intact. In order to determine the transgene copy number, a cellular gene, Xist, of known copy number (1 copy per X chromosome), was compared to the LAT transgene present in transgenic mouse tail DNA. Since Xist is present on the X chromosome, the number of copies of Xist in DNA taken from female mice was divided by 2 to standardize samples, making it possible to compare the LAT transgene to the cellular control for both male and female mice. The average copy number of the LAT transgene was determined to be 0.76 ± 0.24, indicating that only one copy of the transgene is present per mouse genome (data not shown).
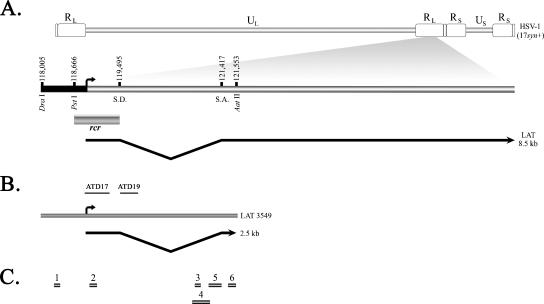
FIG. 1.
Diagram of the HSV-1 LAT region and the LAT transgene insert. (A) HSV-1 genomic map with expanded LAT regions, illustrating the LAT promoter (LAP1; black bar) extending from nt 118,005 to the transcriptional start site at nt 118,803 (arrow); the reactivation critical region (rcr), which encompasses the core LAT promoter (nt 118,666 to 118,803) to the splice acceptor site (S.A.) of the 2.0-kb intron at nt 119,495; and the 8.5-kb primary LAT. The location of the splice donor site (S.D.) is also shown. RL, repeat long region; UL, unique long region; RS, repeat short region; US, unique short region. (B) The region of the LAT included in the LAT 3549 transgenic mouse, from nt 118,005 to 121,553, which encompasses the LAP1 and the first 2.5 kb of the LAT primary transcript. The location of two DNA probes (ATD17 and ATD19) used for Northern blotting and in situ hybridization are shown. (C) Location of products generated by conventional PCR, as described in Materials and Methods (primers shown in Table 1), used to confirm the presence of the transgene.
The presence of the intact transgene was verified using PCR with gene-specific primers (Table 1). Amplification products were obtained for the LAT 5′ exon primers, as well as for the intron primers including the splice acceptor site, indicating that the entire transgene was present in the transgenic mouse line (Fig. 1C).
Abundant LAT expression is seen in a variety of tissues and is not age dependent.
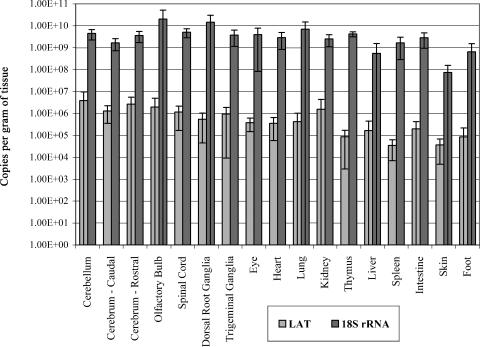
The transgenic mouse was then analyzed for the amount of LAT RNA produced in different tissues by real-time RT-PCR using primers specific for the LAT 5′ exon. Quantities were determined by comparison of tissue sample cycle threshold values to those of a standard curve generated from known quantities of pLAT/LAT plasmid DNA (Materials and Methods). When compared by weight, the spinal cord (P = 0.33), DRG (P = 0.69), TG (P = 0.40), skin (P = 0.17), and foot (P = 0.06) had no significant differences (analysis of variance) in the amount of LAT transgene expressed in 2-month-old mice (Fig. 2). Even when the amount of LAT RNA detected in each tissue was normalized to 18S rRNA, no statistically significant differences existed among the transgenic tissues (data not shown); however, LAT RNA was 1,000 to 10,000 times less abundant than the 18S rRNA present in the tissues examined (Fig. 2). It should be noted that the large standard error present for several of the tissues for both LAT and the 18S RNA is due to one or two samples in each of those groups for which RNA recovery was low.
FIG. 2.
LAT transcription in transgenic tissues. LAT transcription was measured by real-time RT-PCR, as described in Materials and Methods, using primers/probe specific for the LAT 5′ exon. The number of copies of LAT cDNA detected was calculated using a standard curve generated by spiking nontransgenic mouse tissues with 10-fold dilutions of a LAT target plasmid and was normalized per gram of tissue. Transgenic LAT RNA is detectable in both neural and nonneural tissues. Copies of cDNA detected for cellular 18S rRNA is also shown for each tissue. Error bars reflect standard errors (n = 4) for each tissue.
In previous studies with HSV-1 transgenic mice, ICP0 and ICP4 promoter-driven reporter expression levels demonstrated a difference in the two lytic gene promoters as a function of the age of the mice (17, 23). Therefore, our analysis of RNA expression included age as a parameter and examined the amount of LAT RNA present in select tissues at 1 day, 1 month, 2 months, and 18 months of age. No statistically significant differences in LAT transgene expression as a function of age were observed for any of the tissues analyzed and shown in Fig. 2 (data not shown).
The 2.0-kb LAT intron accumulates in sensory ganglia.
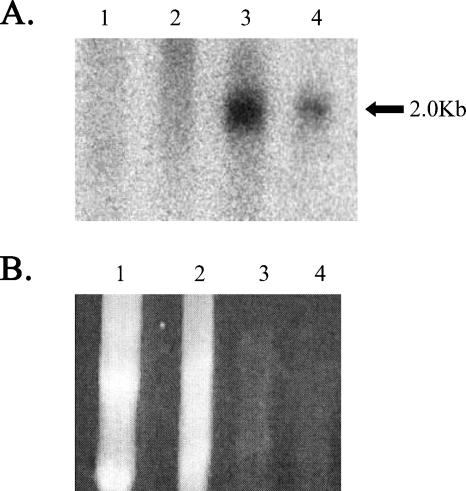
The RT-PCR analyses indicated that the LAT transgene is transcribed in all the tissues of the mouse that were examined. Since a hallmark of LAT expression during HSV-1 latency is the accumulation of the stable 2.0-kb intron within sensory neurons, Northern blot analysis was performed on tissues of both neuronal and nonneuronal origins to determine the tissue distribution of the 2.0-kb LAT intron in the transgenic mouse (Fig. 3). This revealed that the LAT intron accumulates in the DRG but not in the brain or kidney. It should be noted that no LAT intron was detected in the RNA from kidney or brain even when 10-fold more RNA than that in the DRG samples was loaded. The inability to detect the 2.0-kb LAT intron in tissues in which the primary transgene transcript was detected suggested that splicing of the LAT in nonneuronal tissues is inefficient or that the intron is destabilized in those tissues.
FIG. 3.
Detection of the 2.0-kb LAT intron in transgenic mouse tissues by Northern blot analysis. Lanes (representing total RNA from LAT transgenic mouse): 1, kidney; 2, brain; 3, DRG; 4, DRG of nontransgenic mice that were latently infected with HSV-1. (A) Northern blot probed for the 2.0-kb LAT intron. (B) Ethidium bromide staining of the gel. Note that the kidney and brain lanes contain 10-fold more RNA than the DRG lanes, in an attempt to detect the intron. Note that the Northern blot was hybridized with a probe specific for the 2.0-kb LAT intron as described in Materials and Methods.
Splicing of LAT occurs differentially among tissues.
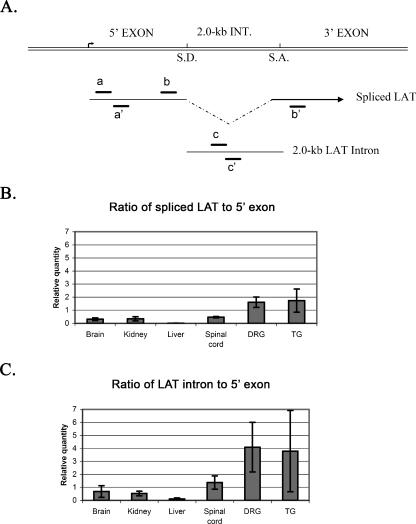
Splicing was further examined by performing real-time PCR on cDNA prepared from transgenic mouse brain, liver, kidney, spinal cord, TG, and DRG. TaqMan primer/probe sets were designed to detect the region corresponding to the LAT intron, the LAT 5′ exon, and the LAT exon 1-plus-exon 2 product formed when the 2.0-kb LAT intron is spliced out (Fig. 4A). As shown in Fig. 4B, compared to the LAT 5′ exon, a region that is present on both the primary transcript and the spliced LAT, the TG and DRG display levels of spliced LAT at ratios greater than or equal to one, suggesting that the LAT is efficiently spliced in these sensory ganglia. In contrast, the brain, kidney, liver, and spinal cord all displayed ratios below one, indicating that the majority of the LAT is unspliced in these tissues. When relative quantities of LAT intron region are compared to the LAT 5′ exon (Fig. 4C), the DRG and TG again show mean ratios well above one, indicating that the LAT intron actually accumulates more than the primary transcript in those tissues. This likely reflects the stability of the 2.0-kb LAT intron lariat relative to the primary or spliced transcript. The ratio of intron to 5′ exon in the spinal cord is significantly higher than that detected in the liver (t test, P = 0.04), indicating that there is some splicing of the LAT intron in the spinal cord but that this splicing is not as efficient as in the TG and DRG. Overall, these results suggest that while LAT is transcribed in most tissues of the transgenic mouse, splicing and accumulation of the 2.0-kb intron occur detectably only in the spinal cord, DRG, and TG and are considerably more efficient in the sensory ganglia.
FIG. 4.
RT-PCR analysis of LAT splicing in transgenic tissues. (A) Diagram of the HSV-1 LAT region illustrating the splice donor (S.D.) and splice acceptor (S.A.) sites. Locations of primers to detect the 5′ exon (a and a′), spliced LAT (b and b′), and the LAT intron (c and c′) are shown below the genome. Real-time (TaqMan) RT-PCR analyses were performed in triplicate and compared to cellular Xist RNA. (B) Ratio of spliced LAT (primers detect only the transcript after the intron is spliced out) to 5′ exon (detection of either the unspliced primary transcript or the spliced transcript). A ratio greater than one indicates efficient splicing in both DRG and TG. (C) Ratio of LAT intron normalized to LAT 5′ exon. Neuronal tissues exhibit a ratio greater than one, indicating accumulation of 2.0-kb LAT intron in these tissues. Error bars reflect standard errors (n = 4) for each tissue.
Expression of LAT is limited to a subset of neurons.
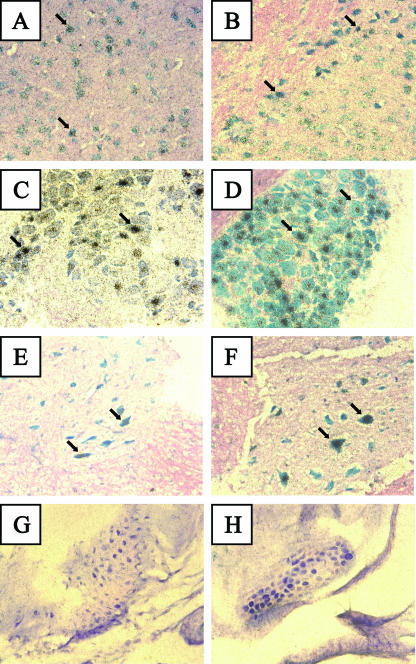
During experimental HSV-1 infection of mice, the LAT is preferentially expressed in a particular subset of neuronal cells of DRG and TG (37). In order to determine if the same specificity occurs in the LAT transgenic mouse, ISH was used to probe for either the LAT 5′ exon or the 2.0-kb LAT intron in DRG, kidney, foot, brain, and spinal cord tissue. Although the transgenic mouse demonstrated a small number of LAT-positive neurons in the thalamic neurons of the brain (Fig. 5A and B), there were considerably more LAT-positive neurons seen in the DRG (Fig. 5C and D). Additionally, a small number of neurons in the spinal cord (Fig. 5E and F) revealed LAT expression levels comparable to those in the DRG. In contrast to the real-time RT-PCR data for nonneuronal tissues, neither the transgenic foot (Fig. 5G and H) nor the transgenic kidney (not shown) displayed any detectable LAT expression by ISH.
FIG. 5.
In situ hybridization for LAT in transgenic tissues. Expression of the LAT transgene in individual cells was determined using a radiolabeled probe for either the 5′ exon (A, C, E, and G) or the LAT intron (B, D, F, and H). Positive hybridization signal, indicated by clusters of black/gray, silver grains, were seen with both probes in thalamic neurons (A and B), DRG neurons (C and D), and spinal cord neurons (E and H). No positive hybridization signal was observed in the feet (G and H). Arrows indicate representative positive cells in tissues in which positive hybridization was detected.
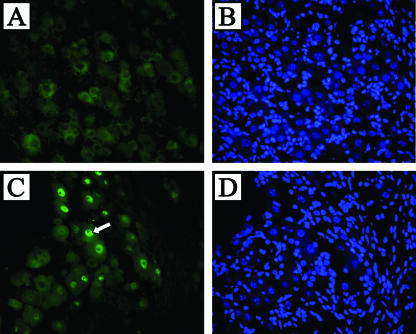
During HSV infection, LAT is detected by ISH in approximately one-third of latently infected neurons (8, 19). However, the LAT intron is readily detectable in approximately 90% of neurons by using FISH analysis of DRG and TG samples taken from the transgenic mouse with a probe transcribed from pATD19 (Fig. 6). Ganglia from four mice were batched and serial sectioned so that any given tissue section would contain a random sampling of ganglionic tissue. Two or three slides were then randomly selected and evaluated for the presence of LAT signal in Hoechst-positive neuronal cross sections. Evaluation of over 400 Hoechst-positive TG neuronal cross sections and 350 Hoechst-positive DRG cross sections revealed that 337/364 (92.5%) neuronal cross-sections with positive nuclear staining by Hoechst also had LAT signal. For TG, we similarly batched ganglia, chose three random slides from the middle of the block, and scored high-quality sections. In doing so, we found 371/414 (89.6%) with detectable LAT signal. This analysis failed to detect LAT hybridization in nonneuronal cells present in the DRG and TG, which are significantly more numerous than the neurons.
FIG. 6.
FISH of transgenic mouse TG detects LAT in the nuclei of most neurons but not glial cells. TG from LAT transgenic mice (C) or nontransgenic littermates (A) were hybridized with a probe to the HSV-1 LAT intron region and visualized by fluorescent microscopy. Nuclear staining was observed in most neurons. Hoechst blue staining of the same slides (B and D) revealed that almost all of the large sensory neuron nuclei present were FISH positive for LAT. Note that none of the glial cell nuclei, which make up the majority of total cells in this field, were positive for LAT. The white arrow denotes the single neuronal nucleus that was stained by Hoechst but was not FISH positive for LAT.
Taken together, these data suggest that while the LAT transgene RNA is transcribed at levels detectable by RT-PCR in most tissues, splicing of the 2.0-kb intron occurs preferentially in the sensory ganglia, and ISH detects its accumulation only in neurons.
Expression of LAT in trans does not detectably alter the course of an acute HSV-1 infection in mice.
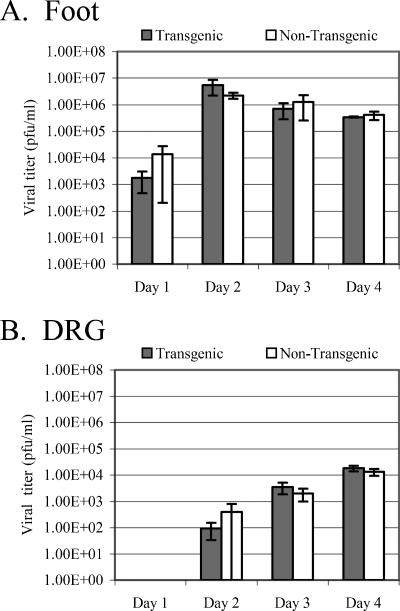
To determine whether expressing LAT in the context of the transgenic mouse would affect the course of HSV-1 infection by altering gene expression, transgenic and nontransgenic mice were infected with wild-type HSV-1 strain 17syn+. Following footpad infection, tissues in the foot and DRG were assayed for amounts of infectious virus present during the acute phase (days 1 to 4) of infection. t test analysis revealed no significant difference between the amounts of infectious virus present in the feet of transgenic and nontransgenic mice at any of the four time points tested (day 1, P = 0.35; day 2, P = 0.24; day 3, P = 0.55; day 4, P = 0.55) (Fig. 7A). Similarly, the amounts of infectious virus in the DRG during the acute infection in transgenic and nontransgenic mice exhibited no significant difference (day 1, not determined; day 2, P = 0.74; day 3, P = 0.43; day 4, P = 0.37) (Fig. 7B).
FIG. 7.
Evaluation of the LAT transgene's effect on acute replication of HSV-1 in vivo. Transgenic and nontransgenic mice were infected on both rear footpads with 1 × 106 PFU of HSV-1 strain 17syn+. Feet (A) and DRG (B) were harvested at the indicated times postinfection. The amount of infectious virus present in each tissue was determined by plaque assay on rabbit skin cells. Error bars reflect standard errors for the time points. Four mice per mouse strain per time point were assayed.
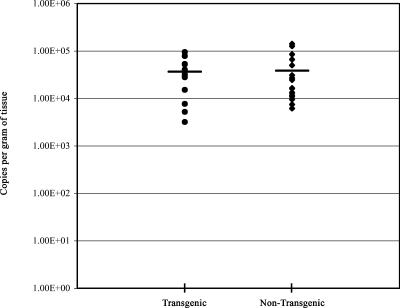
In order to determine whether the expression of the LAT transgene altered the establishment of latency in infected mice, transgenic and nontransgenic mice were infected with HSV-1 17syn+ and sacrificed at least 28 days postinfection. Despite the expression of LAT from the transgenic mouse, paired t test analysis detected no significant difference (t = 0.16, P = 0.87) in the efficiencies of establishing latency in DRG of transgenic and nontransgenic mice (Fig. 8). Thus, the expression of the LAT intron from one integrated copy in trans does not affect the ability of HSV to establish a latent infection.
FIG. 8.
Evaluation of the LAT transgene's effect on establishment of HSV-1 latency. Twenty-eight days postinfection with 500 PFU of HSV-1 strain 17syn+, the number of HSV-1 genomes present in DRG of either transgenic mice or nontransgenic littermates was determined by real-time (TaqMan) PCR. A horizontal bar indicates the average number of genomes in each group. Each symbol represents one mouse (n = 10).
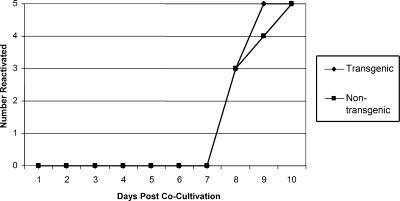
To study the effect of the LAT transgene on the reactivation phase of infection, DRG from latently infected mice were dissected and cultured on a cell monolayer for 14 days, with daily monitoring for CPE. In both the transgenic and nontransgenic mice, virus was evident in all of the cultures by day 10 postcocultivation (Fig. 9). There was also no observed difference in the time frames of reactivation for transgenic and nontransgenic mice.
FIG. 9.
Effect of LAT transgene on reactivation of HSV-1 from latently infected DRG. Transgenic and nontransgenic mice were infected with 500 PFU of HSV-1 strain 17syn+. Twenty-eight days postinfection, six DRG (lumbar ganglia L4, L5, and L6 from each side of the spine) per mouse were removed, pooled, and assayed for reactivation by explant cocultivation of the DRG on monolayers of rabbit skin cells for 14 days. A total of five mice (no. of mice on the y axis) for each group were independently assayed. Reactivation was measured by the appearance of CPE on the cell monolayers.
DISCUSSION
Studies using reporter constructs to examine LAT transcription during acute infection, both in cell culture and in vivo, have suggested that LAT promoter activity varies among different cell types and tends to be more robust within neurons (5, 38, 39). However, the LAT promoter has been shown to be active in nonneuronal cells in the epithelium of the mouse footpad following infection by this route, suggesting that LAT transcription in vivo is not limited solely to neurons (12). During latent infections, LAT expression within neurons seems to be tightly regulated; LAT is detectable only in approximately one-third of all neurons that contain HSV-1 genomes during latency (22), and these HSV-1 LAT-positive neurons tend to colocalize with a subset of sensory neurons that are positive for the SSEA3 and A5 markers (20, 37). This result suggested that LAT expression may be mediated by transcription factors that are more abundant in certain sensory neurons but does not exclude the involvement of other viral factors. While transient assays in cell culture have permitted study of the expression of the LAT promoter in the absence of other viral genes, cell culture analyses do not address differences in LAT expression and/or processing that may occur in the different classes of neurons in sensory ganglia in vivo.
This study represents the first quantitative analysis of HSV LAT expression in different tissues within the mouse. Through the analysis of LAT accumulation patterns by real-time RT-PCR, it was apparent that while LAT RNA could be detected in most tissues of transgenic mice, accumulation to levels detectable by ISH occurred preferentially in nervous tissue. No ISH-positive cells were observed in any of the nonneuronal tissues. At first glance, the finding of the LAT at comparable levels in all tissues appears to contradict the detection of ISH-positive cells only in neurons; however, examination of the amount of LAT present in whole tissue puts these observations in perspective. The RT-PCR analysis detected, at most, 1 × 107 copies of LAT RNA per gram of tissue. Most tissues of the mouse have been estimated to contain 1 × 108 to 1 × 109 cells on a per-gram basis; therefore, the amount of LAT present at the whole-tissue level is very low. This conclusion is confirmed by the amount of LAT detected relative to the cellular RT-PCR controls, in which the amount of LAT was on average 1/10,000 of the 18S rRNA present. This small abundance of LAT expression at the tissue level also applies to the nervous tissue in which LAT was detected by ISH. In this case, even though almost all of the sensory neurons seem to abundantly express the LAT, the neurons make up only approximately 10% of the total cells present within ganglia, and the contribution of additional axon volume to the weight biases the RT-PCR results of nervous tissue against the small population of cells that are actually expressing the LAT. Therefore, our conclusion is that most neurons are capable of high-level LAT expression, but other cell types normally express very little LAT.
The observation that approximately 90% of the sensory neurons express LAT in the context of the transgenic mouse is a significant contrast to the one third of the latently infected neurons that normally express LAT during an HSV-1 infection. This strongly suggests that the bias of LAT expression to a subset of latently infected neurons reflects the activity of viral components outside of the transgene used here or a response of the cells to the viral infection in a way that alters the ability of the LAT to be transcribed. One possibility is that LAT expression may be regulated in an epigenetic manner, in response to genome copy number within a particular cell or to cellular factors present during the establishment of latency.
Another significant finding of this study was that LAT splicing occurs more efficiently in neuronal tissue than in nonneuronal tissue (Fig. 4). Both the TG and DRG levels of spliced LAT occurred at ratios greater than or equal to one relative to the 5′ exon, while the brain, kidney, liver, and spinal cord all displayed ratios below one, indicating that more 5′ exon than spliced product was present. This finding suggests possible differences in splicing machinery or processing of spliced message between sensory ganglia and other tissue. In addition, the ratios of the 2.0-kb LAT intron to 5′ exon were proportionally consistent with the ratio of spliced LAT to 5′ exon, though the magnitudes of product in the spinal cord, DRG, and TG were severalfold higher for the intron than for the 5′ exon. This is likely due to the higher stability of the 2.0-kb intron relative to the splice product (27). It should be noted that while this analysis suggests that the transgene fragment contains those cis elements sufficient to direct accumulation of the 2.0-kb LAT intron primarily within sensory neurons, it does not exclude the possibility that other cis elements located in the HSV-1 genome outside the confines of the LAT transgene may play a role in modulating LAT expression and RNA processing.
Finally, when the transgenic mice were infected with wild-type HSV-1 strain 17syn+, the LAT transgene had no effect on the establishment or reactivation of latent virus. This particular study does not rule out an effect in which one neuron, because of viral copy number, expresses abundant levels of LAT that restrain the lytic cycle. Previous characterization of a transgenic mouse containing an HSV-2 LAT transgene yielded a similar result (35). These findings together suggest that pre-expression of the LAT does not act in trans during establishment or reactivation to alter the course of a wild-type HSV-1 infection.
Acknowledgments
This work was supported by grants AI48633 (D.C.B.), EY10008 (T.P.M.), EY02162 (T.P.M.), and NS40500 (G.F.R.) from the National Institutes of Health, by an Investigator in Pathogenesis Award from the Burroughs Wellcome Fund (D.C.B.), and by an RPB Senior Scientific Investigator Award (T.P.M.). N.V.G. was supported by training grant AI07110 from the National Institutes of Health.
We also acknowledge helpful comments and suggestions from A. Amelio, J. Feller, and J. Hill and thank P. McAnany for technical assistance and help with the figures.
REFERENCES
- 1.Amelio, A. L., N. V. Giordani, N. J. Kubat, J. E. O'Neil, and D. C. Bloom. 2006. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J. Virol. 80:2063-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloom, D. C., J. T. Hill, E. K. Wagner, L. F. Feldman, and J. G. Stevens. 1996. A 348-bp region in the latency associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgin, M. A., R. L. Smith, and C. L. Wilcox. 2001. Inducible cyclic AMP early repressor produces reactivation of latent herpes simplex virus type 1 in neurons in vitro. J. Virol. 75:2912-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson, A. T., T. P. Margolis, W. A. Gomes, and L. T. Feldman. 1995. In vivo deletion analysis of the herpes simplex virus type 1 latency associated transcript promoter. J. Virol. 69:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garber, D. A., P. A. Schaffer, and D. M. Knipe. 1997. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 71:5885-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gressens, P., and J. R. Martin. 1994. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J. Virol. Methods 46:61-83. [DOI] [PubMed] [Google Scholar]
- 9.Haase, A. T., D. Walker, L. Stowring, P. Ventura, A. Geballe, H. Blum, M. Brahic, R. Goldberg, and K. O'Brien. 1985. Detection of two viral genomes in single cells by double-label hybridization in situ and color microradioautography. Science 227:189-192. [DOI] [PubMed] [Google Scholar]
- 10.Hill, J. M., F. Sedarati, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 11.Hogan, B., F. Constantini, and E. Lacy. 1986. Manipulating the mouse genome. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 12.Jarman, R. G., E. K. Wagner, and D. C. Bloom. 1999. LAT expression during an acute HSV infection in the mouse. Virology 262:384-397. [DOI] [PubMed] [Google Scholar]
- 13.Krause, P. R., L. R. Stanberry, N. Bourne, B. Connelly, J. F. Kurawadwala, A. Patel, and S. E. Straus. 1995. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J. Exp. Med. 181:297-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leib, D. A., C. L. Bogard, V. M. Kosz, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loiacono, C. M., R. Myers, and W. J. Mitchell. 2002. Neurons differentially activate the herpes simplex virus type 1 immediate-early gene ICP0 and ICP27 promoters in transgenic mice. J. Virol. 76:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mador, N., E. Braun, H. Haim, I. Ariel, A. Panet, and I. Steiner. 2003. Transgenic mouse with the herpes simplex virus type 1 latency-associated gene: expression and function of the transgene. J. Virol. 77:12421-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maggioncalda, J., A. Mehta, Y. H. Su, N. W. Fraser, and T. M. Block. 1996. Correlation between herpes simplex virus type 1 rate of reactivation from latent infection and the number of infected neurons in trigeminal ganglia. Virology 225:72-81. [DOI] [PubMed] [Google Scholar]
- 20.Margolis, T. P., C. R. Dawson, and J. H. LaVail. 1992. Herpes simplex viral infection of the mouse trigeminal ganglion. Immunohistochemical analysis of cell populations. Investig. Ophthalmol. Vis. Sci. 33:259-267. [PubMed] [Google Scholar]
- 21.Margolis, T. P., F. Sedarati, A. T. Dobson, L. T. Feldman, and J. G. Stevens. 1992. Pathways of viral gene expression during acute neuronal infection with HSV-1. Virology 189:150-160. [DOI] [PubMed] [Google Scholar]
- 22.Mehta, A., J. Maggioncalda, O. Bagasra, S. Thikkavarapu, P. Saikumari, T. Valyi-Nagy, N. W. Fraser, and T. M. Block. 1995. In situ DNA PCR and RNA hybridization of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology 206:633-640. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, W. J. 1995. Neurons differentially control expression of a herpes simplex virus type 1 immediate-early promoter in transgenic mice. J. Virol. 69:7942-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perng, G.-C., E. C. Dunkel, P. A. Geary, S. M. Slanina, H. Ghiasi, R. Kaiwar, A. B. Nesburn, and S. L. Wechsler. 1994. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J. Virol. 68:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rall, G. F., D. M. Lawrence, and C. E. Patterson. 2000. The application of transgenic and knockout mouse technology for the study of viral pathogenesis. Virology 271:220-226. [DOI] [PubMed] [Google Scholar]
- 26.Rall, G. F., L. Mucke, and M. B. Oldstone. 1995. Consequences of cytotoxic T lymphocyte interaction with major histocompatibility complex class I-expressing neurons in vivo. J. Exp. Med. 182:1201-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, M. K., G. B. Devi-Rao, and E. K. Wagner. 1993. Latent phase transcription by alphaherpesviruses, p. 305-324. In K. Adolph (ed.), Genome research in molecular medicine and virology. Academic Press, Orlando, Fla.
- 28.Shibata, S., and J. T. Lee. 2003. Characterization and quantitation of differential Tsix transcripts: implications for Tsix function. Hum. Mol. Genet. 12:125-136. [DOI] [PubMed] [Google Scholar]
- 29.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spivack, J. G., and N. W. Fraser. 1988. Expression of herpes simplex virus type 1 latency-associated transcripts in the trigeminal ganglia of mice during acute infection and reactivation of latent infection. J. Virol. 62:1479-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spivack, J. G., G. M. Woods, and N. W. Fraser. 1991. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT): translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J. Virol. 65:6800-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens, J. G., E. K. Wagner, R. G. B. Devi, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056-1059. [DOI] [PubMed] [Google Scholar]
- 33.Stroop, W. G., D. L. Rock, and N. W. Fraser. 1984. Localization of herpes simplex virus in the trigeminal and olfactory systems of the mouse central nervous system during acute and latent infections by in situ hybridization. Lab. Investig. 51:27-38. [PubMed] [Google Scholar]
- 34.Wang, K., L. Pesnicak, E. Guancial, P. R. Krause, and S. E. Straus. 2001. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J. Virol. 75:8166-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, K., E. Prikhodko, L. Pesnicak, J. I. Cohen, and S. E. Straus. 2002. Presented at the 27th International Herpesvirus Workshop, Cairns, Australia, 20 to 26 July 2006.
- 36.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 102:16055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, L., C. C. Voytek, and T. P. Margolis. 2000. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 74:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zwaagstra, J. C., H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 1991. Identification of a major regulatory sequence in the latency associated transcript (LAT) promoter of herpes simplex virus type 1 (HSV-1). Virology 182:287-297. [DOI] [PubMed] [Google Scholar]
- 39.Zwaagstra, J. C., J. C. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]