Abstract
In this study, we demonstrate that the coactivator-associated arginine methyltransferase 1 (CARM1), which methylates histone H3 and other proteins such as p300/CBP, is positively involved in the regulation of Tax transactivation. First, transfection studies demonstrated that overexpression of CARM1 wild-type protein resulted in increased Tax transactivation of the human T-cell lymphotropic virus type 1 (HTLV-1) long terminal repeat (LTR). In contrast, transfection of a catalytically inactive CARM1 methyltransferase mutant did not enhance Tax transactivation. CARM1 facilitated Tax transactivation of the CREB-dependent cellular GEM promoter. A direct physical interaction between HTLV-1 Tax and CARM1 was demonstrated using in vitro glutathione S-transferase-Tax binding assays, in vivo coimmunoprecipitation, and confocal microscopy experiments. Finally, chromatin immunoprecipitation analysis of the activated HTLV-1 LTR promoter showed the association of CARM1 and methylated histone H3 with the template DNA. In vitro, Tax facilitates the binding of CARM1 to the transcription complex. Together, our data provide evidence that CARM1 enhances Tax transactivation of the HTLV-1 LTR through a direct interaction between CARM1 and Tax and this binding promotes methylation of histone H3 (R2, R17, and R26).
Human T-cell lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia (41, 51) and chronic inflammatory diseases such as HTLV-1-associated myelopathy/tropical spastic paraparesis (14, 36, 39), HTLV-1-associated arthropathy (38), uveitis (35), and Sjögren's syndrome (47). HTLV-1 encodes a 40-kDa protein, Tax, which is critical for viral replication, transformation, and gene regulation (1, 16, 37, 45). Tax drives viral gene expression from three imperfect 21-bp repeat enhancer elements located within the U3 region of the HTLV-1 long terminal repeat (LTR) (22, 29). Tax facilitates transcription through interaction with cellular factors such as CREB, histone acetyltransferase p300/CBP, and P/CAF (3, 12, 15, 17, 18, 20, 26, 30, 34, 46). Our laboratory and others have also demonstrated that Tax interacts with histone deacetylase 1 and regulates binding of the repressor to the HTLV-1 promoter (28, 32).
Coactivator-associated arginine methyltransferase 1 (CARM1) (also known as protein arginine methyltransferase 4 [PRMT4]) is a member of the PRMTs, which catalyze the transfer of methyl groups from S-adenosyl-l-methionine to the guanidine nitrogens of arginine (13). CARM1 preferentially methylates histone H3 in vitro and in vivo (4, 6), and mapping of residues demonstrated specificity for arginine 2 (R2), R17, and R26 (44). A recent study suggested that the CARM1 and histone acetyltransferase CBP or p300 interact synergistically to activate transcription of the estrogen receptor promoter (23, 27). Along these lines, Daujat et al. reported that the cooperation between acetylation and arginine methylation comes from the fact that acetylation at K18 and K23 tethers recombinant CARM1 to the H3 tail, increasing the efficiency of the arginine methyltransferase (11). In addition, it has been shown that CARM1 coactivates p53-dependent transcription and cooperates with β-catenin to enhance transcriptional activation by the lymphoid enhancer factor 1/T-cell factor 4 (LEF1/TCF4) (2, 24).
We now present evidence that the methyltransferase CARM1 plays an important role in Tax transactivation of the HTLV-1 LTR. Tax transactivation was increased significantly in CARM1-overexpressing cells. Expression of the wild type (WT), but not a methyltransferase mutant, enhanced Tax transactivation. Consistent with a positive role of CARM1 in Tax transactivation, a small interfering RNA (siRNA) to CARM1 inhibited CARM1 expression and Tax transactivation. A direct physical interaction between Tax and CARM1 was demonstrated both in vitro and in vivo. Finally, analysis of transcription preinitiation complexes (PICs) and chromatin immunoprecipitation (ChIP) assays provides evidence that Tax facilitates CARM1 binding to the template DNA and that CARM1 is bound to the active HTLV-1 LTR promoter in vivo.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, 293T, Jurkat, Molt-4, and HTLV-1-transformed C81 cells were obtained from the American Type Culture Collection (ATCC). HeLa and 293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine and penicillin/streptomycin and transfected using Effectene transfection reagent (QIAGEN) as described by the manufacturer. Jurkat, Molt-4, and C81 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and penicillin/streptomycin and transfected using Superfect transfection reagent (QIAGEN) as described by the manufacturer. SP cells were obtained from the NIH AIDS Research and References Reagent Program, Division of AIDS, NIAID, NIH, and maintained in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin/streptomycin, and IL-2 (Sigma).
Plasmids and luciferase assay.
The reporter construct HTLV-1 LTR-Luc and Tax wild-type and mutant M29, M30, and M32 expression plasmids were kindly provided by Genoveffa Franchini (NIH) and Warner Greene (University of California, San Francisco), respectively. A Tax internal deletion mutant (del 151-204) was constructed as described previously (40). CARM1 wild-type and catalytically inactive mutant expression plasmids were provided by Michael R. Stallcup (University of Southern California). A GTP binding protein expressed in mitogen-stimulated T cells (GEM)-Luc and the GEM (del CREB) mutant were kindly provided by Marc Montminy (Salk Institute for Biological Studies). The transfection with siRNA for CARM1 (Santa Cruz Biotechnologies) or chloramphenicol acetyltransferase (CAT) (QIAGEN) was performed 12 h prior to DNA transfection using RNAifect transfection reagent (QIAGEN). For luciferase assays, cell lysates were prepared 24 h after transfection following the instructions of the Promega dual luciferase and Tropix GalactoLight assay kits. All transfections included the control plasmid RSV β-Gal (Rous sarcoma virus with β-galactosidase expression) to control for transfection efficiency.
Western blotting.
Cells were prepared by using lysis buffer (50 mM Tris, pH 7.4, 120 mM sodium chloride, 5 mM EDTA, 0.5% Nonidet P-40, 50 mM sodium fluoride, 0.2 mM sodium vanadate). The extracts were incubated on ice for 15 min and centrifuged at 10,000 × g at 4°C, and supernatants were collected. Protein concentrations were determined by Bradford assay (Bio-Rad), and 50 to 100 μg was separated by electrophoresis in a 4 to 20% Tris-glycine gel (Novex). The proteins were then transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon) and probed with antibodies as indicated. Anti-CARM1 (CT) purchased from Upstate and anti-Tab172 monoclonal antibodies were used to detect the expression of Tax protein.
In vitro binding assay.
Five hundred nanograms of glutathione S-transferase (GST)-Tax or GST was incubated with 250 ng of the purified CARM1 protein (Upstate) in 400 μl of binding buffer (50 mM HEPES, pH 7.9, 50 mM NaCl, 0.1% Tween 20, 10% glycerol, 0.2 mM phenylmethlysulfonyl fluoride, 1 mM dithiothreitol, and 1× protease inhibitor cocktail) at 4°C for 1 h. Ten microliters of glutathione-Sepharose was added, and the mixture was incubated for 1 h at 4°C. Complexes were washed four times with the washing buffer (140 mM NaCl, 1 mM EDTA, 0.5% NP-40, 20 mM Tris, pH 8.0, 5% glycerol, 1 mM dithiothreitol, 0.2 mM phenylmethlysulfonyl fluoride) and eluted in sodium dodecyl sulfate sample loading buffer. The eluents were separated by electrophoresis on 4 to 20% Tris-glycine gel (Novex). The proteins were then transferred to PVDF membranes (Immobilon) and analyzed for CARM1 (Upstate) or GST (Santa Cruz).
Coimmunoprecipitation assay.
For analysis of the interaction between Tax and CARM1, nuclear extracts were prepared by using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) as described by the manufacturer. Nuclear extracts (500 μg) from C81 cells or Tax-transfected 293T cells were immunoprecipitated with anti-CARM1 antibody. Immunoprecipitates were denatured, and proteins were separated by electrophoresis on 4 to 20% Tris-glycine gels (Novex). The proteins were then transferred to PVDF membranes and analyzed for Tax or CARM1.
Immunofluorescence.
For immunostaining, C81 cells were cultured on coverslips, fixed with 1% formaldehyde in phosphate-buffered saline (PBS) for 15 min on ice, and permeabilized in cold methanol for 2 min. The permeabilized cells were incubated with 10% normal goat serum in PBS for 1 h, followed by immunostaining with an anti-Tax mouse monoclonal antibody and an anti-CARM1 rabbit polyclonal antibody. Alexa Fluor 488-conjugated anti-mouse immunoglobulin G (IgG) antibody and Alexa Fluor 594-conjugated anti-rabbit IgG antibody were used as secondary antibodies. The immunostained cells were mounted with medium containing DAPI (4′,6′-diamidino-2-phenylindole [Vectashield]; Vector Labs) and were visualized by use of a Leica confocal microscope.
Purification of PICs and analysis of protein components of PICs.
Purification of PICs was carried out as described previously using biotinylated templates (33, 55). Briefly, PICs were assembled by incubating biotinylated HTLV-1 templates (4× TRE G-free cassette) with HeLa nuclear extracts in the absence or presence of the His6-Tax wild type or mutant (del 151-204) and then purified with streptavidin-coated magnetic beads (Dynal Biotech). The protein components of PICs were analyzed by Western blotting with anti-Tab172, -CARM1, -CREB, or -p300 antibody (Upstate).
ChIP assay.
The ChIP assay was carried out using 6 to 10 μg of anti-Tab172, -CARM1, -dimethyl histone H3 (R2, R17, R26, and K9), -histone H3, or -acetyl-K9 antibody following the methods previously described (33). After cross-linking proteins to DNA by 0.5% formaldehyde in SP cells, chromatin was sonicated four times for 10 s each, generating DNA fragments of 100 to 500 bp. The nucleosomes were then precleared with glycogen-coated protein A/G agarose beads (Pierce). The supernatants were diluted 10-fold with ChIP dilution buffer, and the different antibodies indicated above were added. After overnight rotation at 4°C, the immune complexes were collected by addition of protein A-agarose beads. DNA was purified by proteinase K digestion, phenol extraction, and ethanol precipitation and amplified by PCR using primers specific for HTLV-1 LTR (5′-CCACAGGCGGGAGGCGGCAGAA-3′ and 5′-CATAAGCTCAGACCTCCGGGAAG-3′) and primers specific for β-globin (5′-CAATTTGTACTGATGGTATGG-3′ and 5′-GGTGTCTGTTTGAGGTTGC-3′).
RESULTS
Overexpression of CARM1 increases Tax transcriptional activity of the HTLV-1 LTR.
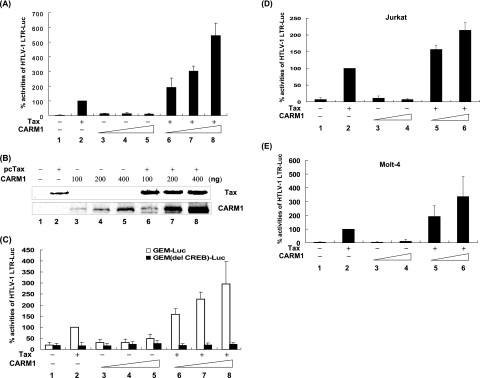
To examine whether CARM1 plays a role in Tax transactivation of the HTLV-1 LTR, we first compared the relative level of Tax transactivation in the presence and absence of exogenous CARM1. HeLa cells were transfected with the HTLV-1 luciferase reporter in the presence or absence of Tax expression plasmid (pcTax) and increasing concentrations of CARM1. At 24 h posttransfection, cell lysates were prepared and luciferase and control β-Gal activities were measured. In the absence of Tax, low levels of basal LTR activity were detected (Fig. 1A, lane 1). In the presence of Tax, a 20-fold increase in LTR transcrption activity was observed (lane 2). Transfection of increasing amounts of CARM1 expression plasmid, in the absence of Tax, resulted in a modest three- to fourfold increase in LTR activity (lanes 3 to 5). In contrast, in the presence of Tax and CARM1, a 130-fold increase in Tax transactivation was observed at the highest level of CARM1 tested (lane 8). Analysis of Tax protein expression in the transfected cells demonstrated that there was a slight increase in the level of Tax in the CARM1-overexpressing cells (Fig. 1B, lanes 2 and 6). Protein quantitation demonstrated that the increase was twofold or less. As the level of CARM1 was increased and LTR transcription activity increased significantly (Fig. 1A), the level of Tax protein stayed constant (Fig. 1B, lanes 6 to 8). These results suggest that CARM1 may play a regulatory role in Tax transactivation. Consistent with these results, we have also observed that Tax transactivation was defective in CARM1−/− compared to CARM1+/+ cells (data not shown).
FIG. 1.
Overexpression of CARM1 increases Tax transcriptional activity of the HTLV-1 LTR. (A to C) HeLa cells were transiently transfected using Effectene transfection reagent (QIAGEN) with reporter construct (0.1 μg) HTLV-1 LTR-Luc (A) or GEM- or GEM(del CREM)-Luc (C), RSV β-Gal (0.05 μg), CARM1 (0.1, 0.2, or 0.4 μg), or Tax (0.1 μg) expression plasmids. At 24 h posttransfection, cells were collected, and luciferase activities were measured. (B) Western blot analysis was performed for Tax (Tab172) and CARM1 (Upstate). (D and E) Jurkat and Molt-4 cells were transfected using Superfect transfection reagent (QIAGEN) with HTLV-1 LTR-Luc (1 μg), RSV β-Gal (0.5 μg), CARM1 (1 or 2 μg), or Tax (1 μg) expression plasmids. All luciferase values were adjusted for transfection efficiency using RSV β-Gal. The graph represents the luciferase activity from three independent experiments. The standard deviation for the three experiments is included.
To examine the effect of CARM1 on Tax transactivation of a cellular CREB-responsive promoter, we chose the GEM promoter recently identified by Conkright et al. (9), which contains the CREB response sequence TGACGTCA. In vivo ChIP experiments carried out in this laboratory have demonstrated that, similar to the HTLV-1 LTR, Tax binds to the GEM promoter along with CREB and CBP (in preparation).
Cotransfection of the GEM reporter plasmid with Tax resulted in a fivefold increase in luciferase activity (Fig. 1C, lane 2). Tax transactivation was dependent upon the CREB response element in the GEM promoter since deletion of this sequence abolished Tax transactivation (lane 2). Overexpression of CARM1 in the absence of Tax did not lead to an increase in GEM promoter activity (lanes 3 to 5). In contrast, overexpression of CARM1 in the presence of Tax resulted in a 15-fold increase in GEM WT promoter activity (lanes 6 to 8). Coexpression of Tax and CARM1 failed to transactivate the CREB-negative GEM promoter (lanes 6 to 8).
We have also tested the effect of CARM1 on Tax transactivation in lymphocyte cell lines. Jurkat and Molt-4 cells were transfected with the HTLV-1 LTR reporter plasmid in the presence of Tax, CARM1, or Tax plus CARM1. Increases in HTLV-1 reporter activity of 14- and 25-fold were observed in the presence of Tax in Jurkat and Molt-4 cells, respectively (Fig. 1D and E). Cotransfection of CARM1 alone led to no increase in promoter activity in Jurkat cells and a modest two- to threefold increase in Molt-4 cells. In the presence of Tax and CARM1, 30- and 84-fold increases in HTLV-1 promoter activity were observed (lanes 5 and 6).
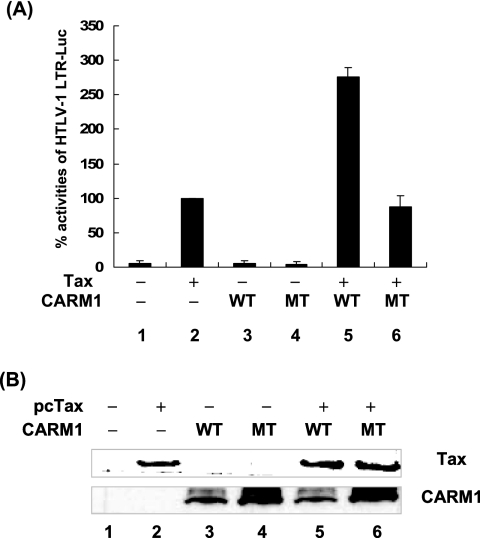
CARM1 methyltransferase activity is required for Tax transactivation.
In another set of experiments, the CARM1 WT or a methyltransferase mutant was transfected in the presence or absence of Tax. At 24 h posttransfection, cell lysates were prepared and luciferase activities driven by the HTLV-1 LTR were measured (Fig. 2A). WT Tax transactivated HTLV-1 LTR activity 20-fold (lane 2). Tax-transactivated HTLV-1 LTR activity was increased to 55-fold in the presence of exogenous CARM1 wild-type protein (lane 5). In contrast, there was no significant change in Tax transactivation in the presence of the CARM1 mutant (lanes 2 and 6). Transfection with the CARM1 wild type or mutant alone had no significant effect on basal HTLV-1 LTR transcription (lanes 3 and 4). The increase in Tax transactivation in the presence of CARM1 was not due to increased Tax expression (Fig. 2B). In these experiments, the levels of Tax protein were the same in the presence or absence of CARM1. These results provide evidence that the CARM1 methyltransferase activity plays a role in transcriptional regulation of the HTLV-1 LTR.
FIG. 2.
CARM1 mutant does not influence Tax transcription of the HTLV-1 LTR. (A) HeLa cells were transiently transfected using Effectene transfection reagent (QIAGEN) with reporter construct HTLV-1 LTR-Luc (0.1 μg), RSV β-Gal (0.05 μg), CARM1 WT or MT (0.1 μg), or Tax (0.1 μg) expression plasmids. At 24 h posttransfection, cells were collected and luciferase activities were measured. Luciferase values were adjusted for transfection efficiency using RSV β-Gal. The graph represents the luciferase activity from three independent experiments. The standard deviation for the three experiments is included. (B) Western blot analysis was performed for Tax (Tab172) and CARM1 (Upstate).
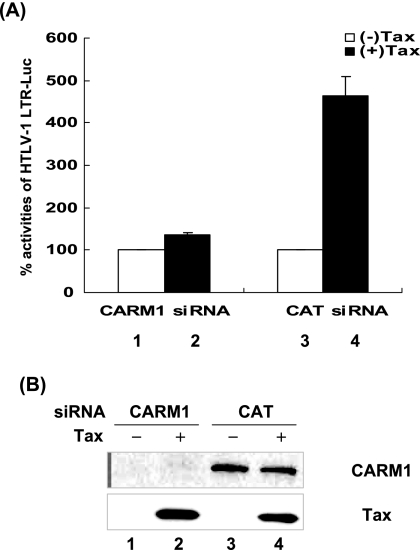
CARM1 siRNA abolishes Tax transactivation.
Another way to analyze the importance of CARM1 in Tax transactivation was to utilize an siRNA to knock out expression of CARM1. Cells were transfected with Tax and CARM1 in the presence or absence of CARM1 siRNA (Fig. 3). In the control experiment, in which a control CAT siRNA was transfected, Tax transactivation was observed (lane 4). The overall level of Tax transactivation is lower in this experiment because Tax protein expression was decreased in the presence of the siRNA in the transfection mix. Importantly, in the presence of the CARM1 siRNA, Tax transactivation was abolished. The Western blot analysis shown in Fig. 3B demonstrates that the CARM1 siRNA, but not the control CAT siRNA, reduced CARM1 expression in the transfected cells. Tax protein levels were equivalent in the control CAT and CARM1 siRNA-transfected cells.
FIG. 3.
Blocking endogenous CARM1 using siRNA prevents Tax activation of HTLV-1 LTR. (A) siRNA (100 nM) for CARM1 (Santa Cruz Biotechnologies) or CAT (QIAGEN) was transfected with RNAifect reagent (QIAGEN) into HeLa cells. After 12 h, cells were transiently transfected using Effectene transfection reagent with reporter construct HTLV-1 LTR-Luc (0.1 μg), RSV β-Gal (0.05 μg), or Tax (0.1 μg) expression plasmids. At 24 h posttransfection, cells were collected, and luciferase activities were measured. Luciferase values were adjusted for transfection efficiency using RSV β-Gal. The graph represents the luciferase activity from three independent experiments. The standard deviation for the three experiments is included. (B) Western blot analysis was performed for Tax (Tab172) and CARM1 (Upstate).
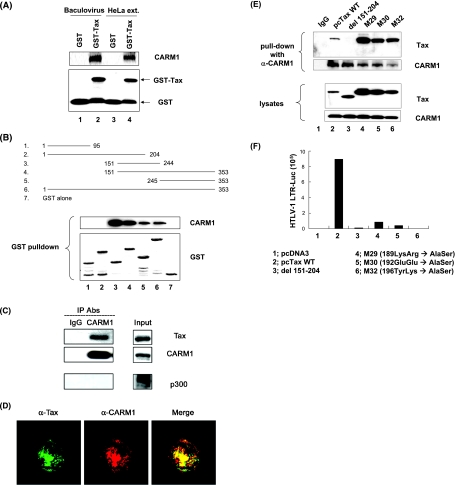
CARM1 and Tax physically and directly interact in vitro and in vivo.
To determine whether Tax protein physically interacts with CARM1, we first performed CARM1 pull-down assays with bacterial GST or GST-Tax. The results of these binding assays demonstrated that GST-Tax, but not GST, interacted with His6-tagged CARM1 protein purified from baculovirus-infected Sf9 cells (Fig. 4A, lanes 1 and 2). Additionally, GST-Tax, but not the control GST, was shown to interact with CARM1 from HeLa cell extracts (lanes 3 and 4). To identify which residues of Tax bind to CARM1, assays were performed with the wild type and Tax deletion mutants as GST fusion proteins (Fig. 4B). The results of this study demonstrated that GST-Tax fusion protein containing amino acids 151 to 244, 151 to 353, and 245 to 353 interacted with CARM1. Of interest, the GST-Tax deletion mutant containing amino acids 151 to 244 binds more strongly to CARM1 than the full-length protein. Since equivalent amounts of the proteins were added to the binding reaction, these results suggest that the CARM1 binding domain may be partially masked in the intact Tax protein. The fact that we observed binding of CARM1 to fragments 151 to 244 and 245 to 353 suggests that there could be multiple binding sites for the protein. Data presented below from in vivo coimmunoprecipitation assays suggest that while the 151-244 CARM1 interaction domain appears to be important in vivo, the 245-353 domain alone is not sufficient for binding. Western blot analysis and GelCode staining of the GST-Tax fusion proteins demonstrated that equivalent amounts of the proteins were added to the binding assays (Fig. 4B) (data not shown).
FIG. 4.
CARM1 and Tax directly interact in vitro and in vivo. (A and B) GST or GST-Tax wild type (A, lanes 1 and 2) and GST-Tax deletion mutants (B) were incubated with purified CARM1 protein. Shown is Western blot analysis of CARM1 levels from GST-Tax pull-down after incubation of HeLa cell nuclear extract with GST or GST-Tax (A, top panel, lanes 3 and 4). Samples were separated by electrophoresis in 4 to 20% Tris-glycine gels, and Western blot analysis for CARM1 or GST was performed. (C) Nuclear extracts from C81 cells were immunoprecipitated (IP) with anti-CARM1 antibody (Ab) and washed extensively, and proteins were separated by electrophoresis in 4 to 20% Tris-glycine gels. Western blot analyses for Tax, CARM1, or p300 were performed. (D) Colocalization of Tax and CARM1. C81 cells were fixed and then immunostained with anti-Tax (α-Tax) and/or anti-CARM1 (α-CARM1) antibodies. (E) 293T cells were transfected with Tax WT or mutant del 151-204, M29, M30, or M32, using Effectene transfection reagent (QIAGEN). At 48 h posttransfection, cell lysates were prepared and immunoprecipated with anti-CARM1 antibody. Immunoprecipitates were separated by electrophoresis in 4 to 20% Tris-glycine gels. Western blot analyses for Tax or CARM1 were performed. The lower panel indicates the levels of expression of Tax and CARM1. (F) Luciferase activities of HTLV-1 LTR were measured in 293T cells transfected with Tax WT or mutants.
In vivo coimmunoprecipitation experiments and confocal analysis of HTLV-1-transformed cells provide further evidence for an interaction between Tax and CARM1. Nuclear extracts from HTLV-1-transformed C81 cells were immunoprecipitated with either anti-IgG or CARM1 antibody (Fig. 4C). Bound proteins were eluted and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the Tax protein was detected by Western blot analysis. The results demonstrated that Tax coimmunoprecipitated with CARM1 but not with control IgG (top panel). Moreover, the interaction of CARM1 with Tax appeared to be stronger than the interaction with p300 as no p300 was coimmunoprecipitated with CARM1. To further verify the interaction of Tax and CARM1 in vivo, HTLV-1-transformed C81 cells were immunostained with antibodies against CARM1 and Tax and analyzed by confocal microscopy. The results of these studies demonstrated that a significant colocalization of the two proteins occurred, as indicated by yellow regions in the merged image between CARM1 (red) and Tax (green) (Fig. 4D).
We have also analyzed the interaction of WT and Tax mutants with CARM1 in transfected cells (Fig. 4E). The Tax mutants included a deletion mutant spanning amino acids 151 to 204, which we identified as a major CARM1 binding site in vitro, and three amino acid substitution mutants within this domain which included M29 (189LysArg→AlaSer), M30 (192 GluGlu→AlaSer), and M32 (196TyrLys→AlaSer). Following transfection of the Tax WT or mutants into the cells, extracts were prepared and immunoprecipitated with anti-CARM1 antibody. The results shown in Fig. 4E demonstrate that CARM1 interacts with WT and M29, M30, and M32 Tax. In contrast, CARM1 fails to interact with the del 151-204 Tax deletion mutant. These results validate our in vitro results, which mapped one CARM1 interaction domain from Tax amino acids 151 to 204 and suggest that the weaker 245-353 domain may not be an important site in vivo. Furthermore, the results from the amino acid substitution mutants suggest that the region from amino acids 189 to 197 within the 151-204 domain is not important for CARM1 interaction. Analysis of additional site specific Tax mutants is in progress.
We also analyzed the effect of the Tax mutations on HTLV-1 transcription (Fig. 4F). While WT Tax significantly increased HTLV-1 LTR transcription (lane 2), the Tax mutant del 151-204 was defective in Tax transactivation (lane 3). Consistent with previous results, Tax mutants M29, M30, and M32 were defective in Tax transactivation (lanes 4 to 6). These studies suggest, therefore, that the CARM1 interaction site closely borders another critical transcription control region.
CARM1 is recruited to the HTLV-1 PICs in the presence of Tax.
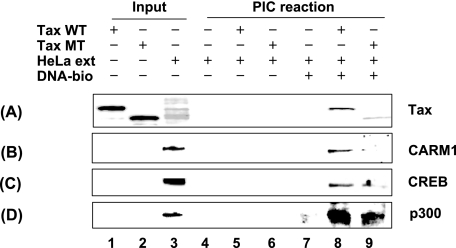
Since CARM1 is important for Tax transactivation of the HTLV-1 LTR and CARM1 directly binds to Tax, we next tested whether Tax recruits CARM1 to the HTLV-1 PICs (Fig. 5). A biotinylated DNA fragment containing four copies of the Tax-responsive 21-bp repeat element and promoter was incubated with HeLa nuclear extract in the absence or presence of Tax protein. Western blot analysis of purified PICs for binding of Tax, CARM1, CREB, and p300 was performed. CREB and p300 were used as positive controls in these experiments. In control reactions containing no DNA, we saw no significant background binding of Tax, CARM1, CREB, or p300 to the streptavidin beads (Fig. 5A to D, lanes 4 to 6). Similarly, when DNA template was added in the absence of Tax, no significant binding of CARM1, CREB, or p300 was observed (Fig. 5B to D, lane 7). In contrast, when Tax was added to the incubation reaction, a PIC containing Tax, CARM1, CREB, and p300 bound to the template DNA (Fig. 5A to D, lane 8). We next examined the composition of PICs formed in the presence of Tax deletion mutant del 151-204. While equivalent amounts of the WT and del 151-204 Tax mutant were added to the reaction (Fig. 5A, lanes1 and 2), a substantial decrease in Tax binding was observed (Fig. 5A, lane 9). Given that the del 151-204 mutant partially overlaps the dimerization domain, the decrease in Tax binding may not be surprising. Concomitant with the decrease in Tax binding, a decrease in CREB and p300 binding was also observed (Fig. 5C and D, lane 9). The binding of CARM1 in the Tax mutant PIC was similar to background (Fig. 5B, lanes 7 and 9). These results are consistent with Tax facilitating the formation of the PIC that includes CREB, Tax, and p300 (15, 20, 26, 50, 54) but does not rule out the possibility that interaction of CARM1 with other factors in the PIC is important. Given the results of the coimmunoprecipitation assay, however, which demonstrates that CARM1 interacts more strongly with Tax than p300, it is tempting to speculate that CARM1 enters the complex through interaction with Tax.
FIG. 5.
CARM1 is recruited to the HTLV-1 PICs in the presence of Tax. HTLV-1 PICs were assembled by incubating biotinylated HTLV-1 templates with HeLa nuclear extracts (ext) in the absence or presence of the His6-Tax WT or mutant (del 151-204) and then purified with streptavidin-coated magnetic beads. The protein components of the purified PICs were analyzed by Western blotting with anti-Tax (A), -CARM1 (B), -CREB (C), or -p300 (D) antibodies. DNA-bio, biotinylated DNA.
CARM1 is associated with active HTLV-1 LTR chromatin in vivo.
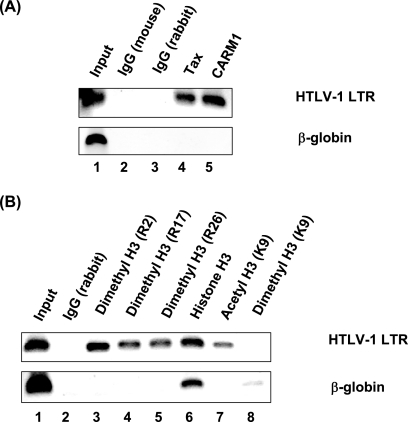
To provide further evidence for the role of CARM1 in HTLV-1 transcription, we performed ChIP assays using SP cells, which contain a single active integrated copy of the HTLV-1 genome and express viral proteins including Tax. Equivalent amounts of cross-linked chromatin were immunoprecipitated with a variety of antibodies, and the precipitated DNA was then subjected to PCR amplification with LTR or control β-globin primers. Figure 6A shows that unlike the control IgG samples, both Tax and CARM1 are associated with the active LTR (lanes 4 and 5). The ChIP analysis further demonstrated that histone H3 associated with the LTR (Fig. 6B, lane 6) and was modified by both acetylation and methylation (Fig. 6B, lanes 3 to 5 and 7). Antibodies to methylated histone H3 R2, R17, and R26, but not lysine 9 (K9), and acetylated H3 K9 precipitated the LTR. These data suggest that Tax and CARM1 are part of the transcription complex bound to the active HTLV-1 LTR promoter in SP cells. As a control for these studies, we used the β-globin promoter, which is repressed in T cells. While histone H3 antibody immunoprecipitated the β-globin promoter, none of the “active” gene markers, including CARM1, methylated histone H3, or acetylated histone H3, were positive.
FIG. 6.
CARM1 is associated with active HTLV-1 LTR chromatin in vivo. SP cells, which contain a single active integrated copy of the HTLV-1 proviral genome, were subjected to ChIP assays. Antibodies specific for Tax, CARM1, dimethyl histone H3 (R2, R17, R26, or K9), histone H3, and acetylated histone H3 (K9) were used for immunoprecipitation. PCRs were carried out to analyze precipitated DNA using primers specific for the HTLV-1 LTR and β-globin promoter region.
DISCUSSION
The importance of protein methylation has become an increasingly significant area of transcription regulation (43). Transcription factors including p53, YY1, and NF-κB have been shown to contribute to the recruitment of PRMTs to eukaryotic promoters (2, 6, 9, 42). Once docked to the transcription complex, the methyltransferases have been shown to methylate histone H3, CBP/p300, SPT5, Tat, and hnRNPs (2, 5, 8, 25, 48, 49). Thus, methylation may play an important role in transcription initiation and elongation as well as mRNA packaging and export (52). Methyltransferases may also target the high-mobility group proteins (HMGs), further regulating transcription and chromatin structure. PRMTs, including CARM1 (PRMT4), share a highly conserved domain encompassing the methyltransferase activity (7). In addition to methyltransferase activity, this homology domain is responsible for formation of homodimers or larger homo-oligomers (53).
As indicated above, CARM1 functions to activate transcription via its methyltransferase activity. Covic et al. reported that CARM1 is a novel transcriptional coactivator of NF-κB and functions as a promoter-specific regulator of NF-κB recruitment to chromatin (10). Moreover, An et al. have provided compelling new evidence that the protein arginine methyltransferases CARM1 and PRMT1 act as coactivators for p53-mediated transcriptional activation (2). In the present study, we demonstrated that histone methyltransferase CARM1 plays a critical role in the activation of HTLV-1 LTR transcription. Overexpression of CARM1 enhanced Tax activation of the HTLV-1 LTR. Of importance, transient transfection with an enzymatically inactive CARM1 mutant failed to rescue Tax transactivation, indicating that methyltransferase activity of CARM1 is important for HTLV-1 LTR transcription. In addition, we transfected with siRNA for CARM1 to knock out the endogenous CARM1. Consistent with results of the CARM1 mutant transfection, CARM1 siRNA blocked the ability of Tax to activate the HTLV-1 LTR.
It has been reported that CARM1 synergistically enhances transcription by nuclear receptors (NRs) through interaction with the activation domain 2 (AD2) of p160 (GRIP1) (5). Recently, Lee et al. reported that CARM1, compared with other PRMTs, has a unique ability to cooperate with p300 and P/CAF and synergistically enhances ER and NR functions with p160, p300, or P/CAF (5, 22). We tested whether CARM1 functionally cooperates with histone acetyltransferase CBP or P/CAF to activate Tax transactivation of the HTLV-1 LTR. The results of these studies suggest that while CBP, P/CAF, and CARM1 all play an essential role in Tax transactivation, the functions of the proteins are independent and additive (data not shown). Consequently, it does not appear, for example, that CARM1 methylates CBP to enhance transcription activity.
Using GST pull-down, coimmunoprecipitation, and confocal microscope assays, we found that CARM1 interacts with Tax in vivo and in vitro. Strikingly, the interaction of Tax with CARM1 facilitates its recruitment to the LTR as part of the preinitiation complex. Consistent with these studies, ChIP assays using SP cells demonstrated that significant levels of Tax, CARM1, histone H3, and dimethyl histone H3 (R2, R17, and R26) were associated with the active HTLV-1 LTR. Of interest, the presence of R2 methylated histone H3 on the HTLV-1 LTR appeared to be higher than the level of methylation at R17 or R26. An in vitro histone methyltransferase assay using recombinant core histone incubated with CARM1 in the presence or absence of Tax suggested that Tax preferentially induced methylation at H3 R2 (data not shown). It will be of interest to determine if this particular modification has an added role in HTLV-1 transcription.
Of interest, Kamoi et al. recently reported that Tax interacts with another histone methyltransferase, SUV39H1, which methylates histone H3 K9 (21). Coexpression of Tax and SUV39H1 represses Tax transactivation of the HTLV-1 LTR. ChIP analysis shows localization of SUV39H1 on the LTR after Tax induction in JPX9 cells containing the HTLV-1 Luc reporter. It will be of interest to determine how these opposing pathways are regulated. Clearly, in the case of HTLV-1-transformed SP cells carrying a single copy of an active LTR, histone H3 K9 methylation was not observed on the LTR. Histone H3 K9 methylation was observed on the repressed β-globin promoter.
In addition to its role as a coactivator of transcription, CARM1 has also been shown to repress CREB-dependent cellular gene expression (49). Methylation of the CBP/p300 KIX domain by CARM1 blocks CREB activation by disabling the interaction between KIX and the KID domain of CREB. Clearly, our studies demonstrate that CARM1 acts as a positive coactivator for the LTR, and we postulate that recruitment of CARM1 to the LTR by Tax may transform CARM1 from a repressor to an activator, providing a mechanism for specific activation of viral gene experession. It will be of interest to determine whether Tax activates cellular CREB-responsive promoters through association and regulation of CARM1 activity.
In conclusion, we have demonstrated that CARM1 plays an important role in HTLV-1 transcription. There is a direct interaction between CARM1 and Tax and Tax-dependent recruitment of CARM1 to the HTLV-1 PICs. The binding of CARM1 to the chromatin template in vivo is accompanied by the presence of methylated histone H3. CARM1 is normally found in a complex of at least 10 proteins called the nucleosomal methylation activator complex (48). It will be of interest to see which of these proteins remain in the Tax-CARM1 complex. Methyltransferase inhibitors have been shown to inhibit replication of hepatitis virus and are under consideration for treatment of prostrate and breast cancers that are hormone dependent (19, 31). The potential of methyltransferase inhibitors for treatment of HTLV-1-associated neuropathies and adult T-cell leukemia may be considered.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. SP cells were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Akagi, T., H. Ono, and K. Shimotohno. 1995. Characterization of T cells immortalized by Tax1 of human T-cell leukemia virus type 1. Blood 86:4243-4249. [PubMed] [Google Scholar]
- 2.An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735-748. [DOI] [PubMed] [Google Scholar]
- 3.Baranger, A. M., C. R. Palmer, M. K. Hamm, H. A. Giebler, A. Brauweiler, J. K. Nyborg, and A. Schepartz. 1995. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature 376:606-608. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, U. M., S. Daujat, S. J. Nielsen, K. Nightingale, and T. Kouzarides. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulanger, M.-C., C. Liang, R. S. Russell, R. Lin, M. T. Bedford, M. A. Wainberg, and S. Richard. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79:124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, D., N. Yadav, R. W. King, M. S. Swanson, E. J. Weinstein, and M. T. Bedford. 2004. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 279:23892-23899. [DOI] [PubMed] [Google Scholar]
- 8.Chevillard-Briet, M., D. Trouche, and L. Vandel. 2002. Control of CBP co-activating activity by arginine methylation. EMBO J. 21:5457-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conkright, M. D., E. Guzman, L. Flechner, A. I. Su, J. B. Hogenesch, and M. Montminy. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11:1101-1108. [DOI] [PubMed] [Google Scholar]
- 10.Covic, M., P. O. Hassa, S. Saccani, C. Buerki, N. I. Meier, C. Lombardi, R. Imhof, M. T. Bedford, G. Natoli, and M. O. Hottiger. 2005. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 24:85-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daujat, S., U. M. Bauer, V. Shah, B. Turner, S. Berger, and T. Kouzarides. 2002. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12:2090-2097. [DOI] [PubMed] [Google Scholar]
- 12.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. [PubMed] [Google Scholar]
- 13.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61:65-131. [DOI] [PubMed] [Google Scholar]
- 14.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed] [Google Scholar]
- 15.Giebler, H. A., J. E. Loring, K. van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grassmann, R., C. Dengler, I. Muller-Fleckenstein, B. Fleckenstein, K. McGuire, M. C. Dokhelar, J. G. Sodroski, and W. A. Haseltine. 1989. Transformation to continuous growth of primary human T lymphocytes by human T-cell leukemia virus type I X-region genes transduced by a Herpesvirus saimiri vector. Proc. Natl. Acad. Sci. USA 86:3351-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C.-Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 19.Hong, H., C. Kao, M. H. Jeng, J. N. Ebie, M. O. Koch, and T. A. Gardner. 2004. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer 101:83-89. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamoi, K., K. Yamamoto, A. Misawa, A. Miyake, T. Ishida, Y. Tanaka, M. Mochizuki, and T. Watanabe. 2006. SUV39H1 interacts with HTLV-1 Tax and abrogates Tax transactivation of HTLV-1 LTR. Retrovirology 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimzey, A. L., and W. S. Dynan. 1999. Identification of a human T-cell leukemia virus type I tax peptide in contact with DNA. J. Biol. Chem. 274:34226-34232. [DOI] [PubMed] [Google Scholar]
- 23.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 24.Koh, S. S., H. Li, Y. H. Lee, R. B. Widelitz, C. M. Chuong, and M. R. Stallcup. 2002. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J. Biol. Chem. 277:26031-26035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak, Y. T., J. Guo, S. Prajapati, K. J. Park, R. M. Surabhi, B. Miller, P. Gehrig, and R. B. Gaynor. 2003. Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11:1055-1066. [DOI] [PubMed] [Google Scholar]
- 26.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 27.Lee, Y.-H., S. S. Koh, X. Zhang, X. Cheng, and M. R. Stallcup. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol. Cell. Biol. 22:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2004. Transcription regulatory complexes bind the human T-cell leukemia virus 5′ and 3′ long terminal repeats to control gene expression. Mol. Cell. Biol. 24:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenzmeier, B. A., E. E. Baird, P. B. Dervan, and J. K. Nyborg. 1999. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291:731-744. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y.-J., M. R. Stallcup, and M. M. C. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, H., C. A. Pise-Masison, R. Linton, H. U. Park, R. L. Schiltz, V. Sartorelli, and J. N. Brady. 2004. Tax relieves transcriptional repression by promoting histone deacetylase 1 release from the human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 78:6735-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, H., C. A. Pise-Masison, T. M. Fletcher, R. L. Schiltz, A. K. Nagaich, M. Radonovich, G. Hager, P. A. Cole, and J. N. Brady. 2002. Acetylation of nucleosomal histones by p300 facilitates transcription from Tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol. Cell. Biol. 22:4450-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mick, J. E., M. A. Colgin, A. Brauweiler, and J. K. Nyborg. 2000. Analysis of CREB mutants in Tax complex formation and trans-activation. AIDS Res. Hum. Retrovir. 16:1597-1601. [DOI] [PubMed] [Google Scholar]
- 35.Mochizuki, M., T. Watanabe, K. Yamaguchi, K. Tajima, K. Yoshimura, S. Nakashima, M. Shirao, S. Araki, N. Miyata, and S. Mori. 1992. Uveitis associated with human T lymphotropic virus type I: seroepidemiologic, clinical, and virologic studies. J. Infect. Dis. 166:943-944. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, F., H. Kajihara, M. Nakamura, H. Sasaki, T. Kumamoto, and K. Okada. 1989. HTLV-1 associated myelopathy in an HTLV-1 and HBV double carrier family: report of a case and the mode of vertical transmission of both viruses. J. Gastroenterol. Hepatol. 4:387-390. [DOI] [PubMed] [Google Scholar]
- 37.Nerenberg, M., S. H. Hinrichs, R. K. Reynolds, G. Khoury, and G. Jay. 1987. The tat gene of human T-lymphotropic virus type 1 induces mesenchymal tumors in transgenic mice. Science 237:1324-1329. [DOI] [PubMed] [Google Scholar]
- 38.Nishioka, K., I. Maruyama, K. Sato, I. Kitajima, Y. Nakajima, and M. Osame. 1989. Chronic inflammatory arthropathy associated with HTLV-I. Lancet i:441. [DOI] [PubMed] [Google Scholar]
- 39.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed] [Google Scholar]
- 40.Park, H. U., S. J. Jeong, J. H. Jeong, J. H. Chung, and J. N. Brady. 2006. Human T-cell leukemia virus type 1 Tax attenuates gamma-irradiation-induced apoptosis through physical interaction with Chk2. Oncogene 25:438-447. [DOI] [PubMed] [Google Scholar]
- 41.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezai-Zadeh, N., X. Zhang, F. Namour, G. Fejer, Y. D. Wen, Y. L. Yao, I. Gyory, K. Wright, and E. Seto. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 44.Schurter, B. T., S. S. Koh, D. Chen, G. J. Bunick, J. M. Harp, B. L. Hanson, A. Henschen-Edman, D. R. Mackay, M. R. Stallcup, and D. W. Aswad. 2001. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40:5747-5756. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, A., C. Takahashi, S. Yamaoka, T. Nosaka, M. Maki, and M. Hatanaka. 1990. Oncogenic transformation by the tax gene of human T-cell leukemia virus type I in vitro. Proc. Natl. Acad. Sci. USA 87:1071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tie, F., N. Adya, W. C. Greene, and C.-Z. Giam. 1996. Interaction of the human T-lymphotropic virus type 1 Tax dimer with CREB and the viral 21-base-pair repeat. J. Virol. 70:8368-8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vernant, J. C., G. Buisson, J. Magdeleine, J. De Thore, A. Jouannelle, C. Neisson-Vernant, and N. Monplaisir. 1988. T-lymphocyte alveolitis, tropical spastic paresis, and Sjogren syndrome. Lancet i:77. [DOI] [PubMed] [Google Scholar]
- 48.Xu, C., and M. F. Henry. 2004. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 24:10742-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 50.Yin, M. J., and R. B. Gaynor. 1996. Complex formation between CREB and Tax enhances the binding affinity of CREB for the human T-cell leukemia virus type 1 21-base-pair repeats. Mol. Cell.Biol. 16:3156-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, M. C., F. Bachand, A. E. McBride, S. Komili, J. M. Casolari, and P. A. Silver. 2004. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 18:2024-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, X., L. Zhou, and X. Cheng. 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, M., M. A. Halanski, M. F. Radonovich, F. Kashanchi, J. Peng, D. H. Price, and J. N. Brady. 2000. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20:5077-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]