Abstract
A new group of nucleocytoplasmic shuttling proteins has recently been identified in the structural proteins encoded by several alphaherpesvirus UL47 genes. Nuclear import and export signals for the bovine herpesvirus type 1 UL47 protein (VP8 or bUL47) have been described previously. Here, we study the trafficking of bUL47 in detail and identify an import signal different from that shown before. It comprises a 20-residue N-terminal peptide that is fully transferable and targets a large, normally cytosolic protein to the nucleus. A conserved RRPRRS motif within this peptide was shown to be essential but not sufficient for nuclear targeting. Using interspecies heterokaryon assays, we further demonstrate that the export activity of the published leucine-rich nuclear export signal (NES) is also transferable to a large protein but is functionally weak compared to the activity of the HIV-1 Rev NES. We show that nuclear export dictated by this bUL47 NES is sensitive to leptomycin B (LMB) and therefore dependent on the export receptor CRM-1. However, nuclear export of full-length bUL47 is fully resistant to LMB, suggesting the presence of an additional NES. We go on to identify a second NES in bUL47 within a 28-residue peptide that is in close proximity to but entirely separable from the N-terminal import signal, and we use fluorescence loss in photobleaching to confirm its activity. This NES is resistant to leptomycin B, and therefore utilizes an export receptor other than CRM-1. As this new sequence bears little similarity to other export signals so far defined, we suggest it may be involved in bUL47 export from the nucleus via a novel cellular receptor.
The alphaherpesvirus UL47 gene encodes a major structural protein of unknown function that is assembled into the tegument of the virus particle (5, 20, 24, 45). Although the role of herpes simplex virus type 1 (HSV-1) UL47 (hUL47, also known as VP13/14) in virus replication has not yet been defined, previous studies of a UL47 deletion mutant in HSV-1 have implicated the protein in the regulation of immediate-early gene expression in infected cells (47). Furthermore, UL47 knockout viruses in pseudorabies virus (PRV) and Marek's disease virus also replicate more slowly than wild-type viruses (8, 20). We have previously shown that the protein encoded by HSV-1 UL47 is targeted specifically to the nucleus in infection and exhibits the properties of a nucleocytoplasmic shuttling protein when expressed in transfected cells (6, 7). The bovine herpesvirus type 1 (BHV-1) homologue of UL47 (bUL47, also known as VP8), which is by far the major structural protein of the BHV-1 virus particle (5), has also been shown to shuttle when expressed in isolation (48). We have recently shown definitively that the virion population of bUL47 is targeted to the nucleus immediately upon infection by BHV-1, and we have demonstrated that bUL47 exhibits nucleocytoplasmic shuttling in infected MDBK cells, the first time this property has been shown during an infection (42).
Within the cell, the movement of proteins between the cytoplasm and nucleus regulates many cellular functions, such as transcription, DNA replication, the cell cycle, and RNA transport. These processes are also vital to virus replication and, as such, a number of virus proteins, such as human immunodeficiency virus type 1 (HIV-1) Rev, HSV-1 ICP27, and influenza virus NS2, were some of the first proteins to be shown to undergo efficient nucleocytoplasmic transport (25, 26, 32, 35). The cellular receptors involved in moving both cellular and viral proteins in and out of the nucleus do so by interacting directly with nucleoporin proteins that are located in the nuclear pore complexes (2, 3, 34). These receptors recognize discrete signals within proteins that specifically dictate either the import into or the export out of the nucleus. Furthermore, in a number of proteins that shuttle between the nucleus and cytoplasm, such as the cellular RNA-binding protein hnRNPA1, the import and export signals are not separable, and in these situations the signals involved have been defined as shuttling signals (27, 28). The majority of cellular nucleocytoplasmic transport receptors belong to the large karyopherin β family of soluble transport factors (30) and recognize specific canonical nuclear transport signals, such as the arginine-rich import signals in HIV-1 Tat and Rev proteins recognized by importin β (18, 33, 34, 41), or the leucine-rich export signal recognized by CRM-1, and found in a wide range of shuttling proteins, such as Rev and the cyclic AMP-dependent protein kinase inhibitor PKI (10, 44). Nuclear transport can also be mediated indirectly by a protein such as importin α, which recognizes the classical lysine-rich import signal as typified by the simian virus 40 (SV40) large T antigen and acts as an adaptor to importin β for nuclear import (13, 14, 18). In all cases, transport by karyopherins is controlled by the small GTPase Ran that regulates both the assembly and the disassembly of transport complexes (15). By contrast, the nonkaryopherin export receptor involved in transporting the majority of mRNA molecules out of the nucleus, known as TAP/NXF1, does not appear to require Ran activity to function (16, 19, 39).
Although the nucleocytoplasmic shuttling behaviors of hUL47 and bUL47 homologues are apparently similar to one another, there is little sequence homology between the two proteins. We have previously shown that nuclear import of hUL47 requires two arginine boxes in the N-terminal 76 residues of the protein (7), while nuclear import of bUL47 has been shown to require a single arginine-rich box with a different sequence but located within the same N-terminal region of the protein (48). In addition, while the nuclear export signal (NES) from hUL47 has not yet been defined, it has been reported that the NES in bUL47 is a leucine-rich motif present in the middle of the open reading frame, which may suggest that this protein is exported from the nucleus via the classical CRM-1 pathway (48). To further address the cellular pathways that the UL47 homologues may utilize in their nuclear trafficking, we have carried out a detailed characterization of the sequences in bUL47 required for nuclear import and export. To define these signals precisely, we have used a system that requires them to be functional when transferred to large proteins that cannot move through the nuclear pore complex without active transport via specific receptors. We show that the nuclear import of bUL47 is dictated by a 20-residue arginine-rich motif that is different from the previously published one (48). We also show that bUL47 contains two definable nuclear export signals—the previously defined leucine-rich signal (48), which we show to be weak and sensitive to leptomycin B (LMB), and a different N-terminal signal that is strong and resistant to LMB. The sequence of the second bUL47 NES bears no similarity to any other NES so far identified; hence, we suggest that it represents a novel NES that may ultimately reveal a new nuclear export pathway.
MATERIALS AND METHODS
Plasmids.
Plasmids pEGFPC1 and pEGFPC2 were obtained from Clontech. The construction of plasmid pMD10, expressing green fluorescent protein (GFP) fused to HSV-1 UL47, has been described previously (7). To construct a plasmid expressing GFP fused to BHV-1 UL47, the UL47 open reading frame was first obtained as a BamH1/Xho1 fragment from plasmid pcVP8 (kindly provided by Vikram Misra, University of Saskatchewan, Canada) and inserted into pcDNAI/Amp to generate pJV1. A BamHI/XbaI fragment was then transferred into pEGFP-C1 (Clontech), generating plasmid pJV2 in which full-length bUL47 is fused to the C terminus of GFP. The plasmid encoding GFP fused to PRV UL47 was generated by inserting the EcoRI/XbaI fragment from pcDNA-UL47 (kindly provided by Thomas Mettenleiter, Friedrich-Loeffler-Institut, Germany) into pEGFP-C2, obtaining plasmid pJV18. The open reading frame of the equine herpesvirus type 1 (EHV-1) UL47 gene was obtained by PCR amplification of genomic DNA from EHV-1 strain RacH (kindly provided by Klaus Osterrieder, Cornell University, Ithaca, N.Y.) with primers containing BglII and EcoRI sites. The resulting PCR product was inserted as a BglII/EcoRI fragment into pEGFP-C1, generating plasmid pJV48. A plasmid containing the KpnI fragment of the varicella-zoster virus (VZV) genome (kindly provided by Valerie Preston, MRC Virology Unit, Glasgow, United Kingdom) was used to amplify the VZV UL47 gene with BglII and HindIII sites at either end. This was then inserted into pEGFP-C1, generating plasmid pJV51.
A number of truncation mutants of bUL47 were constructed as GFP fusion proteins in the following way. To construct GFP fused to residues 17 to 742 (GFP 17-742), pJV2 was digested with BspEI to release the first 50 bases of the bUL47 gene and religated to obtain plasmid pJV3. A BamHI/EcoRI fragment was released from plasmid pJV1 and inserted into pEGFP-C1 digested with BglII/EcoRI to obtain plasmid pJV4, expressing GFP fused to residues 1 to 319 (1-319). Plasmids pJV8, pJV9, pJV10, and pJV11, encoding GFP fused to residues 1-66, 1-131, 1-198, and 1-261, respectively, were constructed by PCR amplification using relevant primers incorporating a BglII site and a HindIII site. The resulting fragments were inserted into BglII/HindIII-digested pEGFP-C1. Plasmid pJV50, encoding GFP fused to residues 65-742, was constructed by PCR amplification using relevant primers incorporating a BglII site and a HindIII site, and the resulting fragments were inserted into BglII/HindIII-digested pEGFP-C1.
Plasmid p3PK, encoding amino acids 17-476 of chicken muscle pyruvate kinase (CMPK), was obtained from J. Frangioni (Beth Israel Hospital, Boston, Mass.) (12). Unique HindIII and BglII sites present at the N terminus of CMPK were used to construct all plasmids expressing bUL47-CMPK fusion proteins. Plasmids pJV23, pJV24, pJV25, pJV28, pJV34, and pJV37, expressing residues 1-66, 1-50, 20-66, 1-30, 1-20, and 10-30, respectively, were constructed by PCR amplification followed by insertion into HindIII/BglII p3PK. Plasmids pJV32, pJV63a, pJV35, pJV42, and pJV47, expressing R box 1 (residues 11-16), R box 2 (residues 47-54), residues 20-31, SV40 NLS, and residues 10-30 Δ R box 1 (arginine-to-glycine mutations), respectively, were constructed by annealing oligonucleotides which were then inserted into p3PK digested with HindIII/BglII. The β-galactosidase (βgal) control plasmids used in heterokaryon assays were kindly provided by M. Dobbelstein (Marburg University, Germany). Plasmid pCFN-βgal encodes β-galactosidase fused to the C terminus of the SV40 NLS, and plasmid pCFNRev-βgal encodes an SV40 NLS/Rev NES β-galactosidase fusion protein (37). To construct plasmids expressing bUL47-β-galactosidase fusion proteins, the β-galactosidase open reading frame was first amplified using primers containing BglII and Xba1 sites. This fragment was inserted in place of CMPK in the plasmid p3PK, generating plasmid pJV53. Plasmids pJV61 and pJV62, expressing residues 1-131 and 1-319 of bUL47 fused to β-galactosidase, were constructed by PCR amplification followed by insertion into HindIII/BglII pJV53. Plasmid pJV68, encoding SV40 NLS fused to residues 67-131 of bUL47 fused to β-galactosidase, was made with a two-step cloning strategy. First, residues 67-131 of bUL47 were amplified as a BglII/BamH1 fragment that was inserted into BglII-digested pJV53. Annealed oligonucleotides encoding the SV40 NLS were then inserted into this plasmid that had been digested with HindIII and BglII. Plasmid pJV72, encoding SV40 NLS fused to residues 95-123 of bUL47, was constructed in the same way. Plasmids pJV69 and pJV70 were constructed by amplifying SV40 NLS fused to residues 67-123 or residues 67-95 from plasmid pJV68 as HindIII/BglII fragments which were then inserted into HindIII/BglII-digested pJV53. Plasmid pJV77, encoding SV40 NLS fused to residues 485-495 of bUL47 as a β-galactosidase fusion protein, was constructed by inserting annealed oligonucleotides for the entire region into HindIII/BglII-digested pJV53. To construct full-length VP8 as a GFP fusion protein in which the four arginine residues in R box 1 had been mutated to glycine residues (plasmid pJV65), the BsrG1/BspE1 fragment from pJV2 was replaced with an annealed oligonucleotide containing these mutations. The ΔNES construct, from which residues 95-123 had been deleted (plasmid pJV76), was constructed by a two-step PCR whereby the BglII/EcoR1 N-terminal fragment of VP8 was amplified lacking these residues and then inserted into BglII/EcoR1-digested pJV2.
Cells and transfections.
HEp-2, COS-1, and NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum. For transfection followed by live-cell fluorescence analysis, HEp-2 cells were plated at a density of 105 cells per well of a two-well cover glass chamber (LabTek). For transfection followed by immunofluorescence, COS-1 cells were plated at a density of 2 × 105 cells per well onto 16-mm glass coverslips in individual wells of a six-well plate. Twenty-four hours after plating, the cells were transfected using the calcium phosphate precipitation technique modified with BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline.
Antibodies.
Polyclonal anti-CMPK antibody (Berkeley Antibody Company) was used for immunofluorescence at a dilution of 1:500. Monoclonal anti-β-galactosidase antibody (Promega) was used for immunofluorescence at a dilution of 1:200. Monoclonal anti-GFP antibody (Clontech) was used for Western blotting at a dilution of 1:10,000.
Microscopy.
Cells expressing GFP-tagged fusion proteins were examined live 20 h after transfection. For immunofluorescence analysis, cells on coverslips were fixed 40 h after transfection in either 100% methanol for 10 min or for 20 min in 4% paraformaldehyde followed by permeabilization for 10 min in 0.5% Triton X-100. The fixed cells were blocked by incubation for 30 min in phosphate-buffered saline (PBS) containing 10% newborn calf serum, and primary antibody was added at the appropriate dilution in the same solution for a further 30 min. Following extensive washing in PBS, fluorescein isothiocyanate-conjugated secondary antibody was added in the same blocking solution, incubated for a further 30 min, and washed extensively in PBS. The coverslips were mounted on glass slides in Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole; Vector Labs). All samples were examined using a Zeiss Axiovert S100 TV inverted microscope. Images were acquired with a Photometrics Quantix digital camera and were processed with Metamorph and Adobe Photoshop software. In some cases, images were acquired using a Bio-Rad MRC600 confocal microscope, and further processing was carried out with Adobe Photoshop software.
Fluorescence loss in photobleaching (FLIP).
HEp-2 cells grown in two-well coverslip chambers were transfected with the relevant GFP-expressing plasmids as described above. Twenty hours later, the cells were transferred to CO2-independent medium (Gibco) and examined live at 37°C with a Deltavision RT imaging system. On individual expressing cells, a small area of cytoplasm was marked out for photobleaching using a line of four photobleaching points. The 488-nm laser module, set at 50% power with a bleach time of 0.05 s, was used to bleach the same line every 10 s for 100 repetitions, and images were acquired following each photobleaching event. The images comprising each time-lapse were then converted to TIFF files and analyzed using NIH Image software.
Heterokaryon assays.
Interspecies heterokaryon assays were performed by transfecting COS-1 cells that had been grown in a 25-cm2 tissue culture flask. One day later, the COS-1 cells were washed three times with trypsin-EDTA and detached from the flask, and 2 × 105 cells were seeded onto coverslips on a six-well plate. The next day, 3 × 105 NIH 3T3 cells were seeded on top of the COS-1 cells in the presence of 50 μg/ml of cycloheximide (Fisher Chemicals) and left to settle for 3 h. Fresh medium containing 100 μg/ml of cycloheximide was then added. After 30 min, coverslips were taken up with tweezers and washed three times by dipping them into prewarmed PBS in a six-well plate. The two cell lines were fused with 50 μl polyethylene glycol (Sigma), made up in medium without serum as a 50% (wt/wt) solution, for exactly 2 min. After fusion, coverslips were washed three times in prewarmed PBS and returned to medium containing 100 μg/ml of cycloheximide. Coverslips were left for 3 h at 37°C and washed three times in PBS, and cells were fixed in 4% paraformaldehyde as described above. Immunofluorescence was carried out as described above, and the nuclei of 3T3 cells were distinguished from the COS-1 nuclei by their speckled appearance with DAPI staining. For experiments carried out in the presence of leptomycin B (Calbiochem), the NIH 3T3 cells were seeded onto COS-1 cells in the presence of 50 μg/ml of cycloheximide and 2.5 ng/ml leptomycin B. The remainder of the experiment was carried out using the same concentration of leptomycin B throughout.
RESULTS
Nuclear localization of five alphaherpesvirus UL47 proteins.
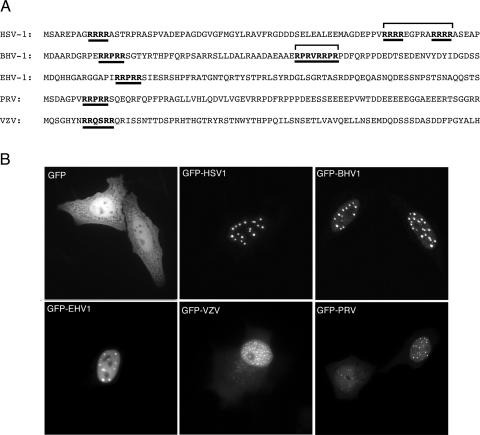
We have previously shown that the herpes simplex virus UL47 protein (hUL47 or VP13/14) is a nucleocytoplasmic shuttling protein, with a steady-state localization in the nucleus and more specifically in nuclear dots (7). Nuclear import of hUL47 requires two R boxes (RRRR) located in the N-terminal 80 residues of the protein (Fig. 1A, HSV-1). However, while these R boxes are not conserved in bUL47, it has previously been shown that nuclear import of bUL47 requires a different R box located in the N terminus of the protein (48) (Fig. 1A, BHV-1). Sequence alignment of the five alphaherpesvirus UL47 proteins revealed that the overall sequence similarity between the homologues was as low as 25%, while alignment of the N termini indicated that neither the hUL47 nor the bUL47 import motifs were conserved in other UL47 proteins (Fig. 1A). However, an R box located within the first 20 residues of hUL47, which is not required for either hUL47 or bUL47 nuclear localization (7, 48), was partially conserved throughout the family. Furthermore, this first arginine box is a highly conserved RRPRRS motif in three of the UL47 proteins (Fig. 1A). As an initial analysis of this group of proteins, we investigated the steady-state localization of all the UL47 homologues by expressing them as GFP fusion proteins by transient expression in HEp-2 cells. In spite of the low sequence conservation between these proteins, they were all targeted efficiently to the nucleus (Fig. 1B). Moreover, they all localized to specific domains within the nucleus that were similar to those observed for hUL47 (Fig. 1B, GFP-HSV1), particularly bUL47 from BHV-1 (Fig. 1B, GFP-BHV1). It should also be noted that both the hUL47 and bUL47 homologues localize to these nuclear domains when examined as native proteins by immunofluorescence (data not shown); hence, this localization is not dictated simply by fusion of GFP to these proteins. Intriguingly, the VZV homologue of UL47 was consistently toxic in HEp-2 cells and had to be examined in COS-1 cells instead (Fig. 1B, GFP-VZV).
FIG. 1.
Subcellular localization of five alphaherpesvirus UL47 homologues. (A) Sequence comparison of the N-terminal 80 residues from the UL47 homologues encoded by HSV-1, BHV-1, EHV-1, PRV, and VZV. The three arginine boxes previously identified for HSV-1 UL47 are underlined. Arginine-rich boxes in the other homologues are also underlined. (B) Each of the full-length UL47 homologues was fused to the C terminus of GFP and transfected into HEp-2 cells. The cells were examined live 20 h later. Note that expression of the VZV homologue consistently resulted in the death of HEp-2 cells, and therefore the image presented here is of a COS-1 cell expressing VZV-UL47.
Nuclear localization of bUL47 requires the N-terminal 17 residues of the protein.
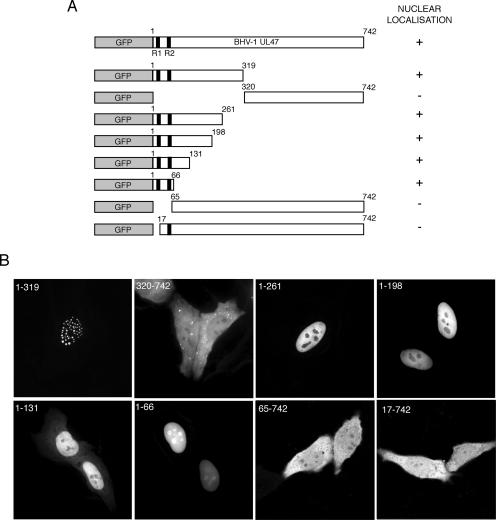
We next decided to examine more closely the regions of bUL47 involved in nuclear targeting, by first generating a series of truncation mutants fused to the C terminus of GFP (Fig. 2A). Initially large truncations of the N terminus and C terminus indicated clearly that the nuclear targeting properties of bUL47 were located in the N-terminal half of the protein (Fig. 2B, compare 1-319 and 320-742). Furthermore, the N-terminal 319 residues also contained the sequences involved in targeting bUL47 to nuclear dots (Fig. 2B, 1-319). Further truncations into the C terminus of residues 1 to 319 revealed that as few as the first 66 residues of bUL47 were required to redirect GFP to the nucleus, in agreement with the location of the previously characterized NLS (48) (Fig. 2B, 1-66). In addition, the deletion of the N-terminal 64 residues from the full-length protein resulted in a protein that could not accumulate in the nucleus, suggesting that the NLS of bUL47 was indeed located at the extreme N terminus (Fig. 2B, 65-742). However, deletion of only the first 16 residues had the same effect on the localization of full-length bUL47, indicating that these extreme N-terminal residues are important for bUL47 nuclear localization (Fig. 2B, 17-742). These 16 residues do not contain the previously defined bUL47 NLS, but do contain the RRPRRS motif that is highly conserved in other UL47 proteins. Interestingly, the deletion of 50 residues from the C terminus of residues 1-319 resulted in a GFP fusion protein that no longer localized to nuclear dots but instead was diffuse in the nucleoplasm (Fig. 2B, compare 1-261 and 1-319). This result demonstrates that the residues between 262 and 319 of bUL47 are involved in targeting this protein to nuclear dots and that nuclear targeting is separable from nuclear dot targeting. It is also noteworthy that, while most of the GFP fusion proteins seem to be absent from the nucleolus, the very smallest one (1-66) was clearly concentrated in the nucleolus (Fig. 2B, compare 1-66 and 1-198). We do not yet understand the basis for this differential localization to the nucleolus.
FIG. 2.
Delineation of the region of bUL47 involved in nuclear localization. (A) A range of bUL47 truncation mutants was fused to the C terminus of GFP. (B) These mutants were transfected into HEp-2 cells and examined 20 h later by live-cell fluorescence. Each was scored for its ability to accumulate in the nucleus of the expressing cell, as summarized in panel A.
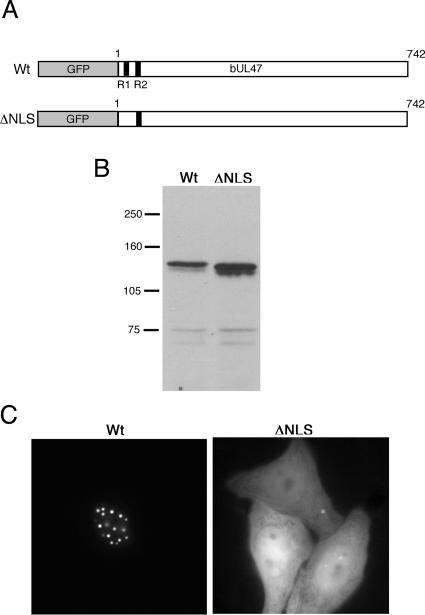
To ensure that the lack of nuclear localization demonstrated by the deletion mutant that expresses residues 17 to 742 reflected a true requirement for the first R box in nuclear localization, we next constructed a variant of the full-length protein in which the four arginine residues in the first R box had been mutated to glycine residues (Fig. 3A). Western blot analysis indicated that this GFP fusion protein was expressed correctly (Fig. 3B). Nonetheless, the subcellular localization of this ΔNLS protein was in striking contrast to the tight nuclear localization of the wild-type (Wt) protein, indicating that mutation of just four arginine residues completely abrogated the efficient nuclear localization of bUL47 (Fig. 3C).
FIG. 3.
Mutation of the arginines in the first R box of bUL47 abrogates nuclear accumulation. (A) The four arginine residues in R box 1 were mutated to glycines in the context of a GFP fusion protein to create ΔNLS. (B) Wt and ΔNLS bUL47 were expressed in COS-1 cells as GFP fusion proteins, and Western blot analysis was carried out with an anti-GFP antibody. Molecular mass markers (in kilodaltons) are given on the left. (C) Wt and ΔNLS bUL47 were expressed in HEp-2 cells as GFP fusion proteins and examined live 20 h after transfection.
The minimal transferable NLS of bUL47 is a 20-residue peptide.
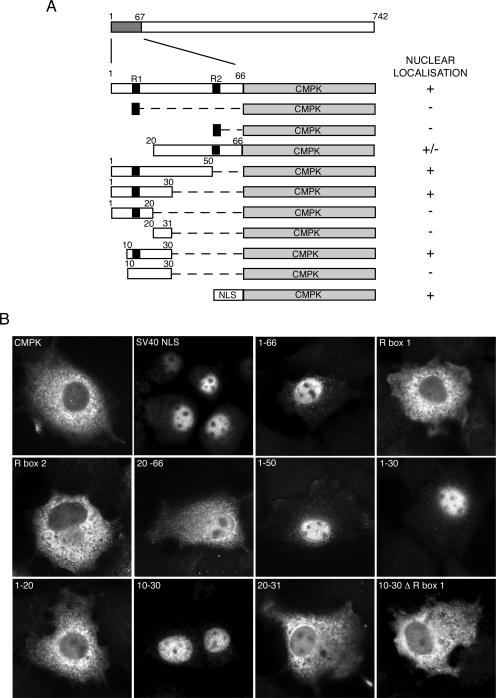
To further characterize the NLS of bUL47, and to determine the relative contribution of the first and second arginine boxes to its activity, we wished to ensure that the 1-66 region of the protein shown to direct GFP to the nucleus could act as a fully transferable NLS and redirect a large cytoplasmic protein to the nucleus. Thus, we fused residues 1-66 of bUL47 to the N terminus of CMPK, a commonly used reporter for defining nuclear localization sequences (12) (Fig. 4A). The localization of this fusion protein was compared to that of CMPK alone and to that of CMPK fused to the SV40 NLS (Fig. 4B) by immunofluorescence of COS-1 cells transfected with the relevant plasmids. As expected, CMPK alone exhibited a cytoplasmic localization, while the SV40 NLS fused to CMPK was almost entirely nuclear (Fig. 4B). Fusion of residues 1-66 of bUL47 to CMPK also caused CMPK to localize to the nucleus (Fig. 4B), confirming that this region contained a NLS that was as efficient as the SV40 NLS. However, neither of the two arginine-rich boxes (see Fig. 1A) present in the first 66 residues of bUL47 was alone sufficient to target CMPK to the nucleus (Fig. 4B, R box 1 and R box 2), suggesting that sequences in addition to these basic stretches were required for nuclear localization. Further truncations from the N or C terminus of residues 1-66 demonstrated that a peptide encompassing the previously characterized NLS of bUL47 (R box 2) was not able to target CMPK to the nucleus with the same efficiency as residues 1 to 66, suggesting that a region required for complete nuclear targeting may be present in the first 20 residues (Fig. 4B, 20-66). However, by contrast, the first 50 residues of the protein containing only R box 1 were fully functional for efficient nuclear import (Fig. 4B, compare 20-66 with 1-50). In addition, fusion of residues 1-30 but not 1-20 to CMPK was sufficient for nuclear targeting (Fig. 4B, compare 1-30 with 1-20), suggesting that residues 20-30 were also involved. These residues are, however, not sufficient for nuclear localization (Fig. 4B, 20-31). The smallest definable NLS present at the N terminus of bUL47 was shown to be a peptide of residues 10-30, which are capable of efficiently relocalizing CMPK to the nucleus (Fig. 4B, 10-30). Furthermore, mutation of the arginines to glycines in R box 1 abrogated the ability of these 20 residues to target CMPK to the nucleus, confirming that although this motif is not sufficient, it is absolutely essential for nuclear targeting (Fig. 3B, 10-30 Δ R box 1).
FIG. 4.
The minimal nuclear import signal in bUL47 as defined by fusions with CMPK. (A) A range of peptides from the N-terminal 66 residues of bUL47 was fused to the N terminus of CMPK. (B) Each of the above fusion proteins was examined by using COS-1 cells transfected with the relevant plasmids. Immunofluorescence was carried out with an anti-CMPK antibody 40 h after transfection, and each fusion protein was scored for its ability to accumulate in the nucleus of the expressing cell, as summarized in panel A. NLS denotes the SV40 nuclear localization signal. The black boxes represent the two arginine-rich motifs as identified in Fig. 1B. Δ R box 1 denotes the mutation of all arginines to glycines in R box 1.
Nucleocytoplasmic shuttling of bUL47.
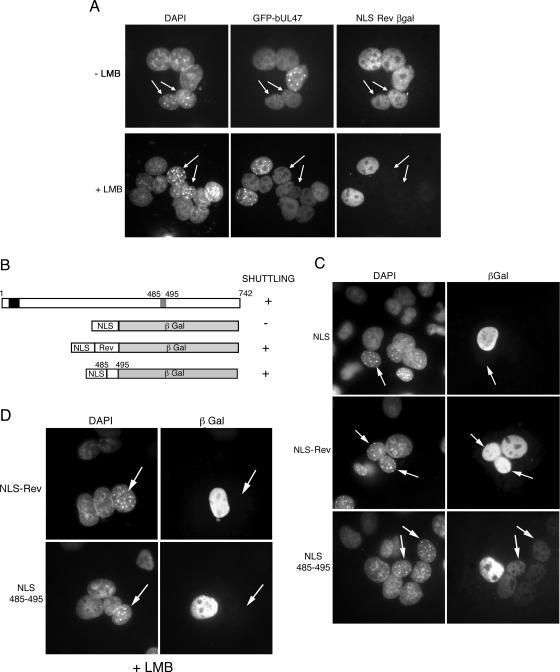
Both hUL47 and bUL47 have previously been shown to be nucleocytoplasmic shuttling proteins (7, 42, 48). While the sequences involved in hUL47 shuttling have not yet been identified, a 10-amino-acid Leu-rich NES has been identified in bUL47 by fusion of this sequence to yellow fluorescent protein (YFP) (48). The majority of proteins that are actively transported from the nucleus do so through the well-characterized CRM-1 receptor protein, which recognizes the classical Leu-rich NES signal, which is similar to that identified for bUL47 (10, 11, 44). The CRM-1 receptor is sensitive to the drug LMB (31); hence, because of the presence of the Leu-rich signal in bUL47, we wished to investigate the sensitivity of its nuclear export to this drug. To examine nucleocytoplasmic shuttling of bUL47, interspecies heterokaryon assays were carried out by fusing transfected COS-1 cells expressing the protein(s) of interest to nonexpressing mouse NIH 3T3 cells and determining if that protein had the ability to move into the nuclei of NIH 3T3 cells within individual heterokaryons. We first cotransfected COS-1 cells with plasmids expressing GFP 1-742 and a fusion protein consisting of the SV40 NLS (to target to the nucleus) and the HIV-1 Rev NES (to dictate shuttling) fused to the N terminus of the large (116-kDa) reporter protein β-galactosidase (NLS-Rev βgal). The latter fusion protein is well characterized as a nuclear-cytoplasmic shuttling protein in heterokaryon assays (37). These coexpressing cells were then used in heterokaryon assays with NIH 3T3 cells either in the absence or in the presence of LMB. In the absence of LMB, both proteins were readily detectable in NIH 3T3 cell nuclei within heterokaryons, as identified by the characteristic DAPI staining of NIH 3T3 cells, confirming that bUL47 exhibits the property of nuclear export (Fig. 5A, nuclei with arrows; Table 1). As expected, leptomycin B blocked the ability of the Leu-rich Rev NES to function as an export signal (Fig. 5A, NLS Rev βgal, nuclei with arrows; Table 2). However, by contrast, full-length bUL47 expressed as a GFP fusion protein in the same cell as the NLS-Rev-βgal fusion peptide was not inhibited from shuttling in the presence of leptomycin B (Fig. 5A, GFP-bUL47, nuclei with arrows; Table 2). This result suggests that either the previously identified Leu-rich NES of bUL47 functions via a pathway other than that of CRM-1, or there is a second, LMB-resistant, NES within bUL47.
FIG. 5.
bUL47 nucleocytoplasmic shuttling is resistant to leptomycin B. (A) COS-1 cells were cotransfected with plasmids expressing GFP-bUL47 and β-galactosidase fused to both the SV40 NLS and the Rev NES. Heterokaryon assays were then carried out between the transfected cells and mouse NIH 3T3 cells in the presence of cycloheximide, with or without LMB. Cells were fixed and stained with DAPI and an anti-βgal antibody. Arrows indicate mouse nuclei within a heterokaryon. (B) Line drawing of constructs used to examine the shuttling capability of the bUL47 NES when transferred to β-galactosidase. (C) COS-1 cells were transfected with plasmids expressing β-galactosidase fusion proteins as shown in panel B, and heterokaryon assays were carried out in the presence of cycloheximide. Cells were fixed and stained with DAPI and an anti-βgal antibody. Each fusion protein was scored for its ability to shuttle in a heterokaryon, as summarized in panel B. Arrows indicate mouse nuclei. (D) COS-1 cells were transfected with NLS β-galactosidase containing either the Rev NES or the bUL47 NES, and heterokaryon assays were carried out in the presence of cycloheximide and leptomycin B. Cells were fixed and stained with DAPI and an anti-βgal antibody. Arrows indicate mouse nuclei.
TABLE 1.
Efficiency of fusion protein shuttling in COS-1/NIH 3T3 heterokaryons
| Fusion protein | No. of heterokaryonsa | % Shuttledb |
|---|---|---|
| GFP 1-742 | 86 | 100 |
| NLS-βgal | 205 | 20.5 |
| NLS-Rev βgal | 107 | 95 |
| NLS-485-495 βgal | 79 | 82.3 |
| 1-319 βgal | 51 | 96 |
| 1-131 βgal | 51 | 96 |
| 1-66 βgal | 49 | 8.5 |
| NLS-67-131 βgal | 125 | 93.7 |
| NLS-67-123 βgal | 177 | 95 |
| NLS-67-95 βgal | 168 | 21.4 |
| NLS-95-123 βgal | 98 | 96 |
| GFP ΔNES2 | 71 | 94.4 |
Number of heterokaryons counted for each fusion protein.
Percentage of heterokaryons containing fusion protein in the nuclei of both COS-1 cells and NIH-3T3 cells.
TABLE 2.
Efficiency of fusion protein shuttling in COS-1/NIH 3T3 heterokaryons in the presence of leptomycin B
| Fusion protein | No. of heterokaryonsa | % Shuttledb |
|---|---|---|
| GFP 1-742 | 82 | 98.7 |
| NLS-Rev βgal | 170 | 19 |
| NLS-485-495 βgal | 116 | 19 |
| NLS-67-131 βgal | 125 | 93.7 |
| NLS-95-123 βgal | 61 | 88.5 |
| GFP ΔNES2 | 52 | 90.4 |
Number of heterokaryons counted for each fusion protein.
Percentage of heterokaryons containing fusion protein in the nuclei of both COS-1 cells and NIH-3T3 cells.
We next wished to determine if the activity of the bUL47 Leu-rich NES would be transferable to the large protein βgal in the same manner as the Rev NES. To compare the activity of the bUL47 NES with that of HIV-1 Rev, we carried out heterokaryon assays with plasmids encoding the SV40 NLS, the SV40 NLS followed by the Rev NES, and the SV40 NLS followed by the bUL47 NES (residues 485-495), all fused to the N terminus of βgal (Fig. 5B). As expected, NLS-βgal localized to the nuclei of transfected COS-1 cells but did not shuttle into the NIH 3T3 cell nuclei in the heterokaryons (Fig. 5C, NLS, nucleus with arrows; Table 1). By contrast, NLS-Rev-βgal was present in the nuclei of COS-1 and NIH 3T3 cells in a large number of the heterokaryons examined (Fig. 5C, NLS-Rev, nuclei with arrows; Table 1). Likewise, the bUL47 NES also enabled the export of βgal into the NIH 3T3 nuclei within heterokaryons, confirming that residues 485 to 495 do indeed constitute a transferable NES (Fig. 5C, NLS 485-495, nuclei with arrows; Table 1). However, it is noteworthy that the bUL47 NES was consistently weaker than the Rev NES (Fig. 5C, compare NLS-Rev with NLS 485-495). To assess the sensitivity of this NES to LMB, we next carried out heterokaryon assays with cells expressing the Rev NES or the bUL47 NES in the presence of that drug. In this case, the activity of both NESs was greatly inhibited by the presence of LMB, suggesting that, like Rev, the bUL47 Leu-rich NES utilizes the CRM-1 pathway for nuclear export (Fig. 5D, nuclei with arrows; Table 2).
The N terminus of bUL47 contains a second nuclear export signal that is separable from its import signal.
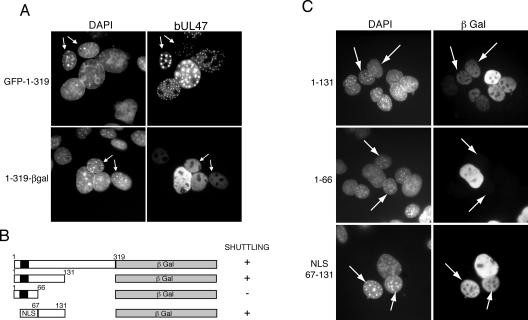
The fact that the bUL47 Leu-rich NES is sensitive to LMB, while nuclear export of full-length bUL47 is resistant to the drug, suggests that the protein may contain a second NES. This is further supported by the fact that full-length bUL47 exhibits efficient nuclear shuttling while the Leu-rich NES alone has only a weak shuttling activity. To assess whether a region of the protein lacking the Leu-rich NES was still able to shuttle between the nucleus and cytoplasm, we tested the ability of the N-terminal half of bUL47 fused to GFP to shuttle in a heterokaryon assay, and found that this protein shuttled efficiently into the nuclei of NIH 3T3 cells within a heterokaryon (Fig. 6A, GFP-1-319, nuclei with arrows). Interestingly, this protein was already localized in its nuclear domains in the cells to which it had shuttled. The molecular weight of the GFP 1-319 peptide is around the minimum size of protein that requires active transport to move out of the nucleus. Thus, to confirm that the N terminus of bUL47 is truly capable of nuclear export, we next fused regions of the N terminus of bUL47 to βgal and determined whether these large fusion proteins could shuttle in a heterokaryon assay (Fig. 6B). Fusion of the first 319 residues of bUL47 to βgal resulted in a protein that was capable of shuttling efficiently into the NIH 3T3 cell nuclei (Fig. 6A, 1-319-βgal, nuclei with arrows; Table 1), confirming that this region of the protein contains a functional NES. It is notable that while GFP 1-319 localizes to nuclear dots, 1-319 βgal is localized in a diffuse nuclear pattern. This difference may be due to the relative presentation of the region required for targeting bUL47 to nuclear dots (residues 261-319), as in the GFP fusion these residues are free at the C terminus of the fusion protein, but in the β-galactosidase fusion, these residues are buried in the center of the open reading frame, just before the β-galactosidase moiety. To further refine the sequences involved in bUL47 shuttling, we truncated the C terminus of the 1-319 region of bUL47 to generate residues 1-131 and 1-66 fused to βgal (Fig. 6B). Heterokaryon assays with these fusion proteins showed that the region 1-131 was able to function efficiently as a shuttling peptide, suggesting that these 131 residues contained both an NLS, as described above, and a NES (Fig. 6C, 1-131, nuclei with arrows; Table 1). By contrast, residues 1-66 containing the bUL47 NLS retained no shuttling activity (Fig. 6C, 1-66, nuclei with arrows; Table 1), implying that the region between residues 67 and 131 is essential for nuclear export. To determine if this region is sufficient for nuclear export, we next replaced the first 66 residues of the 1-131 βgal fusion, containing its natural NLS, with the SV40 NLS (Fig. 6B). When tested in a heterokaryon assay, this fusion protein shuttled efficiently into the nuclei of NIH 3T3 cells (Fig. 6C, NLS 67-131, nuclei with arrows), confirming that residues 67-131 contained a fully transferable nuclear export signal. Furthermore, this transferable NES functioned as efficiently as the Rev NES in the heterokaryon assays (Table 1). Hence, bUL47 contains both an NLS and an NES at the N-terminus of the protein, which although in close proximity to each other are entirely separable.
FIG. 6.
The N terminus of bUL47 contains a second nuclear export signal that is fully separable from its nuclear import signal. (A) COS-1 cells were transfected with plasmids expressing the N-terminal half of bUL47 (1-319) fused to either the C terminus of GFP (GFP-1-319) or the N terminus of β-galactosidase (1-319-βgal). Heterokaryon assays were then carried out between the transfected cells and mouse NIH 3T3 cells in the presence of cycloheximide. Cells were fixed and stained with DAPI and, in the case of the β-galactosidase fusion protein, an anti-βgal antibody. Arrows indicate mouse nuclei. (B) Three peptides from the N-terminal region of bUL47 were fused to the N terminus of β-galactosidase. Black box represents the bUL47 NLS. NLS denotes the SV40 nuclear localization signal. (C) COS-1 cells were transfected with plasmids encoding the βgal fusion proteins described for panel B, and heterokaryon assays were carried out with mouse NIH 3T3 cells in the presence of cycloheximide. The cells were fixed and stained with DAPI and anti-βgal antibody. Each fusion protein was scored for its ability to shuttle in a heterokaryon as summarized in panel B. Arrows indicate mouse nuclei.
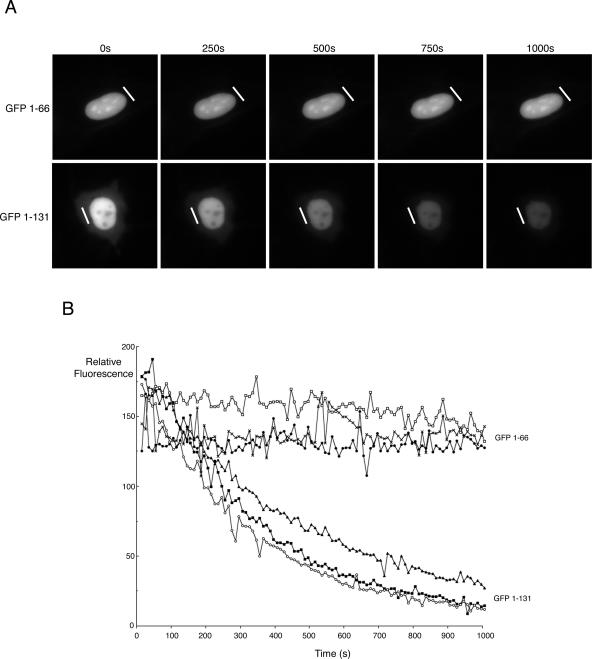
To further confirm that the N terminus of bUL47 is capable of nucleocytoplasmic shuttling, we employed FLIP technology as we had done previously to look at bUL47 shuttling during infection (42). HEp-2 cells were transfected with plasmids expressing GFP fused to either 1-66 of bUL47 or 1-131 of bUL47. Twenty hours later, a small area of cytoplasm in a number of individual expressing cells was photobleached every 10 s for 100 repetitions using the 488-nm laser module of a Deltavision imaging system, with images being acquired after each photobleaching event to produce a time-lapse animation. In the case of GFP 1-66, which lacks the N-terminal NES, repeated bleaching of the cytoplasm had no effect on the intensity of nuclear fluorescence in expressing cells, confirming that this protein does not shuttle between the nucleus and cytoplasm (see example in Fig. 7A). By contrast, in cells expressing GFP 1-131, which contains both the NLS and the N-terminal NES, repeated bleaching of the cytoplasm resulted in a rapid loss of fluorescence in the nuclei of expressing cells, indicating that not only does the region 67-131 of bUL47 constitute a functional NES but that this NES is highly efficient (see example in Fig. 7A). A number of analyses were carried out for each of these GFP fusion proteins, and quantification of nuclear GFP fluorescence during each time-lapse indicated that the results for each fusion protein were highly consistent (Fig. 7B).
FIG. 7.
Analysis of nucleocytoplasmic shuttling by FLIP. (A) HEp-2 cells were transfected with GFP fused to either residues 1-66 or residues 1 to 131 of bUL47. Twenty hours later, the expressing cells were examined using a Deltavision RT imaging system, and individual cells were chosen for FLIP analysis. The laser module was then used to carry out sequential photobleaching events of the area denoted by the white line. This area was exposed to the laser every 10 s for 100 repeats. An image of the field was acquired after each bleach event to determine the loss of fluorescence in the nucleus of the cell. (B) For each fusion protein, the relative fluorescence in the nuclei of three individual cells that had been subjected to FLIP was quantitated using NIH Image software and plotted against time.
Identification of a novel, LMB-resistant NES in bUL47.
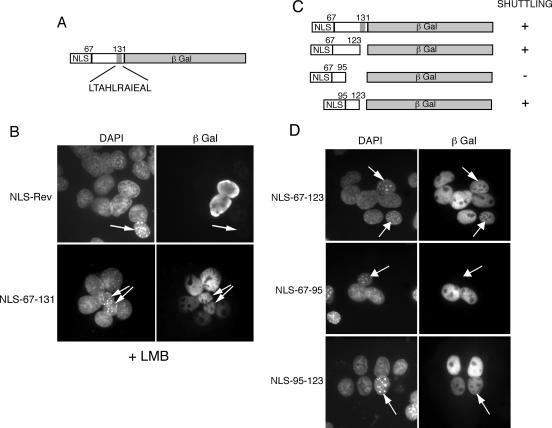
When we analyzed the sequence of the bUL47 NES peptide, we identified a leucine-rich region between residues 123 and 131 (Fig. 8A), and so to determine if this NES was also shuttling through the CRM-1 pathway, we carried out heterokaryon assays in the presence of LMB. We found that, as for the full-length protein, shuttling of βgal dictated by bUL47 residues 67-131 was also resistant to leptomycin B, suggesting that the NES present in the N terminus of bUL47 utilizes an export receptor other than CRM-1 (Fig. 8B, compare NLS-Rev with NLS-67-131, nuclei with arrows; Table 2). To further refine the sequences involved in the nuclear export of bUL47, and to confirm that the leucine stretch between residues 123 and 131 was not involved in nuclear export, we removed these residues from the NES peptide (Fig. 8C). Heterokaryon assays with this fusion protein showed clearly that the peptide was still capable of shuttling efficiently into NIH 3T3 nuclei and, hence, the leucines were not required (Fig. 8D, NLS-67-123, nuclei with arrows; Table 1). We then examined the shuttling potential of the N-terminal or the C-terminal halves of region 67-123 (Fig. 8D, NLS-67-95 and NLS-95-123). Heterokaryon assays with residues 67-95 demonstrated that this peptide no longer retained the ability to export the fusion protein from the COS-1 nuclei (Fig. 8D, NLS-67-95, nucleus with arrows; Table 1). By contrast, residues 95-123 were still able to efficiently export the βgal fusion protein (Fig. 8D, NLS-95-123, nucleus with arrows), exhibiting a shuttling efficiency that was similar to the HIV-1 Rev NES (Table 1). Thus, we have identified a 28-residue peptide at the N terminus of bUL47 that functions as a transferable NES, contains no leucine residues, and is resistant to leptomycin B (Table 2).
FIG. 8.
Delineation of an N-terminal NES in bUL47 that is resistant to leptomycin B. (A) Line drawing of the NES-containing region of bUL47 showing a leucine-rich region (gray box). (B) COS-1 cells were transfected with plasmids expressing the NLS-Rev-βgal control or NLS-67-131 βgal, and heterokaryon assays were carried out in the presence of cycloheximide and leptomycin B. The cells were fixed and stained with DAPI and an anti-βgal antibody, and each fusion protein was scored for its ability to shuttle in a heterokaryon in the presence of leptomycin B. Arrows indicate mouse nuclei. (C) A range of peptides from the N-terminal 131 residues of bUL47 was fused to the N terminus of β-galactosidase. In all cases, the SV40 NLS was fused in front of the bUL47 peptide. (D) COS-1 cells were transfected with plasmids encoding these βgal fusion proteins, and heterokaryon assays were carried out with mouse NIH 3T3 cells 2 days later in the presence of cycloheximide. The cells were fixed and stained with DAPI and anti-β-galactosidase antibody. Each fusion protein was scored for its ability to shuttle in a heterokaryon, as summarized in panel C. Arrows indicate mouse nuclei.
Does bUL47 contain a third NES?
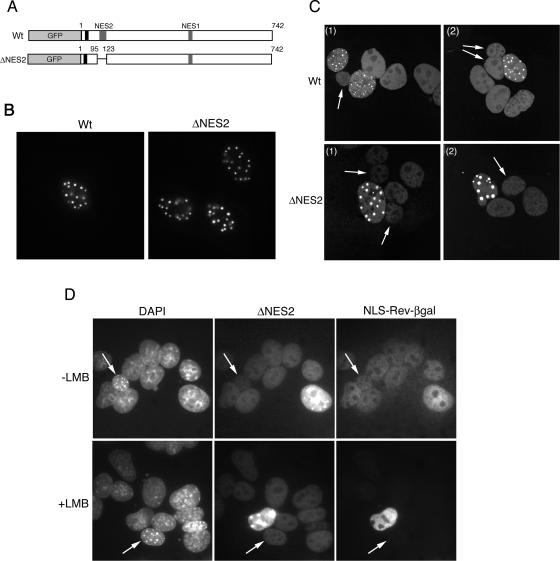
To assess the contribution made by the two NES sequences to nuclear export of full-length bUL47, we constructed a variant of the protein in which NES2, residues 95 to 123, had been deleted (Fig. 9A). Live-cell imaging of this protein demonstrated that its steady-state localization was similar to that of Wt protein, localizing to the nucleus in specific nuclear dots (Fig. 9B). Heterokaryon assays indicated that, although the level of ΔNES bUL47 present in NIH 3T3 nuclei within a heterokaryon was significantly reduced compared to that seen for Wt protein, as might be expected from the relative strength of the signals observed with β-galactosidase fusion proteins, the mutant was still capable of shuttling (Fig. 9C and Table 1). Because the NES remaining in the mutant protein is sensitive to LMB, we therefore expected the ΔNES mutant, which now lacked the LMB-resistant NES, to be sensitive to the drug. To test this, we coexpressed the ΔNES mutant with NLS-Rev-βgal and carried out heterokaryon assays in the presence of LMB. Surprisingly, although Rev nuclear export was inhibited in this experiment, the weak shuttling of the bUL47 ΔNES mutant was still resistant to LMB (Fig. 9D, nuclei with arrows; Table 2). We believe that the most likely interpretation of these data is that a third, as-yet-unidentified NES, one that is also resistant to LMB, is located within the bUL47 protein.
FIG. 9.
Nucleocytoplasmic shuttling properties of full-length bUL47 lacking NES2. (A) The N-terminal NES of bUL47 was mutated by deleting residues 95-123 from the full-length protein (ΔNES2). (B) Wt and ΔNES2 proteins were expressed as GFP fusion proteins in HEp-2 cells and analyzed by live-cell microscopy. (C) Both constructs were transfected into COS-1 cells, and heterokaryon assays were carried out with mouse NIH 3T3 cells in the presence of cycloheximide. Cells were examined live. Two examples of heterokaryons are given for each construct, showing only the GFP channel. Arrows indicate mouse nuclei within a heterokaryon. (D) COS-1 cells were cotransfected with plasmids expressing GFP-bUL47 and the NLS Rev β-galactosidase reporter. Heterokaryon assays were then carried out between the transfected cells and mouse NIH 3T3 cells in the presence of cycloheximide, with LMB (+LMB) or without LMB (−LMB). Cells were fixed and stained with DAPI and an anti-βgal antibody. Arrows indicate mouse nuclei within a heterokaryon.
DISCUSSION
The role that is played by the alphaherpesvirus UL47 gene product during virus infection remains to be defined. Although UL47 is a major structural protein of the tegument of several alphaherpesviruses, it is likely that it has additional contributions to make during the virus replication cycle, as is the case for many other tegument proteins. The UL47 proteins from both HSV-1 and BHV-1 have been shown to undergo nucleocytoplasmic transport, but the exact relevance of this to virus infection is yet to be demonstrated (7, 42, 48). Of all the UL47 proteins, the bUL47 protein is unusual in that it is by far the major structural protein of the BHV-1 virion and is consequently delivered into the cell immediately upon infection in a relatively high concentration. Hence, bUL47 is likely to be a good model for determining the role of UL47 in virus infection. We have recently shown that the virion population of bUL47 is rapidly targeted to the nucleus, suggesting that it may have a role to play there at very early times in infection (42). Although we have also shown that newly synthesized bUL47 shuttles rapidly between the nucleus and the cytoplasm of the infected cell, we do not yet know if the early virion-associated population of the protein also actively shuttles (42).
Before investigating the role of bUL47 nucleocytoplasmic shuttling in infection, and establishing the cellular trafficking pathways used by this protein, we wished to identify the exact signals used by it to enter and exit the nucleus. By using a system that requires a transport signal to be fully transferable to a protein too large to diffuse through nuclear pores, we have refined the major bUL47 NLS to a 20-residue peptide encompassing R box 1 at the N terminus of the protein. We have also shown that the arginine residues are essential for the activity of this nuclear import signal. A previous publication characterizing the signals involved in bUL47 transport identified a NLS different from the one we have shown here (48). In that study, deletion mutagenesis was used to show that R box 2, but not R box 1, was essential for nuclear localization of bUL47. This is in direct contrast to our own results, which show that the mutation of only the four arginine residues in R box 1 in the context of full-length protein is sufficient to abrogate nuclear accumulation (Fig. 3). The previous study also showed that fusion of small peptides encompassing either of the arginine-rich boxes to YFP (Fig. 1A, BHV-1) could result in the accumulation of YFP in the nucleus of expressing cells. However, because YFP is small enough to diffuse into the nucleus, its use as a reporter makes it impossible to determine if the arginine motifs are functioning as true nuclear import signals in these assays by interacting with a cellular receptor or, rather, are effecting nuclear retention by virtue of the basic peptide binding to negatively charged nuclear molecules such as nucleic acid. In our own study presented here, we have used the large cytosolic protein CMPK as a reporter to identify the exact NLS of bUL47. Our results show clearly that neither of the arginine boxes in bUL47 (R box 1 and 2) alone is sufficient to import CMPK into the nucleus, but that R box 1 within the context of a larger, 20-residue peptide functions as an efficient transferable NLS. By contrast, a peptide containing R box 2 exhibits a much weaker ability to import CMPK into the nucleus, suggesting that this peptide probably does not contain a strong nuclear import signal. While it is still possible that the bUL47 contains more than one NLS, it is clear that we have identified the most powerful NLS in our studies, as mutation of just four arginine residues within this signal dramatically alters the exclusively nuclear localization of full-length protein (Fig. 3).
The essential RRPRRS motif that is R box 1 in bUL47 is relatively conserved in the other homologues of UL47 that we examined (Fig. 1A) and is also present in the previously defined NLS from the HTLV1 Rex protein, which was one of the first proteins characterized to bind directly to the import receptor importin β (Fig. 10B) (4, 33). While further in vitro studies are required to confirm the exact cellular receptor that the bUL47 NLS binds, the conserved nature of the RRPRRS motif suggests that it may represent a novel signal that defines nuclear import, albeit in the context of a longer peptide. The fact that residues in addition to this motif are absolutely required for nuclear import may simply reflect the need for the RRPRRS motif to be presented correctly for binding to its receptor. Alternatively, other residues within the NLS peptide may be actively involved in receptor binding. Interestingly, although hUL47 contains an R box at the same position in its sequence as the bUL47 RRPRRS box, we have shown previously that this box is entirely dispensable for nuclear localization of hUL47 (7). Furthermore, it seems to be two additional RRRR motifs between residues 63 and 75 that are crucial for hUL47 nuclear localization (7), implying that hUL47 may have evolved to use a nuclear import receptor different from that used by bUL47. By contrast, the other UL47 homologues that we have examined have a conserved RRPRRS motif but no other R box domains. As they all exhibited strong nuclear targeting, it is tempting to speculate that this N-terminal motif may be required for the nuclear localization of all these proteins (Fig. 1).
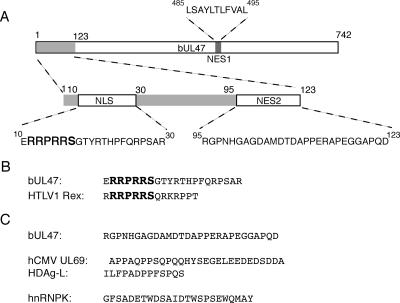
FIG. 10.
The bUL47 nuclear import and export signals. (A) Line drawing of full-length bUL47 with the N-terminal region involved in nuclear shuttling expanded. The peptide sequences of the bUL47 NLS, NES1, and NES2 signals are shown. (B) Comparison between the bUL47 NLS and the NLS from HTLV1 Rex protein. (C) The sequence of the bUL47 NES2 differs from the other nonclassical nuclear export signals so far defined from the virus proteins hCMV UL69, hepatitis delta antigen (HDAg-L), and the cellular protein hnRNPK.
In the study described above by Zheng and coworkers, an export signal which was chosen originally for analysis because it was one of three leucine-rich stretches present in the bUL47 open reading frame was identified by fusion of residues 485 to 495 to YFP (48). By inference, this NES would be expected to use the CRM-1 pathway for export from the nucleus, and in the current study we have now gone on to show that this signal is both transferable to a large protein and sensitive to the CRM-1 inhibiting drug leptomycin B. However, this leucine-rich NES exhibited only weak shuttling activity in our assays compared to the efficient shuttling that was evident for full-length protein. Furthermore, we also show that export of full-length bUL47 is resistant to treatment with leptomycin B and identify a second export signal that is located at the N terminus of the protein and is CRM-1 independent. Although we refined this second NES by demonstrating that the N-terminal region of bUL47, encompassing residues 1-319, is fully functional in heterokaryon shuttling even though it lacks the Leu-rich NES, this result once again conflicts with the result from the previous study, where deletion of the Leu-rich NES totally abrogated the ability of bUL47 to shuttle in a heterokaryon assay. As yet, it is not entirely clear why the two independent studies have produced such different results.
The export signal that we have identified at the N terminus of bUL47 is fully separable from the import signal and appears to represent a novel nonclassical nuclear export signal (Fig. 10A). This signal is completely resistant to treatment with leptomycin B, confirming that it does not function through the well-characterized CRM-1 pathway. As shown in Fig. 10C, it also bears little similarity to other nonclassical export signals that have been previously characterized, such as the serine/acidic signal found in hnRNPK (17) or the positively charged signals found in eIF1 and p50RhoGAP (29). There are, however, four proline residues at the C terminus of this NES, which may suggest a very weak homology with the proline-rich signals found in hCMV UL69 and hepatitis delta antigen (21, 22). While a number of cellular export receptors other than CRM-1 and TAP have now been identified, including specific receptors, such as exportin-t for tRNA transport (1), and more general export factors, such as exportin-7 (29), the receptors for most of the nonclassical export signals remain to be elucidated. Very recently, a novel receptor for the hepatitis delta antigen has been identified and shown to be present predominantly in human liver tissue where the hepatitis delta virus replicates (43). Similarly, the nature of the second bUL47 NES may imply that this protein can use an as-yet-unidentified cell receptor to transport it out of the nucleus, which may be predominant in bovine cells, the natural host for BHV-1. Alternatively, bUL47 may bind to a cellular partner that, in turn, binds to an export receptor. Interestingly, and unlike the situation with the bUL47 NLS, the N-terminal NES is not obviously conserved in any of the other UL47 homologues, including hUL47, which is already known to undergo export from the nucleus (7). Furthermore, the fact that bUL47 exhibited nucleocytoplasmic shuttling (albeit weak activity) under conditions where the activity of both NES1 and NES2 was inhibited suggests that there is likely to be yet another NES present in the bUL47 open reading frame, and further characterization of this as-yet-unidentified NES may help identify NES sequences in the other UL47 proteins. Very recently, the p37 protein of African swine fever virus has also been shown to contain three independent NES sequences (9), and like bUL47, this protein is a major structural protein of the virus that is nuclear at early times in infection but cytoplasmic at late times. It is therefore possible that such viral proteins require multiple NES sequences because they need to interact with different receptors at various times in infection.
The UL47 group of proteins can now be added to a number of other groups of virus-encoded shuttling proteins, including the Rev/Rex grouping of retrovirus proteins and the ICP27/EB2/ORF57 grouping of herpesvirus proteins (38). These proteins are all well defined as being involved in RNA transport, a major property of nucleocytoplasmic shuttling proteins. Although there is no evidence for the involvement of UL47 in RNA transport, hUL47 has been described as an RNA-binding protein (40), and the dynamics of bUL47 shuttling is sensitive to inhibitors of transcription, such as actinomycin D, in the same manner as other RNA-transporting proteins (35, 36, 42). Moreover, most RNA-transporting proteins so far identified use import pathways other than the classical importin α route, and we have now shown that both the hUL47 and the bUL47 proteins contain arginine-rich NLSs that are generally considered indicative of import via importin β (7). Early studies of HSV-1 UL47 have also implicated the protein in the regulation of viral gene expression, as cells infected with HSV-1 lacking the UL47 gene exhibit an 80% reduction in immediate-early gene expression, an effect that could potentially take place at a posttranscriptional level such as RNA transport (23, 46, 47). To date, no UL47 knockout has been described for BHV-1; hence, it is not yet known how essential bUL47 is to the replication cycle of BHV-1.
In summary, the characterization of the viral structural protein as presented here has shown that the nuclear trafficking of bUL47 is highly complex. It has revealed that the protein possesses potentially three nuclear export signals, one of which we have characterized as a novel signal in the N terminus of the protein. As the description of nonclassical nuclear trafficking signals is still developing, it is hoped that further studies on the shuttling properties of all UL47 proteins will aid in the identification of new cellular receptors for nuclear trafficking.
Acknowledgments
We thank Vikram Misra, Thomas Mettenleiter, Klaus Osterreider, and Valerie Preston for providing DNA for the UL47 genes from BHV-1, PRV, EHV-1, and VZV, respectively. We also thank J. Frangioni for the CMPK reporter plasmid and M. Dobbelstein for the βgal control plasmids.
This work was funded by Marie Curie Cancer Care.
REFERENCES
- 1.Arts, G. J., M. Fornerod, and I. W. Mattaj. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305-314. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell 102:99-108. [DOI] [PubMed] [Google Scholar]
- 3.Bednenko, J., G. Cingolani, and L. Gerace. 2003. Importin β contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 162:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., R. A. Fridell, R. E. Benson, J. Hua, and B. R. Cullen. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter, D. E., and V. Misra. 1991. The most abundant protein in bovine herpes 1 virions is a homologue of herpes simplex virus type 1 UL47. J. Gen. Virol. 72:3077-3084. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnelly, M., and G. Elliott. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulalio, A., I. Nunes-Correia, A. L. Carvalho, C. Faro, V. Citovsky, J. Salas, M. L. Salas, S. Simoes, and M. C. de Lima. 2006. Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals. J. Virol. 80:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 11.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 12.Frangioni, J. V., and B. G. Neel. 1993. Use of a general purpose mammalian expression vector for studying intracellular protein targeting: identification of critical residues in the nuclear lamin A/C nuclear localization signal. J. Cell Sci. 105:481-488. [DOI] [PubMed] [Google Scholar]
- 13.Goldfarb, D. S., A. H. Corbett, D. A. Mason, M. T. Harreman, and S. A. Adam. 2004. Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505-514. [DOI] [PubMed] [Google Scholar]
- 14.Görlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 15.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 16.Grüter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649-659. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 18.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 19.Kang, Y., and B. R. Cullen. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, C.-H., S. C. Chang, C. H. H. Wu, and M.-F. Chang. 2001. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 276:8142-8148. [DOI] [PubMed] [Google Scholar]
- 22.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKnight, J. L., P. E. Pellett, F. J. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing factor-dependent activation of α genes. J. Virol. 61:992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLean, G., F. Rixon, N. Langeland, L. Haarr, and H. Marsden. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 71:2953-2960. [DOI] [PubMed] [Google Scholar]
- 25.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology 242:128-137. [DOI] [PubMed] [Google Scholar]
- 26.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 27.Michael, W. M. 2000. Nucleocytoplasmic shuttling signals: two for the price of one. Trends Cell Biol. 10:46-50. [DOI] [PubMed] [Google Scholar]
- 28.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 29.Mingot, J. M., M. T. Bohnsack, U. Jakle, and D. Görlich. 2004. Exportin 7 defines a novel general nuclear export pathway. EMBO J. 23:3227-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosammaparast, N., and L. F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547-556. [DOI] [PubMed] [Google Scholar]
- 31.Nishi, K., M. Yoshida, D. Fujiwara, M. Nishikawa, S. Horinouchi, and T. Beppu. 1994. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 269:6320-6324. [PubMed] [Google Scholar]
- 32.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmeri, D., and M. H. Malim. 1999. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol. Cell. Biol. 19:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 35.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78:3327-3331. [DOI] [PubMed] [Google Scholar]
- 36.Pinol-Roma, S., and G. Dreyfuss. 1991. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science 253:312-314. [DOI] [PubMed] [Google Scholar]
- 37.Roth, J., and M. Dobbelstein. 1997. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J. Virol. 71:8933-8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt, I., and L. Gerace. 2001. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J. Biol. Chem. 276:42355-42363. [DOI] [PubMed] [Google Scholar]
- 40.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol. 19:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhagen, J., I. Hutchinson, and G. Elliott. 2006. Nucleocytoplasmic shuttling of the bovine herpesvirus type 1 UL47 protein in infected cells. J. Virol. 80:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Y. H., S. C. Chang, C. Huang, Y. P. Li, C. H. Lee, and M. F. Chang. 2005. Novel nuclear export signal-interacting protein, NESI, critical for the assembly of hepatitis delta virus. J. Virol. 79:8113-8120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 45.Whittaker, G. R., M. P. Riggio, I. W. Halliburton, R. A. Killington, G. P. Allen, and D. M. Meredith. 1991. Antigenic and protein sequence homology between VP13/14, a herpes simplex virus type 1 tegument protein, and gp10, a glycoprotein of equine herpesvirus 1 and 4. J. Virol. 65:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., D. A. Sirko, and J. L. C. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, C., R. Brownlie, L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2004. Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology 324:327-339. [DOI] [PubMed] [Google Scholar]