Abstract
In Brome mosaic virus (BMV), genomic RNA1 (gB1) and RNA2 (gB2), encoding the replication factors, are packaged into two separate virions, whereas genomic RNA3 (gB3) and its subgenomic coat protein (CP) mRNA (sgB4) are copackaged into a third virion. In vitro assembly assays performed between a series of deletion variants of sgB4 and wild-type (wt) CP subunits demonstrated that packaging of sgB4 is independent of sequences encoding the CP open reading frame. To confirm these observations in vivo and to unravel the mechanism of sgB4 copackaging, an Agrobacterium-mediated transient in vivo expression system (P. Annamalai and A. L. N. Rao, Virology 338:96-111, 2005) that effectively uncouples replication from packaging was used. Cultures of agrotransformants, engineered to express sgB4 and CP subunits either transiently (sgB4Trans and CPTrans) or in replication-dependent transcription and translation when complemented with gB1 and gB2 (sgB4Rep and CPRep), were mixed in all four pair-wise combinations and infiltrated to Nicotiana benthamiana leaves to systematically evaluate requirements regulating sgB4 packaging. The data revealed that (i) in the absence of replication, packaging was nonspecific, since transiently expressed CP subunits efficiently packaged ubiquitous cellular RNA as well as transiently expressed sgB4 and its deletion variants; (ii) induction of viral replication increased specificity of RNA packaging; and most importantly, (iii) efficient packaging of sgB4, reminiscent of the wt scenario, is functionally coupled not only to its transcription via replication but also to translation of CP from replication-derived mRNA, a mechanism that appears to be conserved among positive-strand RNA viruses of plants (this study), animals (flock house virus), and humans (poliovirus).
Packaging of viral genomes by structural protein components leading to the assembly of infectious progeny virions is an essential step in the life cycle of plus-strand RNA viruses pathogenic to humans, animals, insects, and plants (28, 33). The assembly of infectious virions is a carefully orchestrated process that maintains a high degree of precision and specificity, requiring protein subunits interacting with each other and with viral nucleic acids (5). The majority of RNA viruses infecting eukaryotic cells are assembled in the cytoplasm, and therefore an opportunity exists to copackage cellular RNA. However, remarkably mature virions exclusively contain viral RNA, with the exception of a few viruses, such as human immunodeficiency virus, whose virions contain cellular tRNAs. The mechanism and factors that regulate this specificity are not fully understood. However, a high degree of specificity between sequence- and/or structure-dependent interactions of RNA and coat protein (CP) operates during the assembly process (28, 33). Thus, knowledge of detailed mechanisms by which viruses assemble and package their genomes into structurally stable virions is an important prerequisite for understanding overall biology of eukaryotic RNA viruses.
The infection cycle of Brome mosaic virus (BMV), a member of the family Bromoviridae of plant RNA viruses (27), is typical of eukaryotic RNA viruses, and its replication is entirely cytoplasmic (13). The genome of BMV is divided among three RNAs (27). Viral replication is dependent on efficient interaction between nonstructural proteins 1a and 2a, encoded, respectively, by genomic RNA1 (gB1) and RNA2 (gB2) (17, 35). Genomic RNA3 (gB3) is dicistronic. Its 5′ half encodes another nonstructural movement protein (MP) that promotes cell-to-cell spread, while the capsid protein gene (CP) encoded in the 3′ half is translationally silent but is expressed from a subgenomic RNA (sgB4) that is synthesized from progeny minus-strand gB3 by internal initiation mechanisms (22). Therefore, synthesis of sgB4 is contingent on replication of gB3. BMV is a T=3 icosahedral virus whose protein shell is composed of 180 subunits of an ∼20-kDa coat protein (20) packaging all four RNAs (27, 28). Physical and biochemical characterization of BMV virions suggested that gB1 and gB2 are packaged independently into two separate virions, whereas gB3 and sgB4 are copackaged into a third virion (27, 28). Despite variation in configuration of RNA packaging, it is remarkable that all three virions are morphologically indistinguishable and physically inseparable. Consequently, the distribution of four RNAs into three virions is tightly regulated during infection, and the mechanism that controls the proposed packaging scheme is currently obscure.
In BMV, division of function among three different genomic RNAs (gRNAs), together with our ability to reconstitute infectious virus in vitro from dissociated CP subunits and RNA under a variety of physiological conditions (10, 41), has led to an understanding of the mechanism of genome packaging in an RNA virus (7, 10, 28). All four BMV RNAs contain a highly conserved 200-nucleotide (nt) 3′ untranslated region that can be folded to mimic a tRNA-like structure (TLS). TLS has been shown to be an integral part of virus assembly by functioning as a nucleating element of CP subunits (10). However, it is hard to reconcile the specific distribution of four BMV RNAs into three morphologically indistinguishable virions if viral 3′ TLS is the only element that interacts with BMV CP and promotes packaging. Since the entire sequence of sgB4 is located in the 3′ half of gB3, it is envisioned that copackaging is facilitated by a signal located in a commonly shared CP open reading frame (ORF) region. But in vitro assembly assays showed that deletion of the CP ORF has no significant effect on RNA3 packaging (10). A finer deletion analysis of other regions of gB3 revealed that in addition to 3′ TLS, a cis-acting, position-dependent 187-nt region present in the MP ORF is essential for efficient packaging (10). Therefore, the mechanism that regulates copackaging of gB3 and sgB4 is not known. A major limitation in advancing our knowledge to this end can largely be attributed to existing in vivo systems that are unable to dissect replication from packaging. The application of Agrobacterium-mediated synchronized delivery and expression of a desired set of viral mRNAs via transfer DNA (T-DNA)-based constructs (3, 15) has proven to be versatile in unraveling the relationship between replication and packaging (3). Here we extended this approach to systematically evaluate requirements of sgB4 packaging and report that efficient packaging of sgB4 is not only contingent on its synthesis via replication but also requires the translation of CP from replication-derived mRNA.
MATERIALS AND METHODS
Construction of T7-based sgB4 variants.
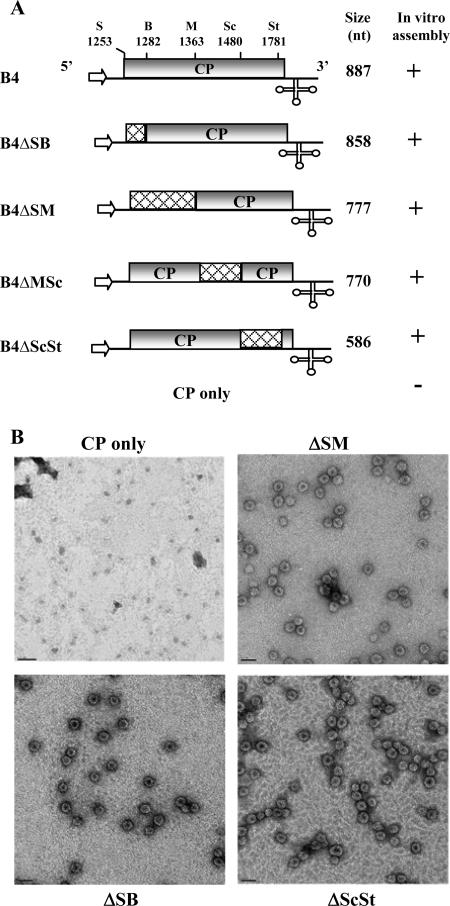
Construction and characterization of pT7B4 plasmid containing a cDNA copy of sgB4 was described previously (Fig. 1A) (8). In vitro transcription of this plasmid with T7 RNA polymerase resulted in synthesis of sgB4 transcripts identical to those found in wild-type (wt) BMV virions (8). Selected deletions spanning the entire CP ORF of pT7B4 were engineered by sequential digestion with specified restriction enzymes and mung bean nuclease (Fig. 1A). Variant sgB4 transcripts derived from these clones were B4/ΔSB (nt 1253 to 1282 deleted), B4/ΔSM (nt 1253 to 1363 deleted), B4/ΔMSc (nt 1363 to 1480 deleted), and B4/ΔScSt (nt 1480 to 1781 deleted). Because in vitro assembly of BMV RNA does not require a 5′ cap structure (41), noncapped transcripts from wt and variant clones of sgB4 were synthesized in vitro using T7 RNA polymerase (7). Procedures used to prepare wt CP subunits and conditions of in vitro assembly assays were as described previously (7).
FIG. 1.
Schematic representation of wild-type (wt) and deletion variants of B4. (A) The organization of wt B4 is shown, with noncoding sequences represented by solid lines and the open box representing the CP ORF region. The arrow at the 5′ end represents the position of T7 RNA polymerase, and the cloverleaf structure at the 3′ end represents a tRNA-like structure (3′ TLS). Restriction sites used for constructing variants of the full-length cDNA clone of B4 are shown above the map. Cross-hatched boxes in each variant clone represent the extent of each engineered deletion. The lengths of in vitro-synthesized RNA transcripts corresponding to wt B4 and its deletion variants are shown. The results of in vitro assembly assays between wt CP subunits and noncapped RNA transcripts (7) of each B4 variant are shown. +, virion assembly; −, no virion assembly. S, SalI; B, BssHII; M, MscI; Sc, SacI; St, StuI. (B) Electron micrographic images of negatively stained virions for selected representative samples from the above-mentioned in vitro assembly assays. Bar, 50 nm.
Electron microscopy.
For negative staining, purified virus preparation was applied to glow-discharged carbon-coated copper grids (1). Grids were washed once with water, stained with 1% uranyl acetate, and air dried. Grids were examined with an FEI Tecnai12 transmission electron microscope operating at 100 kV, and images were recorded digitally.
Wild-type BMV plasmids for agroinfiltration.
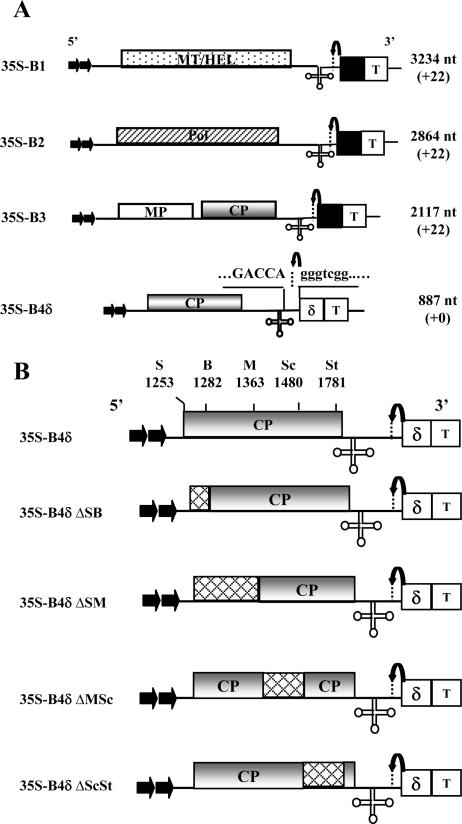
The construction and characterization of T-DNA-based plasmids for efficient transient expression of full-length BMV genomic RNA1 (35S-B1), RNA2 (35S-B2), and RNA3 (35S-B3) were described previously (3). Each plasmid contains, in sequential order, a double 35S promoter, cDNA complementary to respective full-length BMV RNAs, a ribozyme sequence of Satellite tobacco ring spot virus (Rz), and a 35S terminator (Fig. 2A).
FIG. 2.
(A) Characteristics of T-DNA plasmids harboring BMV genomic RNAs used for Agrobacterium-mediated transient expression in plants. The 35S-B1, 35S-B2, and 35S-B3 constructs contain full-length cDNA copies of BMV genomic RNA1 (B1), RNA2 (B2), and RNA3 (B3), respectively (3). Filled arrows at the 5′ end represent the location of double 35S promoters (35S), whereas a filled square and T, respectively, denote ribozyme sequence cassettes derived from Satellite tobacco ring spot virus and the 35S-polyadenylation terminator signals. The bent arrow at the 3′ end represents a ribozyme cleavage site. Characteristics of a T-DNA construct of BMV sgB4 (35S-B4δ) used for transient expression with authentic 5′ and 3′ termini are shown. In 35S-B4δ, the position of a hepatitis delta ribozyme (δ) is shown. All other features are the same those described above. Viral and nonviral nucleotide sequences located at the 3′ end are shown by uppercase and lowercase letters, respectively, and the bent arrow represents the ribozyme cleavage site. The lengths of wt BMV RNAs and the number of nonviral nucleotides left after self cleavage by ribozymes (shown in brackets) are indicated. (B) Characteristic features of agrotransformants of sgB4 and its deletion variants. Characteristic features of each variant are the same as those describe in the legend to Fig. 1. S, SalI; B, BssHII; M, MscI; Sc, SacI; St, StuI.
Construction of 35S-B4δ and its variants for agroinfiltration.
Initially, a variant of pCASS4-RZ (3) was constructed to replace the Satellite tobacco ring spot virus ribozyme (Rz) with hepatitis delta virus ribozyme (δ) in a PCR using forward primer d (AGGGAATTCGGTACCCCATGGGGTCGGCATGGCATCTC; boldface and underline sequences represent, respectively, KpnI and NcoI sites) and reverse primer d (GCTCGGTACGAGCTCTCCTGGCTCTCCCTTAGC; the SacI site is underlined). The resulting fragment was digested with KpnI and SacI and ligated into similarly treated pCASS4-RZ, yielding pCASS4-HDVδ. Full-length wt B4 and each B4 deletion variant sequence was independently amplified in a PCR using forward primer d (TGTCCTAATTCTACGTATTAATAATGTC; SnaBI is underlined) and reverse primer d (ACACACACCATATGGTCTCTTTTAGAG; the NdeI site is underlined). The resulting product was digested with NdeI and blunt ended with mung bean nuclease followed by digestion with SnaBI. The resulting product was subcloned into pCASS4-HDVδ that was digested previously with StuI/NcoI and blunt ended using mung bean nuclease.
Construction of B3 variants for agroinfiltration.
T-DNA-based variant clones 35S-B3ΔSl/St, 35S-B3ΔB/St, and 35S-B3ΔSc/St were constructed by deleting CP ORF sequences located between each unique restriction site. For constructing B3 variants having mutations in the N-proximal region of CP, plasmids pT3B3/10P, pT7B3/13P, and pT7B3/14P (9) were digested with SalI and StuI, and the resulting fragments were subcloned into a similarly treated 35S-B3 clone (3).
Agroinfiltration.
The agroinfiltration procedure was performed as described previously (2, 16). Briefly, Agrobacterium sp. strain EHA105 containing desired transformant was initially streaked on Luria-Bertani (LB) plates containing antibiotics (kanamycin, 50 μg/ml; rifampin, 10 μg/ml) and incubated at 28°C. A single colony was inoculated into 2 ml LB medium with the above antibiotics and grown at 28°C for 48 h with vigorous shaking. One milliliter of the culture was transferred to 50 ml LB medium containing the above antibiotics, 10 mM morpholineethanesulfonic acid (pH 5.6), and 40 μM acetosyringone. After incubation at 28°C for 16 h with vigorous shaking, the optical density at 600 nm of the culture reached 1.0. The bacteria were spun down at 2,000 × g for 10 min, the pellet was resuspended in 50 ml 10 mM MgCl2, and then 125 μl 100 mM acetosyringone was added. To maintain a uniform concentration of Agrobacterium in coinfiltration experiments involving two or more Agrobacterium cultures, the optical density at 600 nm of each culture was adjusted to 1.0. The final inoculum was prepared by mixing equal volumes of each culture. The bacteria were kept at room temperature for at least 3 h without shaking. These cultures were then infiltrated into the abaxial surface of the fully expanded N. benthamiana leaves using a 1-cc syringe without a needle. pCASS4 (3) was used as an empty vector (EV) as a negative control as well as to balance the inoculum concentration.
Progeny analysis and packaging assays.
Total RNA from agroinfiltrated leaves were extracted using a hot phenol method (3), and the RNA pellet was suspended in RNase-free water. Virions were purified from agroinfiltrated leaves as described previously (30). For Northern blot analysis (3), samples of virion RNA (0.5 μg) or plant total RNA (5 μg) were dried in a microcentrifuge and suspended in 10 μl of sample buffer (10× morpholinepropanesulfonic acid buffer-formamide-formaldehyde-H2O in a ratio of 1:1.8:5:2.2, respectively), heated at 65°C for 10 min, and electrophoresed in a 1.2% agarose-formaldehyde gel (32). Following a 3-h electrophoresis, fractionated RNA was transferred to a nylon membrane with a VacuGene XL blotting unit (Pharmacia Biotech). The blot was then processed for prehybridization and hybridization using 32P-labeled riboprobes corresponding to either a 3′ conserved region (29), the MP ORF, the CP ORF, or sequences specific for B1 or B2, as described previously (8). For the preparation of a cellular RNA probe, total nucleic acids were isolated from healthy leaves of N. benthamiana using hot phenol and LiCl precipitation (38). RNAs were decapped with tobacco acid pyrophosphate (Epicenter, Madison, WI), followed by dephosphorylation with alkaline phosphatase prior to end labeling with [γ-32P]ATP using T4 polynucleotide kinase (32). CP samples were analyzed by Western blotting as described previously (24). Progeny accumulation was quantitated as described previously (3).
RESULTS
In vitro packaging of sgB4 is independent of CP ORF sequences.
Previous in vitro assembly assays with gB3 and sgB4 variant transcripts lacking a 3′ TLS demonstrated its importance in packaging (7). Additional in vitro packaging assays with gB3 variants harboring deletions in either the MP ORF or CP ORF revealed that 3′ TLS and a cis-acting 187-nt sequence of MP ORF, but not CP ORF, constitute the core packaging signal for gB3 (10). In light of these observations, the issues that remain to be addressed are how gB3 and sgB4 copackage and whether CP ORF sequences have a specific role in sgB4 packaging. To address this, we engineered a series of deletions into the CP ORF region of the pT7B4 clone (10) using the existing unique restriction sites. Transcripts of wt sgB4 and its deletion variants were incubated under in vitro assembly conditions to form virions with wt CP subunits (10). Final products of each assembly reaction were subjected to electron microscopic examination, and the results are summarized in Fig. 1A; representative examples are shown in Fig. 1B. Irrespective of the extent of an engineered deletion, all sgB4 variants were packaged by CP subunits with efficiency comparable to that of wt sgB4. These observations suggested that CP ORF sequences are dispensable and do not contain any signals that control sgB4 packaging.
In vivo transient expression system and the experimental strategy.
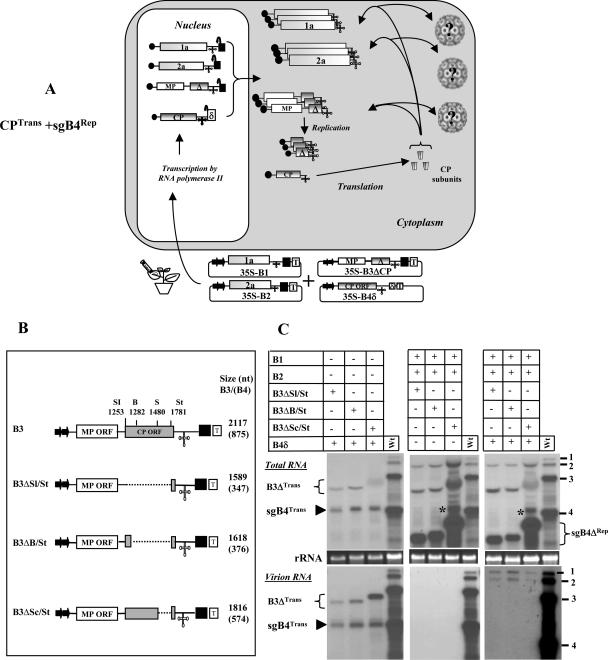
The in vitro data shown above must be validated in vivo. Although sgB4 may package autonomously in vitro (7), its packaging in vivo requires progenitor gB3 (10). Furthermore, since generation of sgB4 is contingent on replication of gB3, it is imperative to analyze whether packaging of sgB4 is coupled to replication. To address this question, we used an Agrobacterium-mediated transient expression system (agroinfiltration) developed in our laboratory for BMV (3). Agroinfiltration allows synchronized codelivery and simultaneous expression of multiple plasmids in a single cell (3, 15, 21). This trait is particularly useful for dissecting replication from packaging. Consequently, in this study each set of inocula was designed to consist of multiple T-DNA-based constructs such that, when agroinfiltrated to N. benthamiana leaves, sgB4 and wt CP subunits are coexpressed in desired combinations involving either transient (i.e., replication-independent mode) or replication-dependent mode expression (i.e., via gB3 replication). To facilitate easy understanding, the following terminology is used throughout. The terminology specifying sgB4Trans and CPRep represents that sgB4 was expressed transiently, whereas CP was translated from an sgB4 that had been generated following replication of its progenitor, gB3. Likewise, sgB4Rep and CPTrans represent synthesis of sgB4 from replicated gB3 and CP from transiently expressed mRNA, respectively. In order to systematically trace the form in which sgB4 and/or CP subunits are required for in vivo packaging, agrotransformants of a desired set of T-DNAs were mixed pair-wise to express sgB4 and CP in the following four combinations: (i) CPTrans and sgB4Trans; (ii) CPTrans and sgB4Rep; (iii) CPRep and sgB4Trans; and (iv) CPRep and sgB4Rep. In assays requiring induction of viral replication to express sgB4Rep and CPRep, the respective inocula were supplemented with T-DNA constructs of gB1 and gB2.
sgB4 packaging mediated by the coexpression of CPTrans and sgB4Trans.
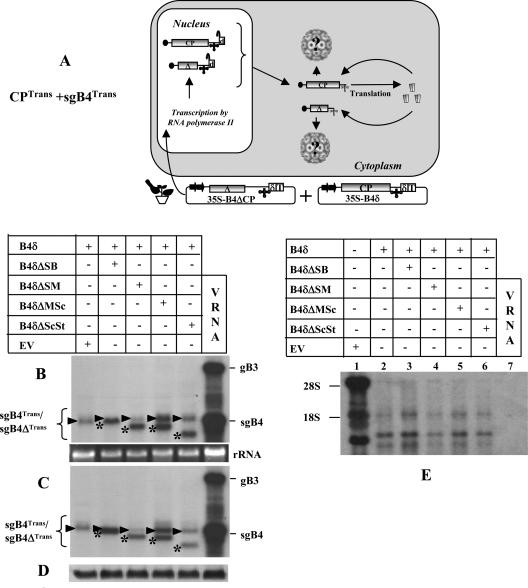
The characteristic features of T-DNA-based constructs of all three wt BMV RNAs amenable for transient expression in N. benthamiana leaves have been reported previously (3) and are schematically shown in Fig. 2A. In our previous study (3), efficient transient expression of CP subunits was achieved by infiltrating 35S-B4.1 agrotransformants. In this construct, the 5′ end of the CP ORF sequence was fused to a translation enhancer leader sequence of Tobacco etch potyvirus, and the 3′ end terminated with a poly(A) tail due to the absence of a self-cleaving ribozyme. We also observed (3) that transiently expressed CP mRNAs having 22 nonviral bases beyond an authentic 3′ end (due to the presence of a self-cleaving ribozyme) failed to translate in vivo due to the intrinsic requirement of a correct 3′ terminus for efficient translation (14). Thus, in this study we engineered a self-cleaving ribozyme from hepatitis delta virus (25) downstream of sgB4 sequence such that the transiently expressed mRNA will terminate precisely with 3′ CCAOH (B4δ; Fig. 2A). N. benthamiana leaves infiltrated with 35S-B4δ resulted in accumulation of translatable mRNA with a size indistinguishable from that of wt sgB4 (see below). Consequently, some agroinfiltration experiments performed in this study used 35S-B4δ as a source vector for CP synthesis.
By infiltrating N. benthamiana leaves with agrotransformants designed to transiently express wt sgB4 (providing CP subunits) and its deletion variants (sgB4Δ) (i.e., CPTrans and sgB4Trans/sgB4ΔTrans; Fig. 3A), we aim to address the specificity and the competitiveness of CPTrans in packaging sgB4Trans/sgB4ΔTrans. Plants infiltrated only with 35S-B4δ served as controls. Results of Northern blot analysis of total and virion RNA preparations (Fig. 3B and C) and Western blot analysis of transiently expressed CP (Fig. 3D) in each infiltrated sample are shown. High-level expression of mRNAs corresponding to sgB4 and each of its deletion variants was clearly detectable, and the relative mobility patterns distinguished wt sgB4 (indicated by an arrowhead in Fig. 3B) from respective deletion variants (indicated by an asterisk in Fig. 3B). In each case, almost all transiently expressed wt sgB4 and its deletion variant RNAs were packaged into virions (Fig. 3B and C). When a duplicate blot containing virion RNA recovered from leaves infiltrated with B4δ and each of its deletion variant was hybridized with a 5′-end-labeled total cellular RNA probe (3), at least three prominent cellular RNAs were clearly detected for each sample (Fig. 3E, lanes 2 to 6). By contrast, virion RNA preparations recovered from control plants infiltrated with wt BMV did not contain any detectable cellular RNA (Fig. 3E, lane 7).
FIG. 3.
(A) Graphic representation of an agroinfiltration scheme designed to analyze sgB4 packaging involving coexpression of sgB4Trans and CPTrans. The composition of the inoculum mixture consisting of two agrotransformants (35S-B4δ and 35S-B4ΔCP) used to infiltrate leaves of N. benthamiana is shown. The double arrowheads represent a cauliflower mosaic virus double 35S promoter. δ, hepatitis delta ribozyme; T, 35S terminator. The filled circle at the 5′ end and the cloverleaf-like structure at the 3′ end, respectively, represent the cap and 3′ TLS. RNA species chosen to verify packaging by CPTrans into virions is indicated by a question mark. (B) Transient expression and packaging in plants of sgB4 (sgB4Trans) and its deletion variants (sgB4ΔTrans). N. benthamiana leaves were infiltrated with indicated Agrobacterium cultures, and the total (B) and virion RNAs (C) recovered 4 days postinfiltration were subjected to Northern blot hybridization. Leaves infiltrated with empty vector (EV) served as a negative control. Approximately 5 μg of total nucleic acid preparation from agroinfiltrated leaves or 0.5 μg of virion RNA was denatured with formamide-formaldehyde and subjected to 1.2% agarose electrophoresis prior to vacuum blotting to a nylon membrane. The blot was hybridized with 32P-labeled riboprobes complementary to a homologous 3′ TLS present on all four BMV RNAs. Conditions of hybridization are as described previously (3). The positions of wt gB3 and sgB4 are shown to the right, and migrating positions of sgB4Trans (arrowhead) and sgB4ΔTrans (asterisk) are indicated by a bracket on the left. wt BMV virion RNA (vRNA) was used as a size marker. (D) Western blot analysis of BMV CP. Total protein extracts from leaves agroinfiltrated independently with either empty vector (EV) or the indicated mixture of agrotransformants were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transferring to polyvinylidene difluoride membrane and probing with antibodies prepared against purified BMV (24). (E) Packaging of cellular RNA by CPTrans. Virion RNAs recovered 4 days postinfiltration from N. benthamiana leaves with the indicated set of Agrobacterium transformant cultures (lanes 2 to 7) were subjected to Northern blot hybridization as described previously (3). Total RNA recovered from leaves infiltrated with empty vector (EV) was used as a marker (lane 1). In each lane, 0.5 μg virion RNA was subjected to Northern blot analysis. The blot was hybridized with a 5′-end-labeled cellular RNA probe (3). The positions of 28S and 18S cellular RNAs are indicated to the left.
Complementation with viral replication inhibited packaging of cellular RNA and sgB4Trans.
Mature virions purified from plants infected with wt BMV have been shown to exclusively contain viral progeny RNA (3). Thus, by complementing viral replication as schematically illustrated in Fig. 4A, we sought to address whether induction of replication would alter packaging specificity. To find an answer to this question, inocula containing agrotransformants of B4δ and each sgB4 deletion variant were complemented with agrotransformants of gB1 and gB2 and coinfiltrated to N. benthamiana leaves (Fig. 4A). Results of Northern blot hybridization of total and virion RNA, packaging competence of cellular RNA, and accumulation levels of CP subunits are shown in Fig. 4B to E. It was observed that compared to absence of viral replication (Fig. 3B to D), its induction significantly altered the packaging competence of CPTrans, since (i) CPTrans packaged replicated progeny of gB1 and gB2 but not sgB4Trans or its deletion variants (Fig. 4B and C), and (ii) RNA packaging of sgB4Trans or its deletion variants was completely inhibited and remained beyond detection, even in the total RNA preparations hybridized with either a commonly shared 3′ TLS probe (Fig. 4B) or by a riboprobe specific for sgB4 (data not shown). Note that the band marked with an asterisk in Fig. 4B was not sgB4, since a duplicate blot hybridized with a riboprobe specific for sgB4 failed to detect this band (data not shown); the origin of this band is unknown. (iii) Finally, packaging of cellular RNAs was also completely inhibited (Fig. 4D).
FIG. 4.
(A) Graphic representation of packaging events examined when leaves coexpressing sgB4Trans or sgB4ΔTrans and CPTrans were supplemented with gB1 and gB2 to induce viral replication. In agrotransformants B1 and B2, a filled square at the 3′ end represents a Satellite tobacco ring spot virus ribozyme cassette. Other features are the same as those described in the legend to Fig. 3. RNA species chosen to verify packaging by CPTrans into virions is indicated by a question mark. The composition of the inoculum mixture consisting of four agrotransformants used to infiltrate leaves of N. benthamiana is shown. (B) Effect of viral replication on packaging of sgB4 (sgB4Trans) and its deletion variants (sgB4ΔTrans) by CPTrans. N. benthamiana leaves were infiltrated with the indicated set of Agrobacterium transformant cultures. Analysis of total (B) and virion (C) RNAs by Northern blot hybridization as described was as described in the legend to Fig. 3. Shown is an analysis of cellular RNA in purified virions (D) and BMV CP (E) in agroinfiltrated leaves by Northern and Western blot hybridization, respectively, as described in the legend to Fig. 3. (B and C) The position of sgB4Trans is indicated by an arrowhead. (B) The asterisk indicates an RNA species of unknown origin migrating slower than sgB4Trans (see the text for details). (D) The position of 28S and 18S cellular RNAs is indicated to the left.
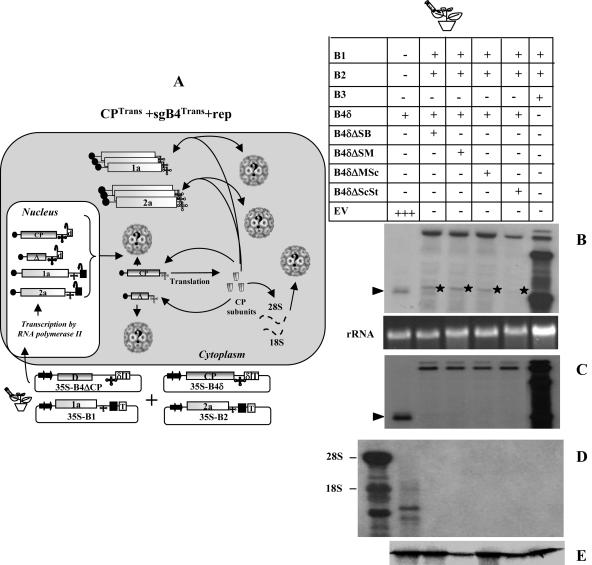
sgB4 packaging mediated by the coexpression of CPTrans and sgB4Rep.
One possible explanation for deficiency of sgB4Trans packaging by CPTrans in the presence of replication (Fig. 4B) is that only replication-derived sgB4 is competent for packaging by CPTrans. To verify this possibility, the experimental strategy shown in Fig. 5A was employed. In this strategy, similar to the previous one (Fig. 4A), CP subunits were supplied in trans but production of sgB4 was linked to replication. To execute this strategy, three deletions were engineered into the CP ORF of agroinfiltratable gB3 transformants. Variants B3ΔSl/St and B3ΔB/St were, respectively, characterized by a deletion of 528 nt and 499 nt located between SalI (at position 1253) and BssHII (located at 1282) and commonly shared StuI (at position 1781), whereas variant B3ΔSc/St was constructed by subcloning the CP ORF region of B4ΔSc/St shown in Fig. 1A. The rationale for engineering these deletions is to generate variants of sgB4 with distinguishable electrophoretic profiles with respect to wt sgB4. It is anticipated that when transcripts of B3ΔSl/St, B3ΔB/St, and B3ΔSc/St enter the replication cycle, each of these variants generates sgΔB4 of 347 nt, 376 nt, and 574 nt (Fig. 5B), respectively, and are readily distinguishable from the wt sgB4 of 887 nt.
FIG. 5.
(A) Graphic representation of packaging events examined in leaves expressing sgB4Rep or sgB4ΔRep and CPTrans. Features of T-DNA constructs are the same as those described in the legend to Fig. 3. RNA species chosen to verify packaging by CPTrans into virions is indicated by a question mark. The composition of the inoculum mixture consisting of four agrotransformants used to infiltrate leaves of N. benthamiana is shown. (B) Characteristics of agrotransformants of wt B3 and its deletion variants. The selected restrictions sites (the same as those specified in the legend to Fig. 1A) used to engineer deletions in the CP ORF are shown. The dotted line in each variant clone represents the extent of an engineered deletion. The lengths (in nucleotides) of wt B3 and its variants are shown. The number shown in brackets represents the predicted nucleotide length of sgB4 synthesized in vivo following replication of respective gB3 variant clones. Other features are the same as those described in the legend to Fig. 3. (C) Packaging competence in vivo of sgB4Rep or sgB4ΔRep by CPTrans. N. benthamiana leaves were infiltrated with an indicated set of Agrobacterium transformant cultures. A column containing the wt represents plants infiltrated with an inoculum containing of all three wt BMV transformants. Total and virion RNAs were subjected to Northern blot hybridization as described in the legend to Fig. 3. The top left image shows a Northern blot hybridization of total RNA preparations showing the accumulation of transiently expressed mRNAs of sgB4 and B3 deletion variants. The bottom left image shows a Northern blot showing the packaging competence of sgB4Trans and B3Trans deletion variants by CPTrans. The middle left panel shows a Northern blot hybridization of total (top) and virion RNA (bottom) preparations showing replicative competence of B3 variants. Note that due to the absence of functional CP, no virion RNA was detected in the bottom panel. The asterisk identifies RNA of unknown origin similar to that in the legend to Fig. 4. The top right panel shows a Northern blot hybridization of total RNA preparations showing replicative competence of B3 variants. The asterisk identifies RNA of unknown origin similar to that described in the legend to Fig. 4. The bottom right panel shows a Northern blot hybridization showing packaging competence of progeny of genomic RNAs and sgB4ΔRep by CPTrans. The conditions of hybridization are similar to those described in the legend to Fig. 3. The positions of four BMV RNAs are indicated to the right. The positions of B3ΔTrans/sgB4Trans and B3ΔRep/sgB4ΔRep are shown to the left of each panel.
Two sets of agroinfiltration assays were performed. The first set (Fig. 5C, left panel) of agroinfiltration assays was designed to verify whether CPTrans is competent to package B3ΔSl/StTrans, B3ΔB/StTrans, or B3ΔSc/StTrans, whereas the second set (Fig. 5C, right panel) was designed to verify the competence of CPTrans to package each sgB4ΔRep generated following replication of respective progenitor gB3. Plants infiltrated with inocula containing B1 and B2 and each B3 variant were used to assess the replicative competence of respective B3 variants in the absence of CP (Fig. 5C, middle panel). Plants with all three wt BMV agrotransformants served as positive controls. Results of Northern hybridization of total and virion RNA are summarized in Fig. 5C.
As was the case with CPTrans and sgB4Trans (Fig. 3), CPTrans was competent to package B3ΔSl/StTrans, B3ΔB/StTrans, and B3ΔSc/StTrans (Fig. 5C, bottom left panel). As expected, when complemented with viral replication (by coinfiltrating gB1 and gB2), B3ΔSl/St, B3ΔB/St, and B3ΔSc/St entered replication cycle, which resulted in the synthesis of the respective sgΔB4 (i.e., sgB4ΔSl/StRep of 347 nt, sgB4ΔB/StRep of 376 nt, and sgB4ΔSc/StRep of 574 nt) with the predicted electrophoretic mobility pattern (Fig. 5C, top right panel). However, the most interesting observations emerged when packaging competence between CPTrans and the replication progeny was analyzed. Surprisingly, only the progeny of gB1 and gB2 was packaged into virions by CPTrans. Neither truncated gB3 progeny nor respective variants of sgB4 (i.e., sgB4ΔSl/StRep, sgB4ΔB/StRep, and sgΔSc/StRep) were packaged (Fig. 5, bottom right panel). Consistent with the data shown in Fig. 4B, sgB4Trans was detected neither in total nor in virion RNA preparations (Fig. 5C, right panel). Despite retaining the required bipartite packaging signal (i.e., MP ORF and 3′ TLS) (10), the reason for the inability of B3ΔSl/St, B3ΔB/St, and B3ΔSc/St progeny to package into virions can be attributed to the lack of sufficient distance between the packaging element (PE) and nucleating element (NE) to maintain optimal structural requirements (10) which appear to be affected by the engineered deletion. Since packaging of sgB4 is linked to prior packaging of gB3 (9), the defective packaging of B3ΔSl/St, B3ΔB/St, and B3ΔSc/St in turn affected packaging of the respective sgB4.
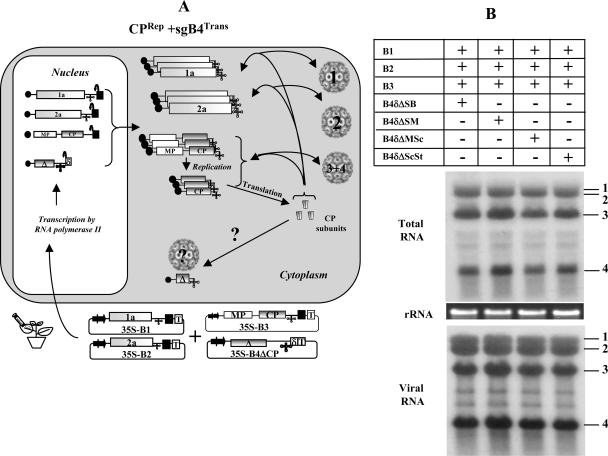
sgB4 packaging mediated by the coexpression of CPRep and sgB4Trans.
In all of the above-described experiments, packaging of either sgB4Trans (Fig. 3 and 4) or sgB4Rep (Fig. 5) was mediated by CPTrans. However, during BMV infection, CP is always translated from a replication-derived sgB4, and therefore the packaging competence of CPRep could be different from that of CPTrans. An experimental strategy, schematically shown in Fig. 6A, was designed to test the competence of CPRep in packaging variants of sgB4 expressed transiently (i.e., sgB4Trans). Consequently N. benthamiana leaves were coinfiltrated with a mixture of agrotransformants containing each sgB4 variant sequence as well as a mixture of all three wt BMV agrotransformants (Fig. 6A). Results of Northern blot analysis of total and virion RNA are shown in Fig. 6B. Although infiltration with all three wt BMV agrotransformants resulted in efficient replication and packaging of four wt BMV RNAs (Fig. 6B), none of the variants of sgB4Trans was found to be packaged (Fig. 6B). Furthermore, as observed above (Fig. 4B), transiently expressed, nonreplicating sgB4Trans variant sequences were not detected even in total RNA preparations when the blots were hybridized with a riboprobe complementary to either 3′ TLS (Fig. 6B) or specific for B4 (data not shown). In these two scenarios where sgB4Trans was not packaged, we speculate that a synergistic interaction between functional replicase and CP selectively filters nonreplicating (as well as cellular) RNAs to maximize packaging of functional viral progeny (see Discussion). Consequently, nonreplicating RNAs such as sgB4Trans are subjected to degradation by host nucleases or silencing.
FIG. 6.
(A) Graphic representation of packaging events examined in leaves expressing sgB4ΔTrans and CPRep. Features of T-DNA constructs are the same as those described in the legend to Fig. 3. The composition of the inoculum mixture consisting of four agrotransformants used to infiltrate leaves of N. benthamiana is shown. RNA species chosen to verify packaging by CPRep into virions are indicated by a question mark. (B) N. benthamiana leaves were infiltrated with the indicated set of Agrobacterium transformant cultures. Total and virion RNAs were subjected to Northern blot hybridization as described in the legend to Fig. 3. The positions of four wt BMV RNAs are indicated to the right.
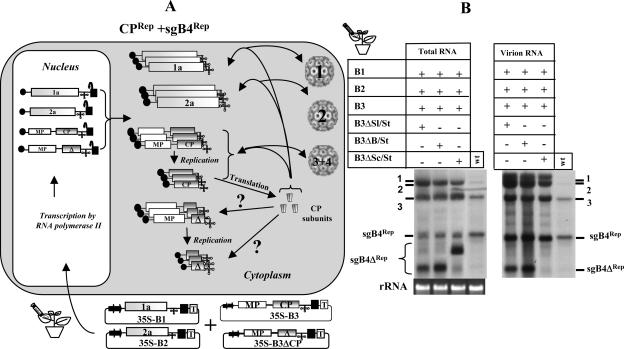
sgB4 packaging mediated by the coexpression of CPRep and sgB4Rep.
Packaging assays from N. benthamiana leaves coexpressing CPTrans and sgB4Rep (Fig. 5) or sgB4Trans and CPRep (Fig. 6) revealed that neither combination resulted in efficient packaging of wt sgB4 or its deletion variants. In wild-type BMV infection, generation of sgRNA and CP is replication contingent (i.e., sgB4Rep and CPRep). Therefore, an experimental strategy reminiscent of wild-type infection, as schematically shown in Fig. 7A, was designed to test whether inherent packaging of sgB4 is not only replication contingent but also requires CP subunits translated from replication-derived mRNAs, i.e., sgB4Rep and CPRep. To verify this, a mixture of inocula containing agrotransformants of all three wt BMV RNAs and either B3ΔSl/St, B3ΔB/St, or B3ΔSc/St (Fig. 5B) was coinfiltrated to N. benthamiana leaves. Unlike in previous assays, in this assay production of CP and synthesis of variants of sgB4 are linked to replication. Results of Northern blot analysis of total and virion RNAs recovered from N. benthamiana leaves infiltrated with each set of inocula are shown in Fig. 7B. Northern blots containing total RNA preparations revealed that, in each case, progeny RNA corresponding to wt B3 and its sgB4 accumulated to detectable levels (Fig. 7B, left panel). In contrast, replicated progeny corresponding to each gB3 variant (i.e., B3ΔSl/St, B3ΔB/St, or B3ΔSc/St) was not detected, whereas the synthesis of respective sgB4 progeny (i.e., sgB4347, agB4376, and sgB4574) with the expected electrophoretic mobility pattern was evident (Fig. 7B, left panel). Although our efforts to detect either plus- or minus-strand progeny of each B3 variant using strand-specific probes were unsuccessful (data not shown), efficient accumulation of respective sgB4 confirmed that each gB3 variant entered the replication cycle. Therefore, we conclude that each B3 variant is not defective in replication as shown in Fig. 5C (middle panel), but in this assay their accumulation levels are severely debilitated due to their inability to complete wt B3 replication.
FIG. 7.
(A) Graphic representation of packaging events examined in leaves coexpressing sgB4Rep or sgB4ΔRep and CPRep. Features of T-DNA constructs are the same as those described in the legend to Fig. 3. The composition of the inoculum mixture consisting of four agrotransformants used to infiltrate leaves of N. benthamiana is shown. RNA species chosen to verify packaging by CPRep into virions are indicated by a question mark. (B) Leaves of N. benthamiana were infiltrated with the indicated set of Agrobacterium transformant cultures. Total and virion RNAs were subjected to Northern blot hybridization as described in the legend to Fig. 3. The positions of four wt BMV RNAs and each of the three sgB4ΔRep are indicated. Virion RNA of wt BMV (wt) was used as marker.
Virion RNA blots revealed interesting packaging profiles (Fig. 7B, right panel). In addition to the expected four wt BMV RNAs, purified virions packaged only sgB4347 and sgB4376 but not sgB4574 (Fig. 7B, right panel). The reasons for the packaging deficiencies exhibited by the progeny sgB4574 are considered below.
DISCUSSION
In this study, in vitro assembly assays performed between CP subunits and transcripts of sgB4 variants harboring defined deletions in the CP ORF showed that packaging of sgB4 is independent of sequences encompassing the CP ORF (Fig. 1). Subsequent in vivo analysis of sgB4 packaging using an agroinfiltration approach designed to express pair-wise combination of agrotransformants involving either a transient (sgB4Trans and CPTrans) or replication-dependent mode (sgB4Rep and CPRep) revealed that (i) a combination involving sgB4Trans and CPTrans resulted in nonspecific packaging, since virions contained not only sgB4Trans but also cellular RNA (Fig. 3); (ii) induction of viral replication enhanced packaging specificity, since packaging of cellular as well as nonreplicating transiently expressed RNAs was completely blocked (Fig. 4 and 5); and finally, (iii) efficient packaging of sgB4 is functionally coupled to replication, since only the pair coexpressing sgB4Rep and CPRep (Fig. 7) resulted in a packaging profile reminiscent of wt infection. These results represent the first demonstration of replication-coupled packaging in a plant RNA virus.
Mechanism of sgRNA packaging in BMV.
Based on results of this study (Fig. 3 to 7), together with previously published data (7, 9, 10), we propose the following likely mechanisms that regulate copackaging of gB3 and sgB4 into a single virion.
(i) RNA-RNA interactions.
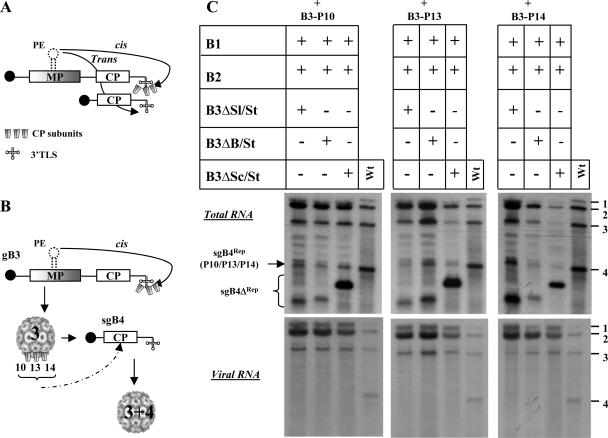
The entire sequence of sgB4 is contained within the 3′ half of gB3. Therefore, a mutually beneficial signal-promoting copackaging is predicted to exist in the commonly shared CP ORF region. Contrary to this assumption, mutational analysis of gB3 sequences revealed that efficient packaging of gB3 requires coordination of bipartite signal consisting of a 187-nt-long cis-acting, position-dependent packaging element (PE) located in the MP ORF and the highly conserved 3′ TLS functioning as a nucleating element (NE), and deletion of CP ORF sequences has no detectable effect on packaging (10). In light of these observations, we propose a packaging mechanism involving RNA-RNA interactions mediated by the PE of gB3 (i.e., MP ORF) and the 3′ TLS of sgB4 which is essential for sgB4 packaging (7). In this mechanism, as schematically shown in Fig. 8A, following initial binding of CP subunits to the 3′ TLS, a cis interaction with PE forms a ribonucleoprotein (RNP) complex consisting of CP and gB3. An interaction between this RNP complex and the 3′ TLS of sgB4 is promoted by PE acting in trans (Fig. 8B). In this scenario, like gB3, packaging of sgB4 also requires a bipartite signal, i.e., trans-acting PE of gB3 and TLS of sgB4. Furthermore, this intrinsic trans-interaction of PE ensures that sgB4 is selectively copackaged with gB3. This proposed mechanism also explains why inhibition of gB3 packaging also inhibits sgB4 packaging (10).
FIG. 8.
(A) Schematic model showing copackaging of gB3 and sgB4 mediated by RNA-RNA interactions. Following nucleation of CP subunits by the 3′ TLS of gB3, its packaging is mediated through a stem-loop structure functioning as a cis-acting packaging element (10). RNA-RNA interaction is mediated by the PE of gB3 functioning in trans, leading to copackaging. (B) Schematic model showing gB3 and sgB4 copackaging mediated by CP-RNA interactions. After initial packaging of gB3 as described for panel A, the N-arginine-rich region of BMV CP is transiently exposed on the surface of the assembled virion, allowing the interaction of sgB4 with a set of amino acid determinants (P10, P13, and P14) specific for this RNA species. (C) Demonstration in vivo of required specific interaction between sgB4 and N-terminal amino acid determinants of BMV CP promoting copackaging. Leaves of N. benthamiana were infiltrated with the indicated set of Agrobacterium transformant cultures. Total and virion RNAs were subjected to Northern blot hybridization as described in the legend to Fig. 3. The positions of four wt BMV RNAs used as size markers are shown to the right of each panel. The positions of sgB4Rep harboring either P10, P13, or P14 and each of the three sgB4ΔRep are shown to the left.
Previously, we demonstrated that BMV CP harboring mutations in the N-terminal arginine-rich region did not affect gB3 packaging but specifically inhibited that of sgB4. By extrapolating these observations to the model discussed above, we want to substantiate the interaction between CP of the RNP complex and the sgB4. Therefore, the experiment shown in Fig. 7 was repeated, except that wt B3 was replaced with variants of B3 having specific mutations in the N-terminal arginine-rich region. In each of these variant clones, an arginine residue located at position 10, 13, or 14 was replaced with a proline residue (9). As observed previously (9), we anticipate that the mutant CP will package gB3 but not sgB4. Northern blot data shown in Fig. 8C confirm this prediction and support the hypothesis that a specific region of CP of the RNA complex interacts with the 3′ TLS region of sgB4, leading to copackaging. If this is true, then why was sgB4574 not packaged by either wt or mutant CP (Fig. 7B and 8C)? Two factors might be responsible for the lack of sgB4574 packaging. First, the extent of the engineered deletion could have induced gross structural changes to the sequence and inhibited interaction of CP with the 3′ TLS of sgB4574. Second, the size limit imposed by the BMV virion could also have influenced copackaging of sgB4574 (see below).
(ii) Packaging controlled by physical constraints of virions.
In addition to specific sequences that promote packaging, another important parameter is the dimension of the capsid that imposes an upper limit on the size or amount of viral nucleic acid that can be accommodated (4, 26). Consequently, nucleic acids larger than a wt genome cannot be packaged, even when they contain appropriate packaging signals, as demonstrated for monocomponent Turnip crinkle virus (26). In BMV, the physical size of the virions has been shown to be controlled by RNA size (11, 19). As shown in this study, packaging of sgB4 is functionally coupled to replication (Fig. 7). Furthermore, since packaging of sgB4 is dependent on prior packaging of gB3 (10) and supplementation of viral replication blocks packaging of transiently expressed sgB4 (Fig. 4), we discount the possibility of autonomous packaging of sgB4 in vivo. Failure to detect B3ΔSl/St or B3ΔB/St progeny by Northern blot analysis (Fig. 7B) suggested that sgB4347 and sgB4376 must have been copackaged only with wt gB3.
Replication-coupled packaging in positive-strand RNA viruses.
Genome packaging in RNA viruses is considered a highly specific process, since mature virions predominantly contain viral RNAs (28, 33). To achieve this process, viral nucleic acid must be distinguished from other cellular RNA molecules present in the compartment where assembly takes place (31). Packaging of virus-specific RNAs into stable virions was shown to be mediated through specific recognition of sequences or structures unique to viral nucleic acids, often termed origins of assembly or packaging signals. These signals strongly enhance RNA packaging, and they were identified in several icosaheral RNA viruses infecting animals (40), insects (42), and plants (10, 12, 26). However, the presence of a packaging signal alone does not guarantee packaging of RNA into virions, since several factors of the packaging signal, such as secondary structure, position with respect to genomic context, and location (cis or trans), also affect packaging efficiency (10, 12). This study revealed active replication as being an important and intrinsic criterion for selective and enhanced packaging of BMV sgRNA. Similar direct coupling between replication and packaging has also been observed in poliovirus (23), flock house virus (37), Kunjin virus (18), and, more recently, Venezuelan equine encephalitis virus (39). Although the general scenario of direct coupling between replication and packaging is commonly shared among these viruses, certain differences exist. For example, in BMV (this study) (Fig. 3) and flock house virus (37), supply of CPTrans resulted in nonspecific packaging of transiently expressed nonreplicating viral RNAs as well as cellular RNAs, whereas in Kunjin virus structural proteins translated from replication-deficient RNAs are incompetent in packaging of any kind of RNAs (18). Interestingly, in BMV the replication-coupled packaging appears to be distinct between the genomic and subgenomic RNAs, since unlike sgB4, whose packaging is replication contingent (Fig. 8), replication-defective TLS-less gB1 and gB2 are efficiently packaged into virions by CPTrans and CPRep (3). In contrast, gB3 lacking the 3′ TLS was not packaged by CPTrans (P. Annamalai and A. L. N. Rao, unpublished data). sgB4 synthesis occurs only when gB3 enters the replication cycle, which explains the differential requirement of replication-coupled packaging among gB1, gB2, and gB3.
Do viral replicase and CP interact synergistically to regulate packaging in BMV?
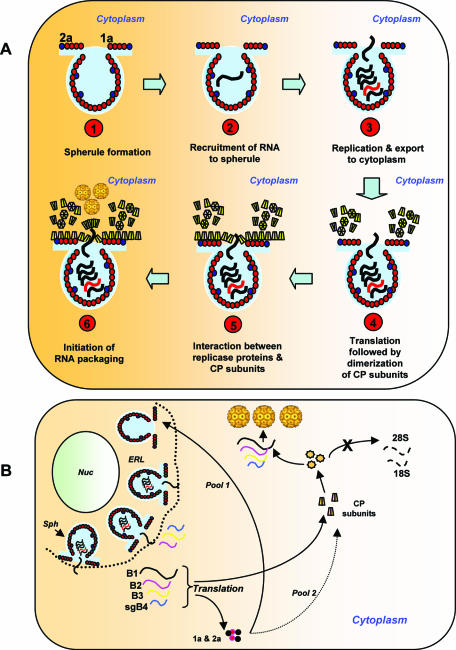
In addition to participating in the synthesis of genomic negative and positive RNAs, BMV replicase has been implicated in other roles, such as cell-to-cell spread (36). Data shown in Fig. 3 to 5 further validate our previous conclusion (3) that induction of viral replication resulting in the formation of active replicase functions as a specificity filter by enhancing packaging of viral RNA but not ubiquitous cellular RNA or nonreplicating viral RNA (Fig. 4 and 6). This suggests that a synergistic interaction between viral replicase proteins and CP may exist to maximize packaging of viral RNAs. We offer two possible mechanisms in support of this hypothesis (Fig. 9). The cytopathological studies of bromoviruses revealed the induction of vesicles (13) or spherules (34) in the perinuclear spaces of infected cells. These specialized structures are induced by replicase protein 1a and have been shown to be the actual sites of RNA synthesis (34). Replicase protein 2a interacts with 1a and recruits RNA to spherules to initiate viral replication (34); the necks connecting spherules serve as channels to export newly synthesized progeny RNA to the cytoplasm for translation. Since CP has been found to copurify with active replicase complex (6), we rationalize that CP must be synthesized near the necks of spherules (Fig. 9A). This scenario provides a transient association between viral replicase proteins and CP, resulting in increased specificity of BMV RNA packaging. In the absence of a functional replicase the specificity of CP is disrupted, resulting in nonspecific packaging of cellular RNAs, as shown in Fig. 3. Alternatively, it is possible that following export of newly synthesized gB1 and gB2 to an unspecified location in cytoplasm, their translation creates a new pool of replicase proteins (Fig. 9B). One portion of this new pool interacts exclusively with CP to enhance specificity, while the other will continue to participate in replication-related events, such as induction of spherules. Thus, it remains to be examined whether synergic interaction between replicase and CP subunits would result in subtle structural alteration of CP subunits to enhance specificity.
FIG. 9.
Schematic diagram illustrating two possible models of BMV RNA packaging mediated by interaction between CP and viral replicase proteins. (A) Viral replicase protein 1a induces spherules (step 1), and replicase protein 2a interacts with 1a and recruits RNA to spherules to initiate viral replication (step 2). The necks connecting spherules serve as channels to export newly synthesized progeny RNA to the cytoplasm for translation (step 3). Since CP has been found to copurify with active replicase complex (6), we hypothesize that CP must be synthesized near the necks of spherules (step 4). This scenario provides a transient association between viral replicase proteins and CP resulting in increased specificity of BMV RNA packaging (steps 4 to 6). (B) As mentioned above, induction of spherules (sph) along the endoplasmic reticulum lumen (ERL) and replication viral RNAs occurs in spherules (34). Transportation of newly synthesized BMV RNAs to cytoplasm results in the synthesis of replicase proteins 1a and 2a and CP at an unspecified location in the cytoplasm. One pool of the replicase proteins continues to induce new spherules along ERL, while the other pool transiently interacts with CP to increase specificity. As a result, packaging of cellular RNA is inhibited, while that of viral progeny RNA is enhanced. Nuc, nucleus.
Acknowledgments
We thank Shou-Wei Ding for the generous gift of pCass4 plasmid and Mellisaane de Wispelaere for helpful discussions.
Research in this laboratory was supported by a grant from the National Institutes of Health (GM 064465-01A2).
REFERENCES
- 1.Annamalai, P., S. Apte, S. Wilkens, and A. L. Rao. 2005. Deletion of highly conserved arginine-rich RNA binding motif in cowpea chlorotic mottle virus capsid protein results in virion structural alterations and RNA packaging constraints. J. Virol. 79:3277-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annamalai, P., and A. L. Rao. 2006. Delivery and expression of functional viral RNA genomes in planta by agroinfiltration, p. 16B.2.1-2.15. In T. Downey (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., Hoboken, N.J. [DOI] [PubMed] [Google Scholar]
- 3.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96-111. [DOI] [PubMed] [Google Scholar]
- 4.Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology 345:532-539. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 6.Bujarski, J. J., S. F. Hardy, W. A. Miller, and T. C. Hall. 1982. Use of dodecyl-β-D-maltoside in the purification and stabilization of RNA polymerase from brome mosaic virus infected barley. Virology 119:465-473. [DOI] [PubMed] [Google Scholar]
- 7.Choi, Y. G., T. W. Dreher, and A. L. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, Y. G., G. L. Grantham, and A. L. Rao. 2000. Molecular studies on bromovirus capsid protein. Virology 270:377-385. [DOI] [PubMed] [Google Scholar]
- 9.Choi, Y. G., and A. L. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging on BMV RNA4 by specific N-terminal arginine residuals. Virology. 275:207-217. [DOI] [PubMed] [Google Scholar]
- 10.Choi, Y. G., and A. L. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 77:9750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, Y. G., and A. L. Rao. 2000. Packaging of tobacco mosaic virus subgenomic RNAs by Brome mosaic virus coat protein exhibits RNA controlled polymorphism. Virology 275:249-257. [DOI] [PubMed] [Google Scholar]
- 12.Damayanti, T. A., S. Tsukaguchi, K. Mise, and T. Okuno. 2003. cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 77:9979-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Francki, R. I. B., R. G. Milne, and T. Hatta. 1985. Atlas of plant viruses: bromoviruses, p. 69-80, vol. II. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 14.Gallie, D. R. 1998. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Gopinath, K., B. Dragnea, and C. Kao. 2005. Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 79:14222-14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 1:91-98. [DOI] [PubMed] [Google Scholar]
- 18.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 75:4633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucas, R. W., S. B. Larson, and A. McPherson. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317:95-108. [DOI] [PubMed] [Google Scholar]
- 21.Marillonnet, S., A. Giritch, M. Gils, R. Kandzia, V. Klimyuk, and Y. Gleba. 2004. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 101:6852-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537-546. [DOI] [PubMed] [Google Scholar]
- 23.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osman, F., G. L. Grantham, and A. L. Rao. 1997. Molecular studies on bromovirus capsid protein. IV. Coat protein exchanges between brome mosaic and cowpea chlorotic mottle viruses exhibit neutral effects in heterologous hosts. Virology 238:452-459. [DOI] [PubMed] [Google Scholar]
- 25.Perrotta, A. T., O. Nikiforova, and M. D. Been. 1999. A conserved bulged adenosine in a peripheral duplex of the antigenomic HDV self-cleaving RNA reduces kinetic trapping of inactive conformations. Nucleic Acids Res. 27:795-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu, F., and T. J. Morris. 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 71:1428-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, A. L. 2001. Bromoviruses, p. 155-158. In O. C. Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Mississauga, Ontario, Canada.
- 28.Rao, A. L. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol., 44:61-87. [DOI] [PubMed] [Google Scholar]
- 29.Rao, A. L., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 86:5335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao, A. L., and G. L. Grantham. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 211:42-52. [DOI] [PubMed] [Google Scholar]
- 31.Roger, H. 2002. Matthews' plant virology. Academic Press, San Diego, Calif.
- 32.Sambrook, J., and D. L. Russel. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schneemann, A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 60:51-67. [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 35.Sivakumaran, K., Y. Bao, M. J. Roossinck, and C. C. Kao. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 74:10323-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traynor, P., B. M. Young, and P. Ahlquist. 1991. Deletion analysis of brome mosaic virus 2a protein: effects on RNA replication and systemic spread. J. Virol. 65:2807-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venter, P. A., N. K. Krishna, and A. Schneemann. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 79:6239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verwoerd, T. C., B. M. Dekker, and A. Hoekema. 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17:2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkova, E., R. Gorchakov, and I. Frolov. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, B., U. Geigenmuller-Gnirke, and S. Schlesinger. 1994. Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, X., J. M. Fox, N. H. Olson, T. S. Baker, and M. J. Young. 1995. In vitro assembly of cowpea chlorotic mottle virus from coat protein expressed in Escherichia coli and in vitro-transcribed viral cDNA. Virology 207:486-494. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, W., R. Dasgupta, and R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is a bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]