Abstract
When the endogenous polypurine tract (PPT) of the Rous sarcoma virus (RSV)-derived vector RSVP(A)Z was replaced with alternate retroviral PPTs, the fraction of unintegrated viral DNA with the normal consensus ends significantly decreased and the retention of part of the PPT significantly increased. If the terminus of the U3 long terminal repeat (LTR) is aberrant, RSV integrase can correctly process and integrate the normal U5 LTR into the host genome. However, the canonical CA is not involved in joining the aberrant U3 LTR to the host DNA, generating either large duplications or deletions of the host sequences instead of the normal 5- or 6-bp duplication.
The RNA genome of retroviruses is copied into double-stranded DNA by reverse transcriptase (RT). First (minus)-strand DNA synthesis is initiated from a host tRNA primer. Removal of this tRNA primer by RNase H defines the right (U5) end of the linear viral DNA. Second (plus)-strand synthesis is initiated from a polypurine tract (PPT) primer generated by specific RNase H cleavages of the RNA genome adjacent to U3. Subsequent removal of this PPT primer by RNase H defines the left (U3) end of the linear viral DNA (12).
The sequence of the PPT is important for the proper generation and removal of the PPT primer by RNase H (2, 4, 5, 8). Mutations in either the 5′ or the 3′ end of the human immunodeficiency virus type 1 (HIV-1) PPT affect RNase H cleavage, reducing viral titer (3, 5, 8). Mutating the second and fifth guanine residues (AAAAGAAAAGGGGGG) of the G tract at the 3′ end of the HIV-1 PPT strongly affected cleavage specificity in vivo (5). Altering the murine leukemia virus (MLV) PPT also affected PPT cleavage (9, 10, 11).
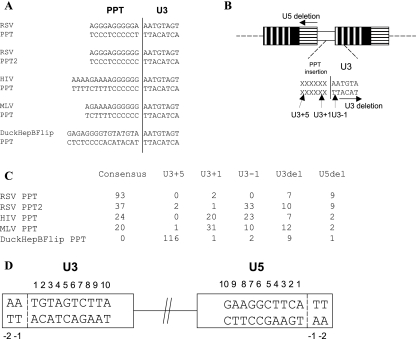
We previously reported that alternate PPTs affected the specific cleavages that generate and remove the PPT in a Rous sarcoma virus (RSV)-derived vector (1) (Fig. 1C). Although RSV RNase H was able to cleave the HIV-1 and MLV PPTs correctly part of the time, the specificity of the cleavage was greatly reduced; RSV RNase H consistently miscleaved the HIV-1 and MLV PPTs, causing the insertion of the first G (U3+1) of the PPT or the deletion of the first residue (A) of U3 (U3−1) at the two-long-terminal-repeat (2-LTR) circle junction. When the 3′ A of the RSV PPT was mutated to G (RSV PPT2), RSV RNase H miscleaved RSV PPT2, leading to the deletion of the first A residue in U3 (U3−1). In the case of the DuckHepBFlip PPT (duck hepatitis B virus PPT in the reverse orientation), the RSV RNase H preferentially cleaved in U3 (U3+5) to cause the insertion of ATGTA; this ATGTA sequence is an exact duplication of the 5′ end of the RSV U3. Replacing the endogenous RSV PPT with the alternate PPTs reduced the relative titer (the titer corrected for the amount of p27). HIV PPT, MLV PPT, DuckHepBFlip PPT, and RSV PPT2 had relative titers of 26%, 32%, 38%, and 73% of the wild-type titer, respectively. The fact that miscleavage by RSV RT generates linear DNAs with aberrant ends raises the question of how the aberrant DNAs are integrated by RSV integrase (IN).
FIG. 1.
Schematic drawing showing the structures of the RSVP vectors. (A) Sequences of the alternate PPTs. (B) Schematic representation of the circle junctions. U3 is indicated by white boxes with vertical black bars; R is indicated by black boxes; U5 is shown by white boxes with horizontal black bars; PPT insertions are indicated by a white box. The terminal sequence of U3 is shown; the PPT sequence is represented by Xs. The sites of the RNase H cleavages are indicated by black arrows. (C) 2-LTR circle junctions isolated from infected cells. (D) Schematic numbering of the nucleotide positions at the ends of the proviruses. The last nucleotide on each end of a normal provirus is number 1.
Recovery of full-length integrated viral DNA.
Full-length integrated viral DNAs were recovered as described previously (6, 7). Recovered plasmids were sequenced, and the chicken genomic sequences were analyzed by BLAT searches (http://genome.ucsc.edu/cgi-bin/hgBlat). Most of the proviruses derived from infections with three of the mutant viruses (HIV PPT, MLV PPT, and RSV PPT2) were integrated normally (Table 1). In two cases, the U3 end was not appropriately processed, but the integration event gave rise to a “normal” provirus. One provirus containing the MLV PPT integrated using an internal CA sequence in U3. In one of the RSV PPT2 proviruses, integrase removed one nucleotide from the U3 end of the linear viral DNA, and the provirus had one nucleotide beyond the canonical CA at the U3/host junction (Table 1).
TABLE 1.
Recovery of full-length integrated viral DNA
| Provirus | Position at indicated enda
|
Duplication (bp) | No. of cases | |
|---|---|---|---|---|
| U3 | U5 | |||
| HIV PPT | 1 | 1 | 6 | 24 |
| 1 | 1 | 5 | 4 | |
| Variable | 1 | Variable | 5 | |
| 1 | 13 | Ambiguous | 1 | |
| MLV PPT | 1 | 1 | 6 | 34 |
| 1 | 1 | 5 | 8 | |
| 11 | 1 | 6 | 1 | |
| 1 | Variable | Variable | 2 | |
| RSV PPT2 | 1 | 1 | 6 | 37 |
| 1 | 1 | 5 | 4 | |
| −1 | 1 | 5 | 1 | |
| Variable | 1 | Variable | 4 | |
| 1 | 19 | Ambiguous | 1 | |
| DuckHepBFlip PPT | −6 | 1 | 6 | 6 |
| −6 | 1 | 5 | 2 | |
| Variable | 1 | Variable | 13 | |
| 6 | 1 | 34b | 1 | |
| −112 | 434 | Ambiguous | 1 | |
The last viral nucleotide at each end of the proviruses is indicated by a number, using the numbering system in Fig. 1D.
Size of deletion.
Many of the aberrant integrations we report here, which involve aberrant U3 ends, are reminiscent of the proviruses generated from linear DNAs with aberrant U5 ends (7). Of the proviruses from infections with three mutant viruses (HIV PPT, MLV PPT, and RSV PPT2), eight proviruses had the canonical CA sequence present only at the U5 LTR terminus and the U3 LTR terminus was deleted (Fig. S1 in the supplemental material). None of these eight aberrant proviruses was flanked by a 5- or 6-bp duplication at the target site. We also recovered a provirus with a complicated junction at the U3 terminus (Fig. S1 in the supplemental material, panel AA). The U5-terminal CA sequence was joined appropriately to chicken chromosome 1. However, there was an insertion of the PPT and a flanking sequence (63 bp) at the end of the U3, immediately followed by part of the pol sequence (542 bp), which was then joined to chicken chromosome 1. The provirus was flanked by a 5-bp duplication of the host DNA. The last nucleotide of the pol sequence was homologous to the first nucleotide of the host sequence. This ambiguous nucleotide was adjacent to a viral CA sequence (Table 2, row AA). Given that the viral DNA is flanked by a 5-bp duplication, it is possible that the viral IN used the CA sequence in pol for integration.
TABLE 2.
Microhomologies at the junctions of unusual integrationa
| PPT designation | 5′ end | 3′ end |
|---|---|---|
| A | AATACG-5′ | 5′-TTCA |
| B | CAGGAC-5′ | 5′-TTCA |
| C | CAGAAC-5′ | 5′-TTCA |
| D | TCCACC-5′ | 5′-TTCA |
| E | ACAT-5′ | 5′-ATGAAG |
| F | ACAT-5′ | 5′-GAAGGC |
| G | ACAT-5′ | 5′-CTGCAT |
| H | AGAATA-5′ | 5′-TTCA |
| I | ATCAGA-5′ | 5′-TTCA |
| J | TTACAT-5′ | 5′-TTCA |
| K | TATGAG-5′ | 5′-TTCA |
| L | ACAT-5′ | 5′-ACCTGC |
| M | ACATCA-5′ | 5′-TTCA |
| N | GTACGG-5′ | 5′-TTCA |
| O | CAGAAT-5′ | 5′-TTCA |
| P | TCAGAA-5′ | 5′-TTCA |
| Q | TACATC-5′ | 5′-TTCA |
| R | AGAATA-5′ | 5′-TTCA |
| S | TTATGA-5′ | 5′-TTCA |
| T | GTTATG-5′ | 5′-TTCA |
| U | TATGAG-5′ | 5′-TTCA |
| V | TCAGAA-5′ | 5′-TTCA |
| W | AGAACG-5′ | 5′-TTCA |
| X | CATCAG-5′ | 5′-TTCA |
| Y | TAATGC-5′ | 5′-TTCA |
| Z | AGACTC-5′ | 5′-ACGCGT |
| AA | CACACG-5′b | 5′-TTCA |
| ATTGCG-5′c | ||
| BB | GTCACC-5′b | 5′-TTCA |
| AGAATTAGTCCTT-5′c |
Four nucleotides from the normal junctions and 6 nucleotides from the unusual junctions are shown. A bold letter indicates matching nucleotides in the viral and host DNAs. The letters A to BB correspond to the letters in Fig. S1 in the supplemental material.
5′ end, second junction.
5′ end, first junction.
Surprisingly, infections with the HIV PPT, MLV PPT, and RSV PPT2 mutants gave rise to four (of a total of 126) proviruses that had defects in U5 (Table 1; also Fig. S1 in the supplemental material). All of these U5-defective proviruses have a normal U3 junction. Given that the fraction of 2-LTR circle junctions with U5 defects was similar to what was found for the wild type, this small fraction (3%) of proviruses with defects at the U5 junction may reflect the percentage of aberrant integrations that arise during infections with wild-type virus.
In 8 of the 22 DuckHepBFlip PPT proviruses, a CA sequence was present at the U3 virus/host junction (Table 1). However, the CA sequence at the end of U3 was not the canonical CA. Instead, the CA sequence (position, −6) in the ATGTA was found at the U3 end in the 2-LTR circle junctions from this mutant (the underlined GT, in the viral minus strand, is base paired with the CA of the plus strand at the end of U3). The sequence for the U3 end of these proviruses indicates that RSV IN was able to remove a single nucleotide from the ATGTA sequence at the U3 terminus and properly integrate the viral DNA (Table 1). However, there are more aberrant U3 integrations than integrations that involve this CA (14 and 8, respectively), suggesting that this sequence is not an efficient substrate for integrase. One apparently normal provirus generated a 218-bp duplication of the host sequence at the target site (Fig. S1 in the supplemental material, panel M). However, we cannot define the exact U3 junction, because the last nucleotide of the U3 LTR terminus was identical to the first nucleotide of the host DNA (Table 2, row M). In the other 13 proviruses, the U3 LTR terminus was usually deleted, and more rarely, there was an insertion of the PPT and flanking sequences. In addition, there were duplications of the host sequences at the target site, ranging in size from a few to hundreds of nucleotides (Fig. S1 in the supplemental material).
These aberrant insertions provide additional support for a model that we proposed in which RSV IN inserts the correct end of the linear DNA normally; however, the aberrant end, if it is not a substrate for processing and/or insertion by RSV IN, is apparently joined to the host DNA by host enzymes (7). This usually generates duplications rather than deletions of the host sequence, and part of the aberrant end of the viral DNA is usually lost.
The aberrant viral end/host DNA junctions often involve microhomology.
When the normal end/host junctions were examined, the microhomologies that were found could be explained by the chance presence of matching nucleotides (Table S1 in the supplemental material). However, when the aberrant viral/host junctions were examined, there were microhomologies involving from 1 to 13 nucleotides between the viral and host sequences (Table 2). Although the frequency of one homologous nucleotide was not statistically significant, the frequencies of two or more homologous nucleotides were statistically significant. The probabilities (P) of obtaining the numbers of 2, 3, 4, 6, and 13 homologous nucleotides by chance were 0.0004, 0.010, 0.0061, <0.0001, and <0.0001, respectively.
Recovery of a provirus flanked by an inversion of host sequences.
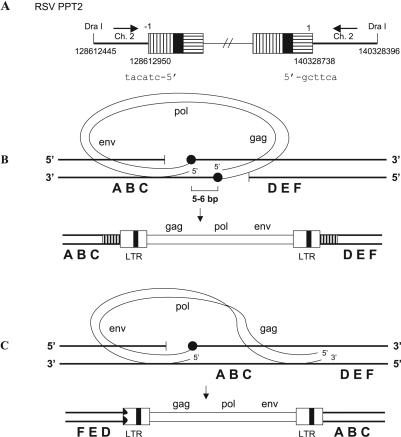
In one provirus derived from an infection by the RSV PPT2 mutant, the U5 LTR terminus was properly joined to chicken chromosome 2. However, only one nucleotide was removed from the U3 LTR terminus, and it was joined to chicken chromosome 2 in the reverse orientation (Fig. 2A) relative to the orientation of chicken chromosome 2 at the U5 LTR junction. The host sequences show that the two host DNA/viral DNA junctions are about 12 kb apart on chicken chromosome 2.
FIG. 2.
Provirus flanked by an inversion of host sequences. (A) Structure of a provirus. U3 is indicated by white boxes with vertical black bars; R is indicated by black boxes; U5 is shown by white boxes with horizontal black bars. Six nucleotides at the viral/host DNA junctions are shown. The orientations of the host sequences are indicated by black arrows. (B) Normal integration event. A 5- to 6-bp duplication is indicated by white boxes with vertical black bars. (C) Model for the generation of a provirus flanked by an inversion of host sequences. The deletion in the 5′ LTR is indicated by a jagged black end. A host DNA is indicated by thick black lines, and a viral DNA is indicated by thin black lines. The joinings between viral DNA and host DNA are indicated by black circles.
A model that explains this inversion of the flanking host sequences is presented in Fig. 2. In the model, the orientation of the aberrant end of the viral DNA is flipped with respect to its “normal” orientation. This appears to be a relatively rare event, suggesting that the “normal” orientation of the viral DNA might be maintained with respect to the host DNA, perhaps by integrase, even if integrase cannot insert the aberrant end. However, the generation of an inversion of the host sequences must also involve a second break/join for restoring the chromosome to a state in which the provirus would be inserted between a centromere and a telomere. For this reason, the recombination event depicted in Fig. 2C may be more common than our data suggest; these insertions may be selected against because they involve two recombination events.
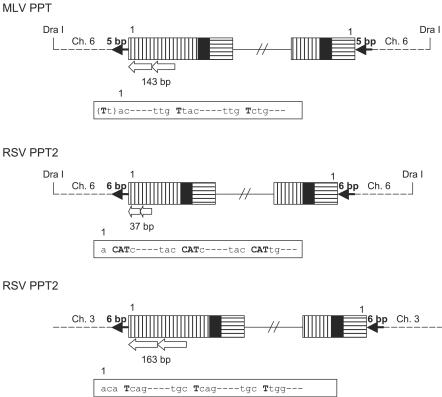
Direct duplications within the U3 of 5′ LTR.
We recovered three proviruses which contained direct duplications ranging in size from tens to hundreds of nucleotides in the U3 segment of the U3 LTR (Fig. 3). The fact that the duplications usually began near the U3 terminus and involved a microhomology at the viral DNA/viral DNA junction suggests that RT may have aborted plus-strand synthesis and then reinitiated it by using a microhomology near the U3 terminus (Fig. 3). These duplications of the U3 terminus did not interfere with the ability of viral integrase to insert these aberrant viral DNAs.
FIG. 3.
Structure of direct duplications of U3 sequences in the U3 LTR. U3 is indicated by white boxes with vertical black bars; R is indicated by black boxes; U5 is shown by white boxes with horizontal black bars. Direct duplications are indicated by white arrows, and the sizes of the duplications are shown. The detailed sequences at the junctions are shown in white boxes, and microhomologies are indicated by bold letters. Normal target duplications are indicated by black arrows.
Supplementary Material
Acknowledgments
We are grateful to Hilda Marusiodis for help in preparing the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Chang, K. W., J. G. Julias, W. G. Alvord, J. Oh, and S. H. Hughes. 2005. Alternate polypurine tracts (PPTs) affect the Rous sarcoma virus RNase H cleavage specificity and reveal a preferential cleavage following a GA dinucleotide sequence at the PPT-U3 junction. J. Virol. 79:13694-13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dash, C. J., W. Rausch, and S. F. Le Grice. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 32:1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 78:13315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 77:11150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh, J., J. G. Julias, A. L. Ferris, and S. H. Hughes. 2002. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J. Virol. 76:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh, J., K. W. Chang, and S. H. Hughes. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integration in vivo. J. Virol. 80:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rattray, A. J., and J. J. Champoux. 1989. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol. 208:445-456. [DOI] [PubMed] [Google Scholar]
- 10.Robson, N. D., and A. Telesnitsky. 1999. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J. Virol. 73:948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robson, N. D., and A. Telesnitsky. 2000. Selection of optimal polypurine tract region sequences during Moloney murine leukemia virus replication. J. Virol. 74:10293-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.