Abstract
Many positive-strand RNA viruses generate 3′-coterminal subgenomic mRNAs to allow translation of 5′-distal open reading frames. It is unclear how viral genomic and subgenomic mRNAs compete with each other for the cellular translation machinery. Translation of the uncapped Barley yellow dwarf virus genomic RNA (gRNA) and subgenomic RNA1 (sgRNA1) is driven by the powerful cap-independent translation element (BTE) in their 3′ untranslated regions (UTRs). The BTE forms a kissing stem-loop interaction with the 5′ UTR to mediate translation initiation at the 5′ end. Here, using reporter mRNAs that mimic gRNA and sgRNA1, we show that the abundant sgRNA2 inhibits translation of gRNA, but not sgRNA1, in vitro and in vivo. This trans inhibition requires the functional BTE in the 5′ UTR of sgRNA2, but no translation of sgRNA2 itself is detectable. The efficiency of translation of the viral mRNAs in the presence of sgRNA2 is determined by proximity to the mRNA 5′ end of the stem-loop that kisses the 3′ BTE. Thus, the gRNA and sgRNA1 have “tuned” their expression efficiencies via the site in the 5′ UTR to which the 3′ BTE base pairs. We conclude that sgRNA2 is a riboregulator that switches off translation of replication genes from gRNA while permitting translation of structural genes from sgRNA1. These results reveal (i) a new level of control of subgenomic-RNA gene expression, (ii) a new role for a viral subgenomic RNA, and (iii) a new mechanism for RNA-mediated regulation of translation.
Recent years have brought an explosion in discoveries of RNAs that regulate gene expression (3, 33, 38, 56). Many different types of noncoding RNAs posttranscriptionally regulate gene expression in trans (16). These include microRNAs, small interfering RNAs, small nucleolar RNAs, small nuclear RNAs, and bacterial small RNAs (5, 8, 27, 33). trans-acting regulatory RNAs are also generated by viruses. The noncoding adenovirus virus-associated (VA) RNAs (34) and Epstein-Barr virus-encoded RNAs (EBER) (7) interfere with host antiviral systems and permit efficient expression of late viral genes. Epstein-Barr virus and other herpesviruses also generatemicroRNAs to downregulate the expression of host and viral genes (38, 44). Red clover necrotic mosaic virus genomic RNA2 directs, in trans, the synthesis of subgenomic mRNA (sgRNA) from genomic RNA1 (52). Flock house virus sgRNA trans activates the replication of a viral genomic RNA (1, 15), which in turn down-regulates synthesis of the sgRNA (57). Here, we provide an example of a different kind of regulation, in which a viral RNA regulates the translation of the other RNAs generated by the virus.
Many families of positive-strand RNA viruses produce nested subgenomic mRNAs during infection (36, 43). These sgRNAs have the same 3′ ends as genomic RNA but have 5′ truncations or deletions relative to the genomic RNA. This places open reading frames (ORFs) that are 5′ distal on genomic RNA near the 5′ ends of sgRNAs, allowing the sgRNAs to serve as mRNAs for translation of these downstream ORFs. Examples of viral pathogens that produce sgRNAs are severe acute respiratory syndrome coronavirus (24), equine arteritis virus (53), Sindbis virus (17), rubella virus (40), tobacco mosaic virus (20), citrus tristeza virus (19), and barley yellow dwarf virus (BYDV) (28, 29). While the control and mechanism of synthesis of the subgenomic RNAs of these and related viruses have been studied (36, 43), little is known about how translation of the viral genomic and subgenomic RNAs is coordinated as they accumulate in the cell (17, 40). The competition among viral RNAs for the host translation machinery and regulation of their translation are likely key control points in viral-gene expression necessary for a successful infection. Here, we provide evidence that translation of BYDV genomic RNA and a subgenomic RNA is regulated in trans by a second, specialized viral sgRNA.
BYDV, a major pathogen of wheat and other cereal crops (32), has a positive-sense RNA genome of 5,677 nucleotides (nt) that encodes six ORFs (Fig. 1) (37). Three 3′-coterminal sgRNAs are generated in infected cells. They are not encapsidated and thus are absent at the initial stage of infection. sgRNA1 is the mRNA for the coat protein (CP) (ORF 3), a putative movement protein (ORF 4), and a C-terminal extension of the coat protein required for aphid transmission (ORF 5). Highly abundant sgRNA2 harbors a small ORF (ORF 6) that encodes a predicted polypeptide of 4.3 to 7.2 kDa, depending on the isolate. The sequence of ORF 6 is poorly conserved among BYDV isolates (9), and it is absent in other members of the genus Luteovirus (13, 49). The product of ORF 6 has not been detected in infected cells, although sgRNA2 can be translated in vitro (55). sgRNA3 comprises the 3′-terminal 332 nt of the BYDV genome (26). It accumulates sporadically, encodes no ORF, and has no known function.
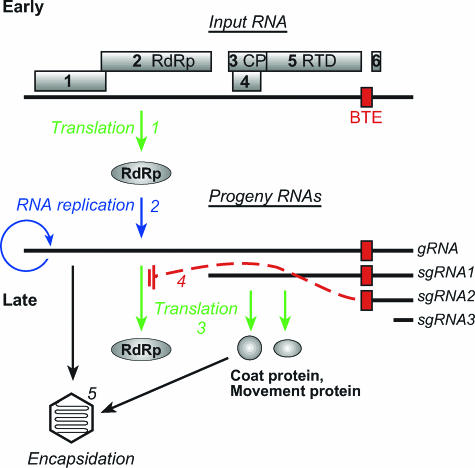
FIG. 1.
trans regulation model of BYDV gene expression. (1) In the early stage of BYDV infection, subgenomic RNAs are absent; thus, the products of ORFs 1 and 2, including the RdRp, are the only proteins produced. (2) Viral genomic-RNA replication and subgenomic-RNA transcription occur. (1, 2, and 3) Viral RNAs accumulate, and viral proteins are produced. (4) The accumulation of sgRNA2 trans inhibits translation of BYDV RdRp from gRNA. (5) However, translation of structural and movement proteins from sgRNA1 is not inhibited. Genomic RNAs are available for encapsidation in the coat protein.
BYDV genomic RNA (gRNA) and sgRNAs have no 5′ cap and no poly(A) tail (2). Highly efficient cap-independent translation of gRNA and sgRNA1 is conferred by a 100-nt BYDV cap-independent translation element (BTE) in the 3′ untranslated region (UTR) (22, 54). To recruit ribosomes or factors to the 5′ ends of the viral RNAs, where translation initiates, the 3′ BTE must base pair with the 5′ UTR (21). The bridging of the 3′ and 5′ ends of the viral RNAs is facilitated by a kissing stem-loop interaction between the 3′ BTE and a 5-base BTE-complementary loop (BCL) sequence present in the 5′ UTRs of both gRNA and sgRNA1 (21, 48).
In addition to conferring cap-independent translation in cis, the BTE inhibits translation of viral genes and nonviral reporter genes in trans (54, 55). In wheat germ extract, sgRNA2, which harbors the BTE at its 5′ end, trans inhibits translation of gRNA more than that of sgRNA1 (55). The inhibition does not require translation of ORF 6, but it requires a functional BTE in sgRNA2 (55). Premature addition of sgRNA2 at the initial moment of infection strongly inhibits BYDV RNA accumulation (50). Based on these data, the following trans regulation model of gene expression was proposed (Fig. 1). Early in BYDV infection, only the replicase genes, ORF 1 and ORF 2 (translated as a fusion with ORF 1 by ribosomal frameshifting) (4), are translated via BTE-mediated cap-independent translation in cis. Once replicase is produced, viral RNA is replicated, and gRNA and sgRNAs accumulate. The highly abundant sgRNA2 would selectively inhibit translation of gRNA relative to sgRNA1 in trans. Structural and movement proteins would then be preferentially translated from sgRNA1, and replicase expression would be shut off. Thus, the BYDV viral life cycle would switch from an early to a late stage of gene expression (Fig. 1) (55). Here, we provide evidence that strongly supports the notion that these events occur in virus-infected cells. Moreover, we show that the different translation efficiencies of the BYDV mRNAs in the presence of sgRNA2 are determined by the proximity to the 5′ end of the mRNA of the stem-loop structure that base pairs to the 3′ translation element to allow cap-independent translation. These observations reveal a novel translational control mechanism by a trans-regulatory RNA and a new function for a viral subgenomic RNA.
MATERIALS AND METHODS
Plasmids and RNA constructs.
The full-length infectious clone of a PAV isolate of BYDV (BYDV-PAV), pPAV6, was used for transcribing infectious BYDV genomic RNA (11). The sgRNA2 knockout mutant clone of BYDV-PAV, pPAV6ΔSG2, was described previously (28) as SG2G/C. It differs from pPAV6 by a G-to-C mutation at position 4810, which prevents sgRNA2 synthesis. pSG2 and pSG2BF allow T7 transcription of sgRNA2 RNA and its mutant sgRNA2BF, respectively (55). sgRNA2BF contains a GAUC duplication at the BamHI site (BF) of sgRNA2 that destroys the in vitro trans inhibition function of sgRNA2 (55).
Clone pGfLUC was described previously (22) as p5′UTR-LUC-TE869-(A)60. GfLUC is the gRNA reporter transcript derived from SmaI-linearized pGfLUC by in vitro T7 transcription. It encodes the firefly luciferase ORF flanked by the UTRs of BYDV. pRenilla-CP393 was cloned by replacing nt 2843 to 4565 of pPAV6 with the Renilla ORF of pRluc (Promega, Madison, WI). pSG1rLUC was cloned by ligating the Bst1107I-BsmI fragment of pRenilla-CP393 into Bst1107I/BsmI-cut pSG1, which was described previously (29). SG1rLUC is the sgRNA1 T7 transcript from SmaI-cut pSG1rLUC. It has the same 5′ UTR as sgRNA1, except for the omission of 14 bases at the extreme 3′ end. pSG1fLuc was constructed by replacing the 5′ UTR of pGfLUC with the 5′UTR of pSG1rLUC.
PAV6-FLAG and sgRNA2-FLAG were constructed by inserting a FLAG tag (amino acid sequence, DYKDDDDK) at the 3′ end of ORF 6 in pPAV6 and pSG2, respectively. The FLAG tag insert was created by a three-step PCR approach by using primers bearing a FLAG tag and pSG1 as templates, as described previously (28). The final PCR fragment was cloned into Acc65I-SmaI-digested pPAV6 or NotI-SmaI-digested pSG2. sgRNA2-LIII CS RNA was in vitro transcribed from a PCR-generated template corresponding to the sgRNA2 sequence. The PCR fragment was amplified using LUC 869-LIII-CS as a template (48) and a forward primer bearing a T7 promoter sequence and a reverse primer corresponding to the 3′ end of sgRNA2.
pA-GfLUC contained a C15A mutation in the loop of stem-loop A (SL-A)and a GAC-to-CUG mutation within the loop of stem loop D (bases 105 to 107) (see Fig. 5A). First, a BYDV 5′-UTR PCR fragment with a GAC-to-CUG mutation within the loop of stem-loop D was generated by three-step PCR and cloned into NotI-BssHI-digested pGfLUC. The clone obtained was then used as a PCR template to generate the additional C15A mutation. pA-GfLUC was then cloned by ligating the NotI-BssHI-digested PCR fragment into NotI-BssHI-cut pGfLUC. pD-SG1fLUC contained an A10C mutation in the loop of stem-loop A and an AGUUA-to-CUGACAA mutation of the loop of stem-loop D (bases 111 to 115) (see Fig. 5A). pD-SG1fLUC was generated by PCR base mutagenesis of SG1fLUC cDNA using the Gene Tailor Site-Directed Mutagenesis System (Invitrogen). All constructs were verified by automated sequencing at the Nucleic Acid Facility of Iowa State University.
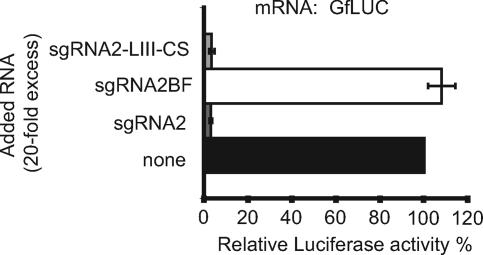
FIG. 5.
Mechanism of trans inhibition by sgRNA2. Shown are the differential effects of 20-fold molar excess of sgRNA2-LIII-CS, sgRNA2, and sgRNA2BF on translation of GfLUC RNA in wheat germ extract. The error bars indicate standard deviations.
In vitro transcription and translation.
All RNAs were synthesized by using T7 MegaScript kits for uncapped RNA or T7 mMESSAGE mMACHINE kits for capped RNA (Ambion, Austin, TX), according to the manufacturer's instructions. Prior to transcription, all constructs were linearized either with SmaI to generate the same 3′ end as the viral RNA or with VspI to include a 60-nt 3′ poly(A) tail. In vitro translation in wheat germ extract (Promega) and a luciferase assay were performed as described previously (50). The reporter RNA transcripts (0.2 pmol), with the indicated molar ratio (n-fold excess) of sgRNA2 or sgRNA2BF transcripts to reporter RNA, were added to the wheat germ translation system with a final reaction volume of 25 μl and translated for 1 h at 25°C prior to luciferase reading. All luciferase assays were performed in at least three independent experiments, each of which was conducted in duplicate or triplicate.
Western immunoblotting.
Western blotting was performed according to the Amersham Pharmacia Biotech protocol. Total protein from inoculated protoplasts was separated on a 10% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. After being blocked overnight with phosphate-buffered saline-Tween buffer (PTB) containing 5% dried low-fat milk, the membrane was incubated with the primary anti-FLAG antibodies (Sigma, St. Louis, MO) in a 1:25,000 dilution in PTB for 2 h, with a 1:600 dilution of fluorescein-conjugated secondary antibodies for 1 h, and then with a 1:2,500 dilution of alkaline phosphatase-conjugated anti-fluorescein antibodies for another hour. The membrane was washed three times with PTB after each incubation. After the final incubation with attophose substrate with a volume of 24 μl/cm2 for less than 20 min, the membrane was air dried and scanned on a STORM 840 chemiluminescence imager (Molecular Dynamics).
In vivo translation.
Oat protoplasts were prepared and electroporated with RNA as described previously (12). For the two-step electroporation method, the voltage was reduced to 280 V. For the in vivo translation assay of the SG2fLUC reporter construct, we included a capped, polyadenylated Renilla luciferase reporter RNA as an internal control to normalize electroporation variation. The Renilla luciferase ORF was flanked by the 5′ and 3′ UTRs of the firefly luciferase gene from pGEMLUC (Promega). Luciferase activity was measured 4 h after electroporation.
In the two-step electroporation, oat protoplasts were inoculated with 1 pmol of infectious BYDV PAV6 or PAV6ΔGS2 RNA and incubated for 24 h at room temperature prior to the second electroporation to allow viral replication and sgRNA accumulation. When transfected directly with the nonreplicative sgRNA2 or sgRNA2BF RNA transcripts, the cells were incubated for 4 h prior to the second electroporation. In the second step, protoplasts were inoculated again with 1 to 2 pmol of GfLUC (A-GfLUC), SG1rLUC, or both, as indicated. In all cases, firefly luciferase and Renilla luciferase were analyzed 4 h after the second electroporation, as described previously (51), and the Promega (Madison, WI) Stop-N-Glo system was used to assay both luciferase activities.
Northern blot analysis.
Total RNAs were extracted from oat protoplasts 24 h postinoculation (p.i.) by using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The RNAs were then analyzed by Northern blotting as described previously (29). A 32P-labeled probe complementary to the 1.5-kb 3′ end of BYDV-PAV genome RNA was used to detect BYDV gRNA and sgRNAs (29).
RESULTS
Like the actual BYDV RNAs, reporter constructs representing gRNA and sgRNA1 are differentially inhibited by sgRNA2 in trans.
To test the selective inhibition of translation hypothesis diagrammed in Fig. 1, we designed reporter constructs that allowed efficient detection of translation from both gRNA and sgRNA1 in vivo. We replaced the viral coding regions with different luciferase genes that could be assayed in the same tube (Promega Dual Luciferase reporter assay). The reporter construct representing gRNA, GfLUC, includes the fireflyluciferase ORF flanked by the BYDV genomic 5′ and 3′ UTRs (Fig. 2A). The reporter construct representing sgRNA1, SG1rLUC, contains the Renilla luciferase ORF in place of ORFs 3 and 4 and most of ORF 5. The 5′ UTR of SG1rLUC contains BYDV nt 2670 to 2842, giving it the same 5′ terminus as sgRNA1. To determine the validity of these two reporter constructs to represent gRNA and sgRNA1, we tested whether GfLUC and SG1rLUC RNAs behave the same as gRNA and sgRNA1 in the presence of sgRNA2 in wheat germ translation experiments (55). When added to translation reaction mixtures containing either GfLUC or SG1rLUC mRNA, sgRNA2 inhibited GfLUC translation more than it inhibited SG1rLUC translation, and this difference increased as the molar ratios of sgRNA2/GfLUC or sgRNA2/SG1rLUC increased (Fig. 2B). The negative control, sgRNA2BF, did not inhibit translation of either reporter RNA (Fig. 2B). sgRNA2BF differs from sgRNA2 only by a GAUC duplication in the BamHI4837 site in the BTE. This mutation abolishes both cap-independent translation in cis and inhibition of translation in trans (2, 54).
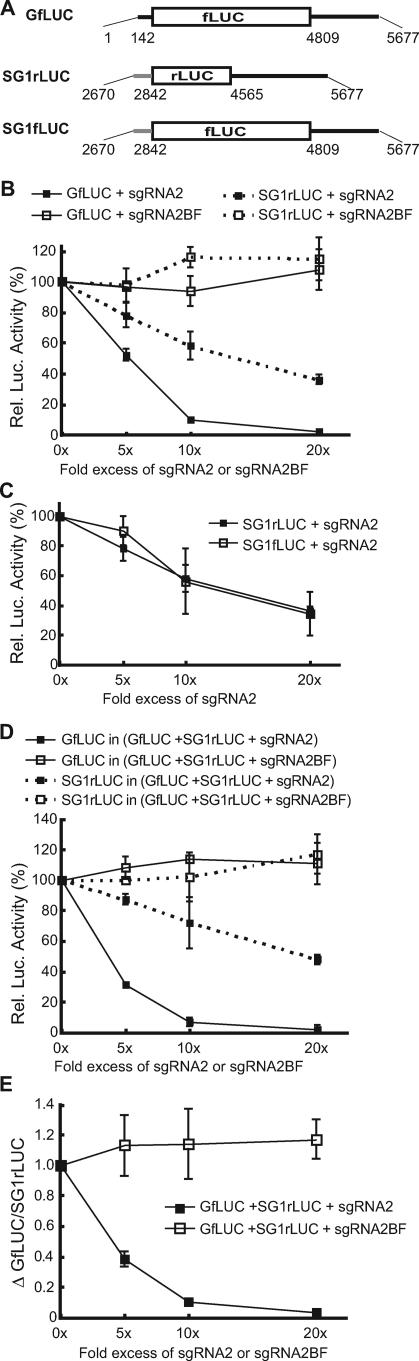
FIG. 2.
Effects of sgRNA2 on translation of reporters in wheat germ translation extracts. In all cases, for each mRNA tested, the relative luciferase (Rel. Luc.) activity in the absence of sgRNA2 is defined as 100%. (A) Maps of reporter RNAs. fLUC, firefly luciferase; rLUC, Renilla luciferase. The ends of the UTRs are numbered as in the full-length BYDV genome. (B) Differential effects of sgRNA2 and sgRNA2BF in trans on translation of GfLUC or SG1rLUC in separate reactions. The error bars indicate standard deviations. (C) Effects of sgRNA2 on different reporters with the same sgRNA1 5′ UTR; relative luciferase activity of SG1rLUC or SG1fLUC in separate reactions in the presence of the indicated (molar) excess of sgRNA2. (D) Differential effects of sgRNA2 or sgRNA2BF on translation of GfLUC and SG1rLUC in the same reaction. The activities of GfLUC and SG1rLUC were plotted individually against the excess of sgRNA2 and sgRNA2BF. (E) Changes in ratios of GfLUC/SG1rLUC activity from panel D. ΔGfLUC/SG1rLUC = (GfLUC/SG1rLUC in the presence of sgRNA2 or sgRNA2BF)/(GfLUC/SG1rLUC in the absence of sgRNA2 or sgRNA2BF).
To determine whether the firefly and Renilla luciferase ORFs caused differences in translation efficiency in the presence of sgRNA2, we constructed an sgRNA1 reporter, SG1fLUC, that differs from GfLUC only in the 5′ UTR (Fig. 2A). SG1fLUC and SG1rLUC behaved indistinguishably in the presence of sgRNA2 (Fig. 2C). Thus, neither the coding regions nor the different lengths of the 3′ UTRs account for the differential effects of sgRNA2 on translation of GfLUC versus SG1rLUC.
To more closely mimic natural infection, gRNA reporter GfLUC, sgRNA1 reporter SG1rLUC, and sgRNA2 were mixed simultaneously in the same wheat germ translation reaction mixture at different ratios. Again, sgRNA2 inhibited translation of GfLUC much more than that of SG1rLUC (Fig. 2D). The difference in translation inhibition by sgRNA2 of GfLUC and SG1rLUC reporters was greater when GfLUC, SG1rLUC, and sgRNA2 were added together in the same translation reaction (compare Fig. 2B and D). The GfLUC/SG1rLUC (gRNA/sgRNA1) expression ratio decreased as the sgRNA2 concentration was increased (Fig. 2E). These results resemble previous observations when gRNA, sgRNA1, and sgRNA2 were all added to a wheat germ translation extract at ratios approximating those in infected cells. In that experiment, gRNA was inhibited by 99% while sgRNA1 was inhibited by only 65%, allowing it to remain a relatively efficient message (55). Thus, the reporter RNAs provide valid representations of the translation of the actual viral RNAs in the presence of sgRNA2. Importantly, these results also reveal that no ORFs or gene products of gRNA or sgRNA1 are necessary for the differential inhibition of translation.
Differential inhibition of GfLUC and SG1rLUC translation in virus-infected cells.
Having validated that reporter constructs GfLUC and SG1rLUC translate like gRNA and sgRNA1 in the presence or absence of sgRNA2 in vitro, we tested the trans regulation model in oat protoplasts by measuring the translation efficiencies of the two reporter constructs in the presence of replicating BYDV RNA. We employed a two-step electroporation method (Fig. 3A) (50). First, oat protoplasts were transfected by electroporation with the infectious transcript of the BYDV genome, PAV6, or with transcript PAV6ΔSG2. PAV6ΔSG2 RNA has a G4810C point mutation that prevents sgRNA2 synthesis but still permits genomic-RNA replication (28). After a 24-hour incubation to allow viral replication and sgRNA accumulation (Fig. 3B), the protoplasts were electroporated again, this time with GfLUC or SG1rLUC reporter RNA. Then after another 4-hour incubation to allow translation of these RNAs, firefly luciferase and Renilla luciferase activities were measured (Fig. 3C and D).
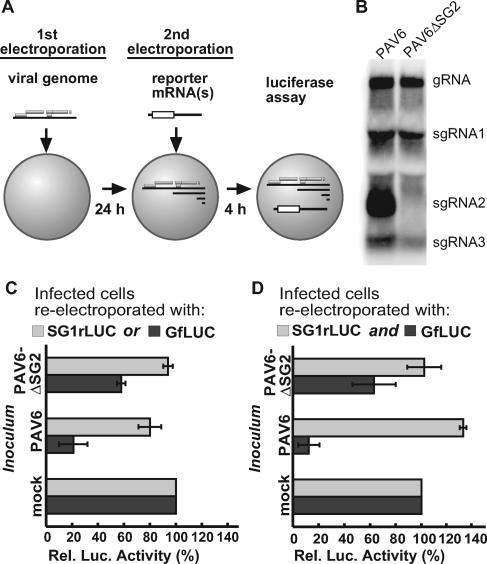
FIG. 3.
Differential effects of PAV6 and PAV6ΔSG2 replication on translation of GfLUC and SG1rLUC in oat protoplasts. (A) Diagram of the two-step electroporation method. First, oat protoplasts were inoculated with full-length infectious BYDV PAV6 or PAV6ΔSG2 transcripts. After a 24-h incubation to allow viral replication and sgRNA accumulation, the cells were electroporated again with 1 pmol GfLUC, 1 pmol SG1rLUC (C), or both (D). Luciferase activities were measured 4 h later. The error bars indicate standard deviations. (B) Northern blot hybridization showing replication of PAV6 and PAV6ΔSG2 at 24 h p.i. (C) Luciferase activities in cells first transfected with PAV6 or PAV6ΔSG2 RNAs and then reelectroporated with the indicated reporter RNA. The luciferase activity of GfLUC (or SG1rLUC) in mock-transfected cells was defined as 100%. (D) Same as in panel C, but 1 pmol GfLUC and 1 pmol SG1rLUC were coelectroporated together into the same batch of transfected protoplasts.
Wild-type and mutant BYDV genomes accumulated to similar levels in protoplasts, with the conspicuous absence of sgRNA2 in protoplasts infected with PAV6ΔSG2 (Fig. 3B). After the second electroporation to introduce reporter mRNA, we observed that the presence of replicating PAV6 RNA caused nearly an 80% drop in the translation of the gRNA reporter GfLUC compared to uninfected cells but caused only a 20% reduction in the translation of the sgRNA1 reporter SG1rLUC (Fig. 3C, PAV6). In cells transfected with PAV6ΔSG2 RNA, translations of GfLUC and SG1rLUC (Fig. 3C, PAV6ΔSG2) were reduced by only 40% and less than 5%, respectively. Thus, infection with PAV6ΔSG2 (which makes no sgRNA2) inhibited the translation of GfLUC less than did wild-type PAV6 RNA.
When GfLUC and SG1rLUC were coelectroporated in the second step in cells previously transfected with PAV6 RNA, GfLUC translation dropped by 88%, while translation of SG1rLUC actually increased slightly, compared to that of uninfected cells (Fig. 3D, PAV6). SG1rLUC translated at least 10 times more efficiently than GfLUC relative to uninfected controls. Thus, the differential inhibition effects of the replicating PAV6 on translation of GfLUC and SG1rLUC were greater when both reporter RNAs were present simultaneously. PAV6ΔSG2 had a less inhibitory effect on translation of GfLUC than did wild-type PAV6 infection (Fig. 3D, PAV6ΔSG2), strengthening the concept that sgRNA2 is the major influence on the differential inhibition of GfLUC and SG1rLUC translation. The 40% inhibition of GfLUC translation in the presence of replicating PAV6ΔSG2 likely results from the presence of the BTE at the 3′ ends of gRNA and sgRNA1, which, being much less abundant than sgRNA2, would be expected to inhibit reporter gene expression to a lesser extent, as was observed.
BYDV sgRNA2 alone selectively trans inhibits translation in vivo.
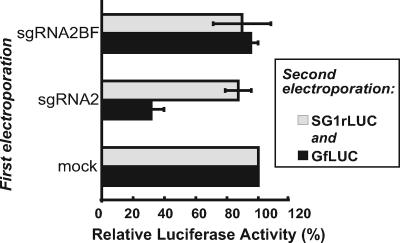
It is possible that the differential inhibition of reporter gene expression in infected cells is controlled by viral proteins or by host proteins whose expression or function is affected by viral infection, as well as by sgRNA2. To avoid the complicating effects of a viral infection, we observed the effects of sgRNA2 or sgRNA2BF RNA alone on GfLUC or SG1rLUC translation in cells in another two-step electroporation assay. Oat protoplasts were first electroporated with either sgRNA2 or sgRNA2BF transcripts or mock transfected. Four hours later, the same protoplasts were electroporated with GfLUC and SG1rLUC RNAs. Both luciferase activities were measured after another 4-hour incubation to allow translation of the reporter RNAs. In cells preelectroporated with sgRNA2, GfLUC translation dropped by 70% compared to that in cells initially mock transfected or preelectroporated with sgRNA2BF (Fig. 4). In contrast, the presence of sgRNA2 caused only a 10% reduction in translation of SG1rLUC RNA. These data show that sgRNA2 alone is sufficient to mediate selective trans inhibition of gRNA. It requires a functional BTE but does not require viral replication or infection in vivo.
FIG. 4.
Differential trans inhibition by sgRNA2 alone in oat protoplasts. The two-step electroporation assay was employed, with 4 h between electroporations. The graph shows the luciferase activities of GfLUC and SG1rLUC measured 4 h after they were coelectroporated into cells previously electroporated with sgRNA2 or sgRNA2BF. The activities of GfLUC and SG1rLUC in cells that were preelectroporated with no RNA were defined as 100%. The error bars indicate standard deviations.
The mechanism of trans inhibition does not rely on base pairing of the BTE to the 5′ UTR of gRNA.
A possible mechanism of trans inhibition is base pairing of loop III of the BTE in sgRNA2 with the BCL in the 5′ UTR of the gRNA and effective competition with the kissing between the BTE and BCL that occurs in cis to facilitate cap-independent translation. To test this hypothesis, we observed the trans inhibition of the translation of the gRNA in wheat germ extract by a mutant sgRNA2 containing a different sequence in loop III of the BTE that bears no complementarity to the 5′ UTR of gRNA. BTE loop III in sgRNA2 was changed from UGUCA to GGCAUAUUGA(sgRNA2-LIII-CS) and is not complementary to the UGACA in the BCL or any other sequence within the gRNA. sgRNA2-LIII-CS trans inhibited the translation of GfLUC as efficiently as wild-type sgRNA2 (Fig. 5A). In a reciprocal experiment, sgRNA2 with the BF mutation (Fig. 2) did not inhibit in trans, even though it retained complementarity to the gRNA 5′ UTR. Thus, the trans inhibition effect of sgRNA2 does not rely on base pairing to the mRNA that it inhibits.
The 5′ UTRs of gRNA and sgRNA1 determine the differential trans inhibition effects.
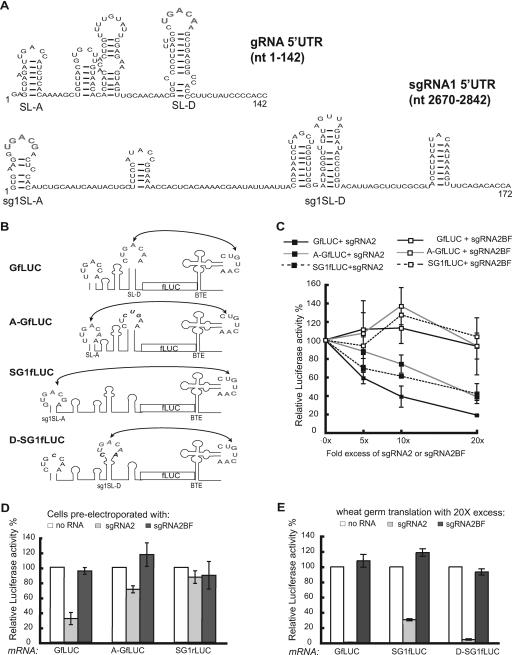
The previous results led us to wonder what property of the viral RNA determines differential inhibition. Because neither the ORFs nor the 3′ UTRs affect inhibition by sgRNA2 (Fig. 2C), we conclude that features of the different 5′ UTRs determine the ability to be inhibited in trans. A striking difference between the two 5′ UTRs is that the BCL is in the 5′-proximal stem-loop in sgRNA1 but in the fourth stem-loop (SL-D) from the 5′ end in the gRNA (Fig. 6A). The loop of SL-D is located 104 to 109 nt from the 5′ end of the gRNA, whereas the sgRNA1 BCL is just 10 nt from the 5′ end of its RNA (Fig. 6A, sg1SL-A). To determine the effect of the distance of the BCL from the 5′ end on cap-independent translation in the presence of sgRNA2, we engineered GfLUC so that the 5′-proximal stem-loop of its 5′ UTR (SL-A) was able to kiss (base pair to) the BTE, and the natural kissing bases of SL-D were mutated so that it could no longer interact with the 3′ BTE (Fig. 6B). This construct, A-GfLUC, translates at an efficiency similar to that of GfLUC (48). To determine whether this relocation of the BCL to the 5′-proximal stem-loop of the gRNA influences the selectivity of trans inhibition by sgRNA2, we tested the translation efficiency of A-GfLUC relative to GfLUC and SG1fLUC in the presence of sgRNA2 in wheat germ extract (Fig. 6C). A-GfLUC behaved similarly to SG1fLUC in the presence of various ratios of excess sgRNA2 and thus was inhibited less by sgRNA2 than was GfLUC (Fig. 6C).
FIG. 6.
Features of the 5′ UTRs of gRNA and sgRNA1 that determine the differential trans inhibition by sgRNA2. (A) The known secondary structure of the BYDV gRNA 5′ UTR (21) and the MFOLD-predicted (58) secondary structure of the sgRNA1 5′ UTR. The kissing BCL bases that participate in the long-distance interaction with the 3′ BTE are in gray. (B) Schematic diagram of the 3′ BTE-5′ UTR interactions in the indicated reporter constructs. In A-GfLUC, the 5′-proximal loop of SL-A was converted by a single C-to-A change at position 15 (italics), which made SL-A complementary to the 3′ BTE at five consecutive bases (boldface gray). The endogenous SL-D kissing bases were mutated to prevent base pairing with the 3′ BTE (GAC to CUG; black italics). In D-SG1fLUC, the loop of sg1SL-D was converted from AGUUA to CUGACAA (bases 110 to 116). The modified D-SG1LUC also contained an A-to-C change at position 10 in the loop of sg1SL-A, which prevented base pairing to the 3′ BTE. (C) Differential effects of sgRNA2 on translation of A-GfLUC, GfLUC, and SG1fLUC in wheat germ extract. The activity of GfLUC, A-GfLUC, or SG1fLUC in the absence of sgRNA2 was defined as 100%. The error bars indicate standard deviations. (D) Differential inhibition by 40-fold excess of sgRNA2 or sgRNA2BF of GfLUC, A-GfLUC, and SG1rLUC translation in oat protoplasts. The two-step electroporation assay was employed as in Fig. 4. GfLUC and SG1rLUC or A-GfLUC and SG1rLUC (2 pmol each) were coelectroporated in oat protoplasts 4 h after the indicated RNAs were electroporated into the same cells. SG1rLUC levels differed little in the presence of GfLUC or A-GfLUC, so average SG1rLUC readings are shown. For each reporter RNA, luciferase readings were normalized to the amount detected in the absence of RNA in the first electroporation. (E) Differential effects of 20-fold molar excess of sgRNA2 or sgRNA2BF on translation of GfLUC, SG1fLUC, and D-SG1fLUC in wheat germ extract. The readings were normalized to the amount detected in the absence of sgRNA2 or sgRNA2BF RNA.
To examine in vivo the effect of relocating the kissing loop, a two-step electroporation assay was performed (Fig. 6D). The wild-type (GfLUC) or the modified (A-GfLUC) genomic reporter transcripts were coelectroporated with SG1rLUC RNA in cells previously transfected with sgRNA2 or sgRNA2BF, and the translation efficiencies of both reporter RNAs were compared. While GfLUC translation was inhibited by more than 70% in the presence of sgRNA2, translation of A-GfLUC and SG1fLUC dropped by only 25% and 10%, respectively, compared to mock-transfected cells (Fig. 6D). Thus, unlike the in vitro result (Fig. 6C), A-GfLUC was slightly more susceptible than SG1rLUC to sgRNA2 trans inhibition effects in vivo. Most importantly, A-GfLUC was inhibited far less than GfLUC, from which it differs by only four base changes in the 5′ UTR that allow the BTE to base pair to stem-loop A and not stem-loop D. As expected, sgRNA2BF had little or no inhibitory effect on the translation of any of the reporter RNAs in vitro or in vivo (Fig. 6).
To further analyze the effect of the position of the BCL on selective trans inhibition by sgRNA2, we moved the BCL of SG1fLUC 100 nt downstream to the fourth stem-loop (sg1SL-D) (Fig. 6A) from the 5′ end (construct D-SG1fLUC) (Fig. 6B). We tested the translation efficiency of D-SG1fLUC relative to SG1fLUC and GfLUC in the presence of 20-fold-excess sgRNA2 and sgRNA2BF in wheat germ extract (Fig. 6E). While the 5′-distal position of the BCL in D-SG1fLUC did not affect cap-independent translation in the absence of sgRNA2, D-SG1fLUC was inhibited to a level similar to that of GfLUC in the presence of sgRNA2. As expected, sgRNA2BF did not inhibit the translation of either reporter RNA (Fig. 6E). Taken together, the effects of moving the BCL in either gRNA or sgRNA1 reporters demonstrate that the proximity of the kissing loop to the 5′ end of the RNA is the major determinant of susceptibility to inhibition of translation by sgRNA2.
Lack of a role for sgRNA2 as an mRNA.
We next determined whether sgRNA2 functions as an mRNA in cells. sgRNA2 encodes a small, poorly conserved ORF (ORF 6) that is translatable in vitro (55). Unlike gRNA and sgRNA1, sgRNA2 harbors the BTE in its 5′ UTR. No significant sequence complementarity between the 3′ and 5′ UTRs is predicted in sgRNA2. Thus, sgRNA2 is unlikely to be circularized by 5′-3′ base pairing. Because circularization of eukaryotic mRNAs is generally required for efficient translation in vivo but not in vitro (35, 45), we speculated that sgRNA2 may not be translatable in vivo (in protoplasts).
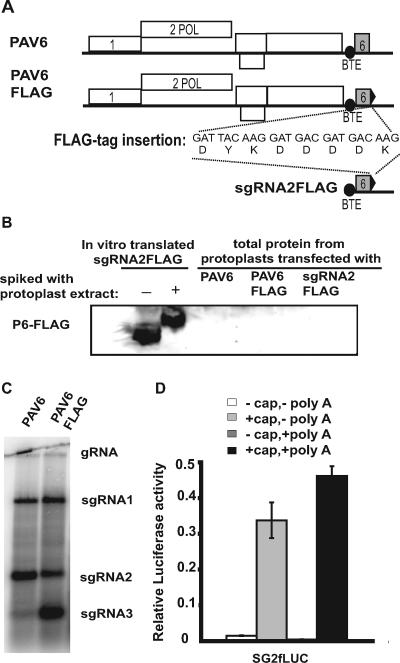
To test the translatability of sgRNA2 in vivo, we fused a FLAG tag to ORF 6 and attempted to detect the P6-FLAG fusion using anti-FLAG antibodies (Fig. 7A). In immunoblots of total protein from cells inoculated with the infectious transcript of the BYDV genome bearing the FLAG tag fusion (PAV6-FLAG), no protein corresponding to P6-FLAG was detected (Fig. 7B), despite the production of abundant sgRNA2 by the PAV6-FLAG virus (Fig. 7C). To determine whether ORF 6 could be translated directly from sgRNA2 in the absence of viral replication, transcript corresponding to sgRNA2 with a FLAG tag (sgRNA2-FLAG) was electroporated into oat protoplasts. As a positive control, this RNA was translated in vitro and P6-FLAG was detected by Western blotting (Fig. 7B). The immunoblot of total protein from protoplasts electroporated with sgRNA2-FLAG revealed no detectable P6-FLAG, even after varied times of sample collection, long exposures, or heavy gel loading (Fig. 7B and data not shown). Thus, sgRNA2 is either not translated or its product (P6) is highly unstable in cells. To test the latter possibility, the in vitro translation product of sgRNA2-FLAG was spiked in an extract of uninfected cells prepared by the same method that was used for attempted detection of P6-FLAG from electroporated cells. The cell extract affected mobility of in vitro-translated P6-FLAG, but P6-FLAG was clearly stable enough to be detected (Fig. 7B). Thus, the cell extract does not degrade FLAG-tagged P6 significantly.
FIG. 7.
Attempted detection of ORF 6 translation in vivo. (A) Maps of BYDV genomic PAV6 and PAV6-FLAG and the subgenomic sgRNA2-FLAG transcript. Nucleic acid and amino acid sequences of the FLAG tag inserted at the 3′ end of ORF 6 (black triangle) are shown. (B) Western blot using anti-FLAG antibodies on total protein from oat protoplasts inoculated with infectious BYDV PAV6, PAV6-FLAG, or nonreplicative sgRNA2-FLAG RNA 24 h posttransfection. As a positive control, the wheat germ translation product of sgRNA2-FLAG is shown in the absence (−) or presence (+) of protoplast extract, which retarded protein mobility in the gel. (C) Northern blot hybridization of total RNA from virus-infected cells showing replication of infectious BYDV PAV6 and PAV6-FLAG. (D) Translation in oat protoplasts of SG2fLUC transcript with ORF 6 fused to the firefly luciferase ORF. The presence (+) or absence (−) of a cap and/or a poly(A) tail on this transcript is indicated. Relative luciferase activity was normalized to that of a capped, polyadenylated, nonviral Renilla reporter construct.
A second approach to determine the translatability of sgRNA2 in vivo was to fuse ORF 6 to a luciferase ORF and measure luciferase expression in protoplasts. The 1,800-nt ORF 6-LUC fusion rendered the full-length transcript PAV6 noninfectious, so we examined translation of a transcript representing sgRNA2 containing the ORF 6-fLUC fusion (SG2fLUC) in cells directly. No luciferase activity was detected in cells transfected with the SG2fLUC transcript that resembled sgRNA2 by lacking a 5′ cap and a 3′ poly(A) tail (Fig. 7D, − cap, − poly A). Only the addition of a cap (+ cap, − poly A), or both a cap and a poly(A) tail (+ cap, + poly A), rendered SG2fLUC translatable in vivo (Fig. 7D), indicating that sgRNA2 does not translate cap independently in vivo. Taken together, these findings are consistent with a lack of a role for sgRNA2 as an mRNA.
DISCUSSION
BYDV sgRNA2 is a riboregulator that preferentially trans inhibits translation of gRNA versus sgRNA1 in vitro and in vivo.
Previously, we reported that sgRNA2 is a riboregulator of viral-gene expression and that the premature presence of sgRNA2 inhibits BYDV replication (50). Here, we demonstrate that its mechanism of action is via selective inhibition of the translation of BYDV genomic RNA. Reporter ORFs can replace viral ORFs in gRNA and sgRNA1, and both reporter RNAs respond the same as viral gRNA and sgRNA1, respectively, in the presence or absence of sgRNA2 in vitro (Fig. 2) (55). This leads to the noteworthy conclusion that neither BYDV coding sequences nor the protein products of the coding regions are necessary for the selective trans inhibition of translation. This allowed us to discover that BYDV sgRNA2 trans inhibits translation of gRNA but has little or no effect on the translation of sgRNA1 in vivo (Fig. 3 and 4). trans inhibition depends on the functional BTE in sgRNA2, as illustrated by the inability of sgRNA2BF, which differs from sgRNA2 by only a 4-base duplication in an essential region of the BTE, to trans inhibit translation (Fig. 2 to 6). In contrast, trans inhibition does not involve base pairing between the BTE of sgRNA2 and the 5′ UTR of the BYDV reporter RNAs (Fig. 5).
Interestingly, the difference in inhibition by sgRNA2 of translation of gRNA and sgRNA1 reporters was greater in vivo than in vitro. Moreover, these differences were augmented when GfLUC, SG1rLUC, and sgRNA2 were all present simultaneously in vitro and in vivo (compare Fig. 2B and C, Fig. 3C and D). This mixture most closely mimics natural infection and reveals a level of gene regulation in which BYDV RNAs are well coordinated.
A new mechanism of subgenomic-mRNA gene expression control.
The data strongly support the trans regulation model in Fig. 1. Early in BYDV infection, when sgRNAs are absent, only ORF 1 and ORF 2 (replicase genes) are translated via BTE-mediated, cap-independent translation to express the RNA-dependent RNA polymerase. This in turn replicates viral genomic RNA and generates sgRNA1 and particularly large amounts of sgRNA2 (Fig. 3B). The abundant sgRNA2 trans inhibits translation of gRNA only. Unlike in wheat germ extract, where sgRNA1 was inhibited somewhat but to a lesser extent than gRNA, sgRNA1 translation was inhibited only very slightly or not at all in vivo. Thus, the expected large quantity of coat protein can be translated from sgRNA1 as the viral life cycle enters a later stage. This model predicts that the presence of abundant sgRNA2 at the moment of inoculation will block virus replication by prematurely preventing translation of the polymerase, and indeed that was observed previously (50).
We now conclude that there are at least two levels of temporal control of viral-gene expression via subgenomic mRNAs. First and foremost is synthesis of the subgenomic RNAs. They are absent initially; thus, only ORFs 1 and 2 can be translated early in infection from gRNA. sgRNA1 synthesis positively controls the expression of ORFs 3, 4, and 5. The trans regulation of translation by sgRNA2 provides the second level of control. It acts negatively to turn off translation of gRNA, favoring translation of sgRNA1 only. Not only does this control the level of RNA-dependent RNA polymerase (RdRp) produced, we propose that this selective trans inhibition could also free the gRNA of ribosomes, making it available for replication and encapsidation.
Note that shutoff of gRNA translation by sgRNA2 is not absolutely required for RNA replication in protoplasts, as indicated by the accumulation (at 24 h p.i.) of viral RNAs in isolated protoplasts inoculated with mutant BYDV RNA (PAV6ΔSG2). Thus, sgRNA2 may serve as a fine-tuning device to maximize replication and/or its effects may be seen more clearly later in infection or in whole plants, where CP and movement proteins are necessary.
The proximity of the kissing stem-loop in the 5′ UTR to the 5′ end determines sensitivity to translation inhibition by sgRNA2.
The abilities of gRNA and sgRNA1 to be differentially inhibited by sgRNA2 are attributable to their different 5′ UTRs (Fig. 2C). Thus, neither the reporter gene nor the distance of the BTE from the 5′ UTR affected the efficiency of translation in the presence of sgRNA2. In contrast, the mutations that alter the location of the BCL relative to the 5′ end did change the response of the reporter RNA to sgRNA2 (Fig. 6).
The favored translation of sgRNA1 over gRNA in the presence of sgRNA2 may be explained by different structures of the 5′ UTRs per se or by differential requirement for a host factor(s). The positive correlation of proximity of the 5′ UTR kissing stem-loop to the 5′ end and competitiveness of the mRNA in the presence of sgRNA2 (Fig. 6) supports a simple scanning efficiency mechanism. Given evidence that BTE-mediated translation requires 5′-end-dependent ribosome scanning (21), a longer tract, with significant secondary structure, between the 5′ terminus and the BCL may require more translation factors to facilitate ribosome scanning (48). In the absence of sgRNA2, the factors may be in sufficient supply to allow efficient translation of gRNA. Only in the presence of competing sgRNA2 would the factors be reduced enough to hinder translation of gRNA, whereas RNA with the 5′-proximal BCL (sgRNA1) would have a lower factor dependence and hence be less inhibited by competing sgRNA2 (48).
Selective translational control by viral 5′ UTRs in cis has been observed in other viruses. For example, subgenomic RNA 4 of brome mosaic virus has a translational competitive advantage over the other three viral RNAs (46). The 5′ UTR of the coat protein-encoding sgRNA of turnip crinkle virus mediates translation more efficiently than the gRNA 5′ UTR (47). This is not surprising, because the coat protein is needed and expressed at orders of magnitude greater levels than the replication proteins translated from the gRNA. The 5′ UTRs of influenza virus mRNAs (18, 41) and the 5′ end of the capsid ORF of Sindbis virus subgenomic mRNA (17) mediate selective translation of viral mRNAs when translation of host mRNAs is shut off. In contrast to the above-mentioned RNAs, BYDV sgRNA translation is regulated in trans.
A role for viral proteins in selective translation of BYDV sgRNA1 in the presence of sgRNA2 has been ruled out, but host proteins may participate, as is the case for other viruses that use different mechanisms to control translation. The 5′ UTR of brome mosaic virus RNA 2 confers a specific requirement for the host translation factor DED1 in Saccharomyces cerevisiae(39). The cellular protein GRSF-1 participates in selective translation of influenza virus mRNAs (25, 42). With the exception of turnip crinkle virus, all of the above-mentioned translation regulation involves capped viral mRNAs. In contrast, the selective translation mediated by BYDV 5′ UTRs is between two uncapped mRNAs dependent on a 3′ cap-independent translation element and is mediated by a third viral RNA (sgRNA2).
Potential mechanism(s) of trans inhibition of translation of gRNA by sgRNA2.
Regulatory RNAs inhibit gene expression by at least two mechanisms. One mechanism is by base pairing of the regulatory RNA to the target RNA(s) to block translation or to recruit an inhibitory protein(s) to the target RNA(s). Examples include the microRNAs, small interfering RNAs, and bacterial small RNAs (8, 33, 38). This mechanism is unlikely for trans inhibition by BYDV sgRNA2 because the sequence within the BTE of sgRNA2 that is complementary to the 5′ UTR of the gRNA is not necessary for its trans inhibition activity (Fig. 5). Moreover, it stimulates translation in cis, and also it trans inhibits translation of nonviral mRNAs to which it has no sequence homology (54, 55).
The more likely mechanism is that BYDV sgRNA2 is a molecular decoy that competes for translation initiation factors. In support of this, addition of eukaryotic translation initiation factor 4F (eIF4F) restored translation of mRNA in extracts inhibited by the addition of BTE RNA (54). Indeed, recently eIF4F has been found to interact directly with the BTE and not with the nonfunctional mutant with the filled BamHI site (E. P. Kneller, K. Treder, E. Allen, and W. A. Miller, unpublished data). Thus, eIF4F binding correlates with the trans inhibition function. It is highly unlikely that translation of sgRNA2 is necessary for its function. Not only is no translation product detectable (Fig. 7), but mutant sgRNA2 containing a frameshift mutation that disrupts ORF 6 still selectively inhibited translation of gRNA in vitro (55).
Other trans-regulatory RNAs from viruses have very different functions. Red clover necrotic mosaic virus RNA2 has a 34-nt trans activator sequence, which is required for transcription of sgRNA from RNA1 (52) and for encapsidation (6). This trans activator base pairs to RNA1 to facilitate sgRNA synthesis. Flock house virus sgRNA3, but not its translation product, trans activates replication of viral genomic RNA2 (14, 15), which then down-regulates the synthesis of sgRNA3 from genomic RNA1 (57). Epstein-Barr virus EBER RNAs may function similarly to VA RNAs in blocking the host protein kinase RNA-activated antiviral response, because they can rescue replication of adenovirus lacking VA RNAs (7, 10). The herpesvirus microRNAs downregulate both viral and host gene expression at different stages of the viral infection (38, 44). Among the above-mentioned viral regulatory RNAs, VA RNAs, EBER RNAs, and most likely BYDV sgRNA2 are not mRNAs. Others function as both a coding RNA and a noncoding regulatory RNA.
It remains to be investigated whether other viruses employ this type of selective, negative regulation of translation. Competitive inhibition of translation among RNAs of other multi-RNA viruses has been observed primarily in vitro (30, 31, 46). The RNA elements that confer translational competitiveness of viral subgenomic RNAs have been mapped to both the 5′ (17, 31, 40) and 3′ (23) UTRs. However, (i) selective trans inhibition of translation of one viral RNA but not another, (ii) trans inhibition of cap-independent translation, and (iii) inhibition by an apparently untranslated RNA all are properties that so far are known only in BYDV infection. It will be particularly interesting to know whether other viruses that generate multiple subgenomic mRNAs, such as severe acute respiratory syndrome coronavirus and other members of the Nidovirales, or the Closteroviridae, are also controlled by a subgenomic RNA in trans. By extension, it is possible that a host mRNA with particularly high affinity for translation factors could negatively regulate translation of other host mRNAs by this mechanism.
Acknowledgments
This journal paper of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, was supported by NIH grant GM067104 and by Hatch Act and State of Iowa Funds. A.M.R. was funded by fellowships from the Fulbright Foundation and Pioneer Hi-Bred, Johnston, Iowa.
We thank Sang Ik Song for construction of pRenilla-CP393 and Sondra Schlesinger and Steve Whitham for critical reading of the manuscript and for providing insightful comments.
REFERENCES
- 1.Albarino, C. G., L. D. Eckerle, and L. A. Ball. 2003. The cis-acting replication signal at the 3′ end of Flock House virus RNA2 is RNA3-dependent. Virology 311:181-191. [DOI] [PubMed] [Google Scholar]
- 2.Allen, E., S. Wang, and W. A. Miller. 1999. Barley yellow dwarf virus RNA requires a cap-independent translation sequence because it lacks a 5′ cap. Virology 253:139-144. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia, S., and E. G. Wagner. 2000. Switching on and off with RNA. Proc. Natl. Acad. Sci. USA 97:9824-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry, J. K., and W. A. Miller. 2002. A programmed −1 ribosomal frameshift that requires base-pairing across four kilobases suggests a novel mechanism for controlling ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. USA 99:11133-11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 6.Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology 345:532-539. [DOI] [PubMed] [Google Scholar]
- 7.Bhat, R. A., and B. Thimmappaya. 1983. Two small RNAs encoded by Epstein-Barr virus can functionally substitute for the virus-associated RNAs in the lytic growth of adenovirus 5. Proc. Natl. Acad. Sci. USA 80:4789-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 9.Chaloub, B. A., L. Kelly, C. Robaglia, and H. D. Lapierre. 1994. Sequence variability in the genome-3′-terminal region for 10 geographically distinct PAV-like isolates of barley yellow dwarf virus: analysis of the ORF6 variation. Arch. Virol. 139:403-416. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, P. A., N. A. Sharp, J. R. Arrand, and M. J. Clemens. 1990. Epstein-Barr virus gene expression in interferon-treated cells—implications for the regulation of protein synthesis and the antiviral state. Biochim. Biophys. Acta 1050:167-173. [DOI] [PubMed] [Google Scholar]
- 11.Di, R., S. P. Dinesh-Kumar, and W. A. Miller. 1993. Translational frameshifting by barley yellow dwarf virus RNA (PAV serotype) in Escherichia coli and in eukaryotic cell-free extracts. Mol. Plant-Microbe Interact. 6:444-452. [DOI] [PubMed] [Google Scholar]
- 12.Dinesh-Kumar, S. P., and W. A. Miller. 1993. Control of start codon choice on a plant viral RNA encoding overlapping genes. Plant Cell 5:679-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domier, L. L., N. K. McCoppin, R. C. Larsen, and C. J. D'Arcy. 2002. Nucleotide sequence shows that Bean leafroll virus has a Luteovirus-like genome organization. J. Gen. Virol. 83:1791-1798. [DOI] [PubMed] [Google Scholar]
- 14.Eckerle, L. D., C. G. Albarino, and L. A. Ball. 2003. Flock House virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 317:95-108. [DOI] [PubMed] [Google Scholar]
- 15.Eckerle, L. D., and L. A. Ball. 2002. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 296:165-176. [DOI] [PubMed] [Google Scholar]
- 16.Erdmann, V. A., M. Z. Barciszewska, M. Szymanski, A. Hochberg, N. de Groot, and J. Barciszewski. 2001. The non-coding RNAs as riboregulators. Nucleic Acids Res. 29:189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolov, I., and S. Schlesinger. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfinkel, M. S., and M. G. Katze. 1993. Translational control by influenza virus—selective translation is mediated by sequences within the viral messenger RNA 5′-untranslated region. J. Biol. Chem. 268:22223-22226. [PubMed] [Google Scholar]
- 19.Gowda, S., M. A. Ayllon, T. Satyanarayana, M. Bar-Joseph, and W. O. Dawson. 2003. Transcription strategy in a closterovirus: a novel 5′-proximal controller element of citrus tristeza virus produces 5′- and 3′-terminal subgenomic RNAs and differs from 3′ open reading frame controller elements. J. Virol. 77:340-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grdzelishvili, V. Z., S. N. Chapman, W. O. Dawson, and D. J. Lewandowski. 2000. Mapping of the Tobacco mosaic virus movement protein and coat protein subgenomic RNA promoters in vivo. Virology 275:177-192. [DOI] [PubMed] [Google Scholar]
- 21.Guo, L., E. Allen, and W. A. Miller. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7:1103-1109. [DOI] [PubMed] [Google Scholar]
- 22.Guo, L., E. Allen, and W. A. Miller. 2000. Structure and function of a cap-independent translation element that functions in either the 3′ or the 5′ untranslated region. RNA 6:1808-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hann, L. E., A. C. Webb, J. M. Cai, and L. Gehrke. 1997. Identification of a competitive translation determinant in the 3′ untranslated region of alfalfa mosaic virus coat protein mRNA. Mol. Cell. Biol. 17:2005-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain, S., J. Pan, Y. Chen, Y. Yang, J. Xu, Y. Peng, Y. Wu, Z. Li, Y. Zhu, P. Tien, and D. Guo. 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 79:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kash, J. C., D. M. Cunningham, M. W. Smit, Y. Park, D. Fritz, J. Wilusz, and M. G. Katze. 2002. Selective translation of eukaryotic mRNAs: functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis. J. Virol. 76:10417-10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly, L., W. L. Gerlach, and P. M. Waterhouse. 1994. Characterisation of the subgenomic RNAs of an Australian isolate of barley yellow dwarf luteovirus. Virology 202:565-573. [DOI] [PubMed] [Google Scholar]
- 27.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 28.Koev, G., and W. A. Miller. 2000. A positive strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 74:5988-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 73:2876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak, M. 1986. Regulation of protein synthesis in virus-infected animal cells. Adv. Virus Res. 31:229-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon, C. S., and W. I. Chung. 2000. Differential roles of the 5′ untranslated regions of cucumber mosaic virus RNAs 1, 2, 3 and 4 in translational competition. Virus Res. 66:175-185. [DOI] [PubMed] [Google Scholar]
- 32.Lister, R. M., and R. Ranieri. 1995. Distribution and economic importance of barley yellow dwarf, p. 29-53. In C. J. D'Arcy and P. Burnett (ed.), Barley yellow dwarf: 40 years of progress. APS Press, St. Paul, Minn.
- 33.Majdalani, N., C. K. Vanderpool, and S. Gottesman. 2005. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 40:93-113. [DOI] [PubMed] [Google Scholar]
- 34.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268-32276. [DOI] [PubMed] [Google Scholar]
- 36.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Miller, W. A., S. Liu, and R. Beckett. 2002. Barley yellow dwarf virus: Luteoviridae or Tombusviridae? Mol. Plant Pathol. 3:177-183. [DOI] [PubMed] [Google Scholar]
- 38.Nair, V., and M. Zavolan. 2006. Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14:169-175. [DOI] [PubMed] [Google Scholar]
- 39.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pappas, C. L., W. P. Tzeng, and T. K. Frey. 2006. Evaluation of cis-acting elements in the rubella virus subgenomic RNA that play a role in its translation. Arch. Virol. 151:327-346. [DOI] [PubMed] [Google Scholar]
- 41.Park, Y. W., and M. G. Katze. 1995. Translational control by influenza virus—identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 270:28433-28439. [DOI] [PubMed] [Google Scholar]
- 42.Park, Y. W., J. Wilusz, and M. G. Katze. 1999. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA 96:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 44.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 45.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 46.Pyne, J. W., and T. C. Hall. 1979. Efficient ribosome binding of brome mosaic virus (BMV) RNA4 contributes to its ability to outcompete the other BMV RNAs for translation. Intervirology 11:23-29. [DOI] [PubMed] [Google Scholar]
- 47.Qu, F., and T. J. Morris. 2000. Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol. 74:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakotondrafara, A. M., C. Polacek, E. Harris, and W. A. Miller. Oscillating kissing stem-loop interactions mediate 5′ scanning-dependent translation by a viral 3′ cap-independent translation element. RNA, in press. [DOI] [PMC free article] [PubMed]
- 49.Rathjen, J. P., L. E. Karageorgos, N. Habili, P. M. Waterhouse, and R. H. Symons. 1994. Soybean dwarf luteovirus contains the third variant genome type in the luteovirus group. Virology 198:571-579. [DOI] [PubMed] [Google Scholar]
- 50.Shen, R., and W. A. Miller. 2004. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by Barley yellow dwarf virus subgenomic RNA 2. Virology 327:196-205. [DOI] [PubMed] [Google Scholar]
- 51.Shen, R., and W. A. Miller. 2004. The 3′ untranslated region of tobacco necrosis virus RNA contains a barley yellow dwarf virus-like cap-independent translation element. J. Virol. 78:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated transactivation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 53.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, S., K. S. Browning, and W. A. Miller. 1997. A viral sequence in the 3′-untranslated region mimics a 5′ cap in facilitating translation of uncapped mRNA. EMBO J. 16:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, S., L. Guo, E. Allen, and W. A. Miller. 1999. A potential mechanism for selective control of cap-independent translation by a viral RNA sequence in cis and in trans. RNA 5:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamore, P. D., and B. Haley. 2005. Ribo-gnome: the big world of small RNAs. Science 309:1519-1524. [DOI] [PubMed] [Google Scholar]
- 57.Zhong, W., and R. R. Rueckert. 1993. Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J. Virol. 67:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]