Abstract
Cytomegaloviruses carry the US22 family of genes, which have common sequence motifs but diverse functions. Only two of the 12 US22 family genes of murine cytomegalovirus (MCMV) are essential for virus replication, but their functions have remained unknown. In the present study, we deleted the essential US22 family genes, m142 and m143, from the MCMV genome and propagated the mutant viruses on complementing cells. The m142 and the m143 deletion mutants were both unable to replicate in noncomplementing cells at low and high multiplicities of infection. In cells infected with the deletion mutants, viral immediate-early and early proteins were expressed, but viral DNA replication and synthesis of the late-gene product glycoprotein B were inhibited, even though mRNAs of late genes were present. Global protein synthesis was impaired in these cells, which correlated with phosphorylation of the double-stranded RNA-dependent protein kinase R (PKR) and its target protein, the eukaryotic translation initiation factor 2α, suggesting that m142 and m143 are necessary to block the PKR-mediated shutdown of protein synthesis. Replication of the m142 and m143 knockout mutants was partially restored by expression of the human cytomegalovirus TRS1 gene, a known double-stranded-RNA-binding protein that inhibits PKR activation. These results indicate that m142 and m143 are both required for inhibition of the PKR-mediated host antiviral response.
Cytomegaloviruses (CMVs) are prototypes of the β subfamily of the Herpesviridae. Their genomes span 230 kb and are the largest among the herpesviruses. Of the approximately 170 genes on a CMV genome, about 46 are conserved among the herpesviruses (42). An additional 34 genes are characteristic of the β-herpesviruses (12, 21, 44, 58), and the remaining genes are unique to a particular virus.
A typical property of β-herpesvirus genomes is the presence of gene families, which probably arose by duplication from ancestral genes. One of the largest, the US22 gene family, was first identified in human cytomegalovirus (HCMV) and was later also found in other β-herpesviruses (12, 21, 44, 51, 58). It is characterized by four conserved sequence motifs consisting of hydrophobic residues interspersed with charged amino acids. HCMV and murine cytomegalovirus (MCMV) both include 12 members of the US22 gene family on their genomes (12, 51). The rat CMV and human herpesviruses 6 and 7, two other human β-herpesviruses, also possess up to 11 US22 family genes (20, 21, 44, 58).
Little is known about the functions of US22 gene products. However, 5 of the 12 US22 gene products of MCMV affect the virus' ability to replicate in macrophages: M36, M43, m139, m140, and m141 (28, 29, 32, 41). The molecular mechanism of action for only one of these has been elucidated. The M36 gene encodes an antiapoptotic protein that binds to procaspase 8 and inhibits death receptor-mediated induction of apoptosis (41). The positional and sequence homolog of M36 in HCMV, UL36, was originally proposed to function as a transcriptional transactivator (16). However, a later study showed that UL36 encodes a viral inhibitor of caspase 8 activation (vICA), a function it shares with its counterpart in MCMV (40, 41, 54).
A comprehensive mutational analysis of all 12 US22 gene family members of MCMV identified m142 and m143 as the only two genes of this family that are essential for virus replication in cell culture (41). These two genes are transcribed with immediate-early kinetics and lack motif II, one of the four conserved motifs (26, 27). The functions of these two proteins, however, remain unknown. The assumption that immediate-early proteins are likely to function as transcriptional transactivators—as has been shown for the HCMV TRS1 and IRS1 gene products (52, 55)—could not be confirmed for m142 and m143 (26).
In the present study, we investigated the functions of m142 and m143 during MCMV infection. Viral deletion mutants of these genes replicated only on complementing cells and were unable to express viral late proteins and to amplify viral DNA in normal fibroblasts. They failed to inhibit activation of the double-stranded RNA (dsRNA)-dependent protein kinase R (PKR) and shutdown of protein synthesis, suggesting that this antiviral response prevents replication of the deletion mutants. Consistent with our results, Child et al. report that m142 and m143 form a dsRNA-binding complex and that the two proteins can jointly rescue a vaccinia virus deficient for the dsRNA-binding protein E3L (13).
MATERIALS AND METHODS
Plasmids and genes.
The TRS1, m142, and m143 genes were amplified by PCR from the HCMV AD169 and the MCMV genomes, respectively, using the following primers: 5′-A AAG AAT TCC ACC ATG GCC CAG CGC AAC GGC ATG TCG-3′ (TRS1-fw), 5′-AAA CTC GAG TCA AGC GTA GTC TGG GAC GTC GTA TGG GTA TTG AGC ATT GTA ATG GTA GT-3′ (TRS1-rev), 5′-A AAG AAT TCC ACC ATG GAC GCC CTG TGC GCG GC-3′ (m142-fw), 5′-AAA CTC GAG TCA AGC GTA GTC TGG GAC GTC GTA TGG GTA GTC GTC ATC GTC GGC GTC CGC-3′ (m142-rev), 5′-AAA GGA TCC ACC ATG TCT TGG GTG ACC GGA GAT-3′ (m143-fw), and 5′-A AAG AAT TCA AGC GTA GTC TGG GAC GTC GTA TGG GTA CGC GTC GGT CGC TCT CTC GTC-3′ (m143-rev). Suitable restriction sites (in italics) for cloning in pcDNA3 (Invitrogen) and a hemagglutinin (HA) epitope sequence (underlined) at the 3′ end were introduced by the primers. It is noteworthy that the m143 gene sequence differed from the published sequence (51) by the presence of an additional guanosine residue between positions 201402 and 201403 of the MCMV sequence (GenBank accession number NC_004065). This leads to a frameshift compared to the published sequence analysis and a termination of the m143 open reading frame (ORF) at nucleotide position 200963 of the MCMV genome. The same difference was independently detected by others (13), suggesting that the published sequence is incorrect.
Retroviral transduction.
The genes of interest were excised from pcDNA3 and inserted into the retroviral vector plasmid pLXSN (TRS1 and m142) or pLXRN (m143). Production of Moloney murine leukemia virus-based retroviral vectors using the Phoenix packaging cell line and transduction of NIH 3T3 cells was done as in a previous study (8). Transduced cells were selected with 700 μg/ml G418 and grown as bulk cultures without clonal selection.
Mutagenesis.
All mutant viruses were constructed on the basis of an MCMV variant expressing the green fluorescent protein (MCMV-GFP), which has been used in previous studies (7, 8, 31). The MCMV-GFP genome, cloned as a bacterial artificial chromosome (BAC), was modified in the Escherichia coli strain DY380 as described previously (8, 9). For the construction of the deletion mutants, a zeocin resistance gene (zeo) was PCR amplified with primers that contained 50-nucleotide (nt) sequences homologous to positions 199621 to 199670 and 200749 to 200798 (for Δm142) or 201121 to 201170 and 202594 to 202643 (for Δm143). For construction of the m142 revertant genome, the plasmid pBSo142 was constructed, which is based on pBluescriptII KS(+) (Stratagene) and contains a synthetic oligonucleotide encoding 50-nt sequences up- and downstream of the m142 ORF, spaced by EcoRI, EcoRV, XhoI, and HpaI restriction sites. The HA-tagged m142 gene and a kanamycin resistance gene (kan) were inserted into pBSo142 using the EcoRI/XhoI and HpaI sites, respectively. The entire cassette was excised with SacI and ApaI and used for repair of the m142 deletion. The kan gene (which was flanked by FRT sites) was subsequently removed using FLP recognition target (FLP) recombinase as described previously (8). The m143 revertant genome was constructed by the same strategy with plasmid pBSo143, containing the 50-nt homologies up- and downstream of the m143 ORF. To replace m142 and m143 with TRS1, we inserted the TRS1 gene into pBSo142 and pBSo143, respectively, as had been done for the revertants. Alternatively, TRS1 was also inserted at an ectopic position, replacing the nonessential ORFs m02 to m06. This was done using the pReplacer plasmid as described previously (31). Wild-type and recombinant MCMV BAC DNAs were isolated from E. coli using NucleoBond PC100 columns (Macherey-Nagel). The viral genomes were digested with suitable restriction endonucleases and separated on 0.6% agarose gels. The DNA was subsequently transferred to a nylon membrane using a TurboBlotter device (Schleicher & Schuell). Hybridization with a digoxigenin-labeled probe directed against the m142 and m143 genes and chemiluminescent detection were performed using a DIG-High Prime DNA-labeling and detection kit (Roche) according to the manufacturer's recommendations.
Cells and viruses.
MCMV was propagated on NIH 3T3 cells (ATCC CRL-1658) according to standard procedures (6). To reconstitute recombinant viruses from mutant genomes, BAC DNA was transfected into normal or complementing NIH 3T3 cells. Virus stocks were produced on the same cells and titered using the 50% tissue culture infectious dose (TCID50) method (39). For growth kinetics, cells were seeded in six-well plates and infected with MCMV at the indicated MOI. Two hours after infection, the cells were washed with phosphate-buffered saline, and fresh medium was added. The medium was replaced at the indicated time points, and the virus content in the supernatant was determined by titration. All growth kinetics experiments were done in triplicate. To analyze viral-DNA replication, total DNA was extracted from infected NIH 3T3 cells, blotted onto a nylon membrane, and detected with a digoxigenin-labeled probe against the MCMV M45 gene as described above.
Protein detection.
For Western blot analysis, cells were lysed in RIPA buffer containing 20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 1% Na deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, and a protease inhibitor cocktail (Roche). Denatured protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Amersham). For immunological detection, the following antibodies were used: CROMA101 against MCMV IE1 and CROMA103 against E1 (both provided by Stipan Jonjic, University of Rijeka, Rijeka, Croatia), and 2E8.21A against the MCMV envelope glycoprotein B (gB) and 3B9.22A against M44 (both provided by Lambert Loh, University of Saskatchewan, Saskatchewan, Canada). Monoclonal antibodies against the HA epitope tag (16B12 [Covance Research Products] and 3F10 [Roche]), β-actin (A5316; Sigma), and murine PKR (B-10; Santa Cruz) and polyclonal rabbit antibodies against phosphorylated and total murine eIF2α (the α subunit of eukaryotic translation initiation factor 2; Cell Signaling) were purchased from suppliers as indicated. Horseradish peroxidase-coupled secondary antibodies (Dako and Cell Signaling) and enhanced chemiluminescence reagents (Amersham) were used to develop the blots. For immunofluorescence, cells were grown on coverslips, fixed with 3% paraformaldehyde, and permeabilized with 0.3% Triton X-100. Proteins were detected using an anti-HA primary antibody and an Alexa Fluor 488- or Alexa Fluor 594-coupled secondary antibody (Molecular Probes). Nuclei were stained with propidium iodide or 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI). Fluorescent images were generated with a Zeiss LSM 510 confocal microscope.
For metabolic labeling, NIH 3T3 cells were infected at a multiplicity of infection (MOI) of 5 TCID50/cell. Twenty-four hours after infection, the cells were incubated for 1 h in medium containing [35S]methionine and [35S]cysteine (143 μCi total radioactivity). Protein lysates of labeled cells were separated by polyacrylamide gel electrophoresis. The protein concentration for each lysate was determined using a Bicinchoninic Acid Protein Assay kit (Pierce), and equal amounts of total protein were loaded onto each lane. The gel was fixed, dried, and exposed to an X-ray film (Kodak).
Real-time reverse transcription (RT)-PCR.
Total RNA was isolated with Trizol reagent (Invitrogen), purified with an RNeasy kit (QIAGEN), and subsequently digested with DNAse I to remove contaminating DNA. The quality of the RNA preparations was confirmed by denaturing agarose gel electrophoresis. First-strand cDNA synthesis was performed with Superscript III reverse transcriptase (Invitrogen) using 125 ng random-hexamer-primed total RNA. The reverse-transcribed total RNA was digested with RNAse H. For each sample, a background control without the addition of reverse transcriptase was made.
Real-time PCR analysis was performed using a Lightcycler 1 instrument (Roche). Each reaction mixture was made with the SYBR Green FastStart Kit (Roche) in 15 μl total volume containing 2.5 mM MgCl2, 7.5 pmol of each primer, and 1 ng reverse-transcribed total RNA. As a housekeeping gene control, 18S rRNA was chosen, because previous experiments had shown that its level of expression is not significantly affected by MCMV infection. Each run was performed with 45 cycles, followed by a melting-curve analysis to confirm the identities of the PCR products. For the detection of viral- and housekeeping gene transcripts, the following primers were used: 5′-TTA TGG TTC CTT TGG TCG CTC G-3′ (18s-fwd), 5′-CAC CGG GTT GGT TTT GAT CTG A-3′ (18s-rev), 5′-GCG ATG TCC GAG TGT GTC AAG-3′ (gB-fwd), 5′-CGA CCA GCG GTC TCG AAT AAC-3′ (gB-rev), 5′-TCG TTC GTG AAC ATC GTG GTG-3′ (gM-fwd), 5′-GAT CGC GTT GTA CAT CGT CAG G-3′ (gM-rev), 5′-TGC ACC AGG CGC TCT GTA AC-3′ (M44-fwd), and 5′-CGC TGA GGA AGT TCT CGA TGG-3′ (M44-rev). Relative quantification was carried out according to the method of Pfaffl (48).
RESULTS
Construction of MCMV m142 and m143 deletion mutants.
To analyze the functions of m142 and m143 in the context of an MCMV infection, we constructed deletion mutants of these genes. MCMV-GFP was used as a basis for the construction of mutant viruses, because GFP expression by infected cells facilitates the monitoring of potentially growth-defective viruses.
A BAC clone of MCMV-GFP was modified by homologous recombination in E. coli. The m142 and m143 ORFs were replaced individually with a zeocin resistance gene (Fig. 1A). In a second step, revertant genomes were constructed by replacing the zeocin resistance gene of the deletion mutants with an HA epitope-tagged m142 or m143 gene, respectively (Fig. 1A). A kanamycin resistance gene flanked by FRT sites was inserted downstream of the respective gene to serve as a selectable marker for homologous recombination. This cassette was subsequently removed by FLP recombinase, leaving only a single FRT site behind. The restriction patterns of these genomes showed the expected changes (Fig. 1B). Transfection of the wild-type and the revertant genomes into NIH 3T3 fibroblasts yielded replication-competent virus, but the Δm142 and Δm143 deletion mutants did not grow. Individual green fluorescent cells were obtained after transfection of the genomes, but these cells disappeared after repeated passaging of the cells.
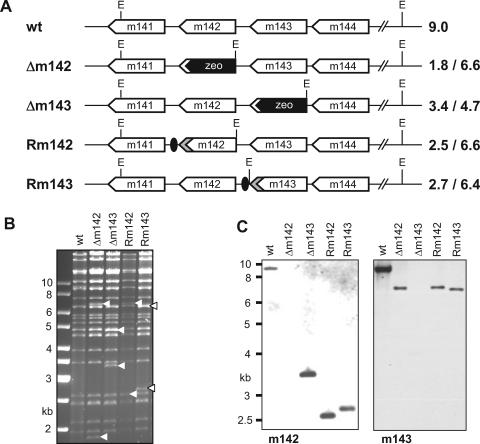
FIG. 1.
Construction of MCMV deletion mutants. (A) The m142 and m143 open reading frames were replaced by a bacterial zeocin resistance gene (zeo), using recombinant BAC technology. To obtain the revertants, Rm142 and Rm143, the HA-tagged open reading frames, together with an FRT-flanked kanamycin resistance gene, were inserted into the MCMVΔm142 and Δm143 genomes, respectively. The kan gene was subsequently removed using FLP recombinase, leaving a single FRT site (black oval) behind. EcoRI (E) restriction sites and the expected fragment sizes are indicated. wt, wild type. (B) EcoRI-digested MCMV BACs were separated on an ethidium bromide-stained agarose gel. The arrowheads indicate altered fragments. (C) Southern blot of EcoRI-digested viral DNA from MCMV-infected cells hybridized with m142- and m143-specific probes, respectively.
To overcome this problem, NIH 3T3 cells were transduced with retroviral vectors carrying m142 or m143, respectively, and a G418 resistance gene for selection of the transduced cells. As shown in Fig. 2A and B, the cells expressed the HA-tagged proteins. They could be detected by Western blotting and immunofluorescence. When these complementing cells were transfected with the genomes of the MCMV deletion mutants, the mutant viruses could be reconstituted. To confirm the identities of the recombinant viruses, viral DNA was extracted from virus preparations and analyzed by Southern blot hybridization. All mutants showed the expected band patterns (Fig. 1C). Moreover, the revertant viruses expressed HA-tagged m142 or m143 proteins, respectively (Fig. 2C and D).
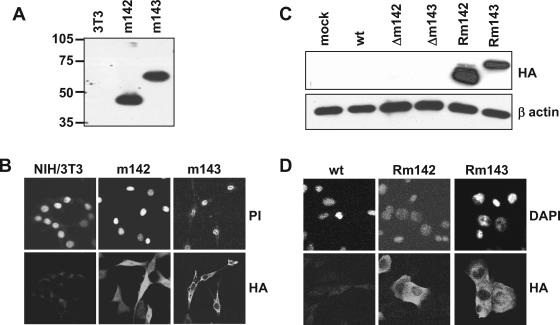
FIG. 2.
Expression of m142 and m143 in transduced and infected cells. (A) Western blot and (B) indirect immunofluorescence analysis of NIH 3T3 cells stably transduced with retroviruses encoding HA-tagged m142 and m143 proteins, respectively. (C) Detection of HA-tagged m142 and m143 proteins expressed by the revertant MCMVs Rm142 and Rm143, respectively, by Western blotting and (D) immunofluorescent staining of infected NIH 3T3 cells. Nuclei were stained with propidium iodide (PI) or DAPI. mock, mock infected.
Growth properties of the Δm142 and Δm143 deletion mutants.
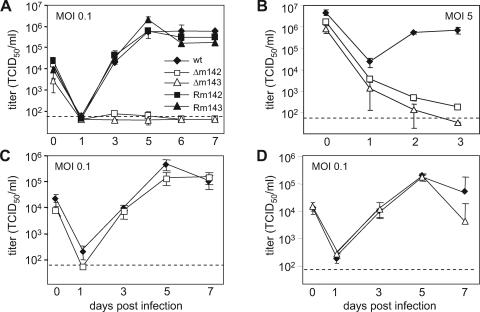
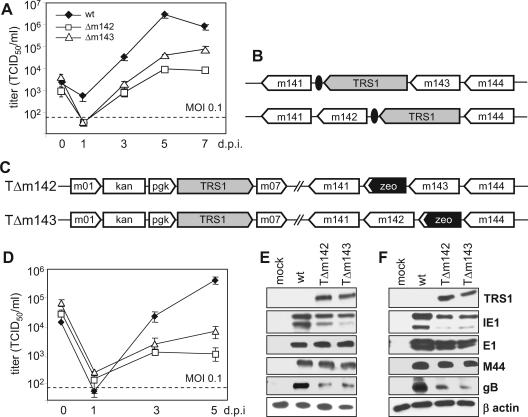
It is conceivable that transfection of an MCMV genome into fibroblasts provides less favorable conditions for initiation of the viral replication cycle than an infection. Hence, the failure to regenerate a virus from a mutant BAC, as shown in a previous study (41), provided only indirect evidence for essential roles of the m142 and m143 genes. Moreover, it is possible that a gene deletion prevents virus replication during an infection at a low MOI but that this defect can be overcome by infecting cells at a high MOI. This has been shown, for instance, for the HCMV IE1 gene, which is required for efficient growth at a low MOI but is dispensable for replication at a high MOI (22, 43). Therefore, we infected murine fibroblasts at low and high MOIs and analyzed virus replication. The growth curves in Fig. 3A and B show that the Δm142 and Δm143 viruses did not grow on noncomplementing fibroblasts, either at a low or at a high MOI. However, the viruses grew to almost normal titers on complementing cells (Fig. 3C and D). This indicated that each of the two genes is essential for virus growth in cell culture at both low and high MOIs.
FIG. 3.
Growth kinetics of recombinant MCMVs. Replication of the Δm142 and Δm143 deletion mutants and revertants in NIH 3T3 cells was analyzed upon infection at (A) a low or (B) a high MOI. (C) Replication of the Δm142 mutant in m142-expressing NIH 3T3 cells and (D) of the Δm143 mutant in m143-expressing cells. The error bars represent the standard errors.
m142 and m143 are required for late-protein synthesis and DNA replication.
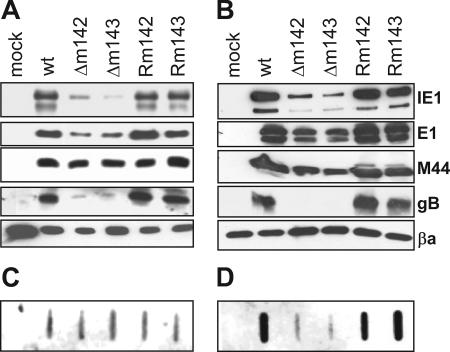
To identify a block in the cascade of viral-gene expression as a potential reason for the inability of the deletion mutants to replicate, we analyzed the expression of immediate-early (α), early (β1), early-late (β2), and late (γ) MCMV proteins by Western blotting. To exclude the possibility that gene expression by the mutant viruses is delayed, we analyzed the expression of these proteins not only at 24, but also at 72 h postinfection (p.i.). Although expression of the α gene IE1 (34) was reduced in Δm142- and Δm143-infected cells, the levels of immediate-early proteins were apparently sufficient to activate expression of the β1 gene E1 (10) and the β2 gene M44 (38), as their levels were not dramatically altered compared to the wild type and the revertants. A major difference was seen in the levels of the late protein gB (37, 50), which was barely detectable in cells infected with the deletion mutants (Fig. 4A and B). Moreover, viral DNA was present in comparable amounts in these cells at 24 h p.i., but an amplification to high levels at very late times (72 h p.i.) apparently did not occur (Fig. 4C and D).
FIG. 4.
Viral-gene expression and DNA replication. NIH 3T3 cells were infected with recombinant MCMVs at an MOI of 0.5 TCID50/cell. Viral-gene products were detected with specific antibodies 24 (A) and 72 (B) h p.i. Detection of viral-DNA amplification in infected cells by slot blotting and hybridization with an MCMV-specific probe 24 (C) and 72 (D) h p.i. βa, beta actin; wt, wild type; mock, mock infected.
m142 and m143 are required to prevent PKR activation and shutdown of protein synthesis.
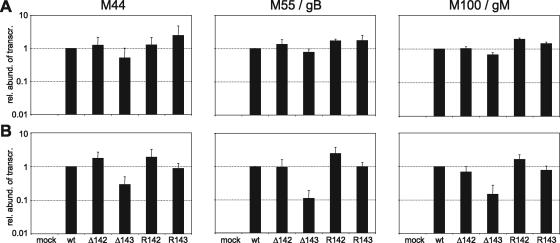
Earlier studies identified a transcriptional transactivating function of the HCMV US22 gene family proteins TRS1, IRS1, and UL36 in assays using transient transfection of expression and reporter plasmids (16, 52, 55). Although m142 and m143 failed to show similar transactivating activities in a more recent study (26), we wanted to test whether m142 and m143 are required for late-gene transcription. The mRNA levels of the early-late gene M44 and the late genes M55/gB and M100/gM (36) were measured by real-time PCR. At 24 h p.i., the mRNA levels of these genes were not significantly altered in the deletion mutants compared to wild-type or revertant viruses (Fig. 5A). Only at very late times (72 h p.i.) were the mRNA levels reduced about 10-fold in cells infected with Δm143 (Fig. 5B). This suggested that the defect in late-protein expression is based primarily on a posttranscriptional block. Indeed, when global protein synthesis was analyzed by metabolic labeling with [35S]methionine and [35S]cysteine, it was found to be markedly reduced in cells infected with the Δm142 or Δm143 mutant (Fig. 6A).
FIG. 5.
Real-time RT-PCR quantification of MCMV late-gene transcripts. NIH 3T3 cells were infected at an MOI of 1 TCID50/cell with wild-type (wt) and mutant MCMV. Total RNA was harvested (A) 24 and (B) 72 h p.i. and quantified by real-time RT-PCR. Two RNA preparations from cells infected with different virus stocks were made, and from each preparation, two to four independent quantitative PCRs per sample were done. The transcript abundance in MCMV wt-infected cells was defined as 1. The abundances of transcripts from mutant viruses are shown relative to the wt. The error bars represent the standard errors. mock, mock infected.
FIG. 6.
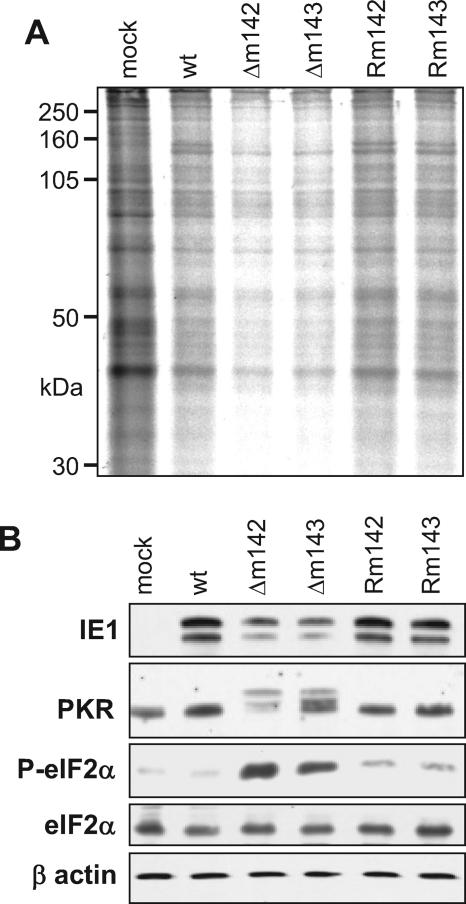
Shutdown of protein synthesis in Δm142- and Δm143-infected cells. (A) NIH 3T3 cells were metabolically labeled with [35S]methionine and [35S]cysteine 24 h p.i. at an MOI of 5 TCID50/cell. Cell lysates (10 μg total protein per sample) were separated by gel electrophoresis and analyzed by autoradiography. (B) Detection of phosphorylated PKR and eIF2α (P-eIF2α) in Δm142- and Δm143-infected cells by Western blotting. Total eIF2α and β-actin were used as controls. mock, mock infected; wt, wild type.
Global shutdown of protein synthesis is a well-known innate defense mechanism by which infected cells can inhibit the production of progeny virus (47, 53). This process usually involves activation of the double-stranded-RNA-dependent PKR and PKR-mediated phosphorylation of eIF2α, an essential cofactor for translation of mRNAs into polypeptides. When PKR and eIF2α were analyzed in infected cells by Western blotting, a more slowly migrating form of PKR was detected only in cells infected by the deletion mutants, but not in cells infected with the wild-type or the revertant virus (Fig. 6B). The more slowly migrating band most probably represents phosphorylated (i.e., activated) PKR, because treatment of NIH 3T3 cells with poly(I · C), a known inducer of PKR phosphorylation, resulted in a comparable band shift (data not shown). Unfortunately, an antibody specifically recognizing phosphorylated murine PKR was not available, and thus, the identity of the more slowly migrating band could not be formally proven. However, phosphorylation of the PKR target protein eIF2α was clearly detected in these cells (Fig. 6B), suggesting that the shutdown of protein synthesis is indeed mediated by PKR-dependent phosphorylation of eIF2α.
HCMV TRS1 can restore the growth of Δm142 and Δm143.
The TRS1 and IRS1 proteins of HCMV have recently been shown to rescue a vaccinia virus (VV) lacking the dsRNA-binding protein E3L (14, 15). Similarly, TRS1 and IRS1 were able to reverse the PKR-mediated protein synthesis shutoff induced by a recombinant herpes simplex virus type 1 (HSV-1) lacking the γ134.5 gene (11). In addition, TRS1 was shown to possess dsRNA-binding activity (24). These results suggested that TRS1 and IRS1 are responsible for inhibiting a shutdown of protein synthesis during an HCMV infection, although this has not yet been formally demonstrated.
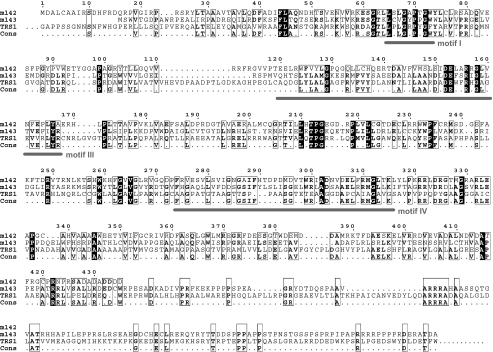
TRS1 and IRS1 share a number of properties with m142 and m143 of MCMV. They are expressed at immediate-early times, are members of the US22 gene family, and lack one of the four sequence motifs characteristic of this gene family (27). However, the sequence similarity of TRS1 to m142 and m143 is noticeable, but not striking. The m143 protein has 18% identical and 30% similar amino acids to both m142 and TRS1. The m142 protein shares only 11% identity and 21% similarity with TRS1 (Fig. 7). Hence, we asked if TRS1 could rescue the growth defect of the MCMV Δm142 and/or Δm143 virus. To investigate this, we infected NIH 3T3 cells stably transduced with a TRS1-expressing retrovirus and studied the growth of the Δm142 and Δm143 deletion mutants. Both mutants grew on TRS1-expressing fibroblasts (Fig. 8A), albeit to 10- to 100-fold-lower titers than the wild-type virus. We also inserted the TRS1 gene into the genomes of the Δm142 and Δm143 mutants in order to test if TRS1 expression in cis would also rescue virus growth. Surprisingly, insertion of TRS1 in place of the deleted m142 and m143 genes (Fig. 8B) did not rescue virus growth (data not shown). However, when TRS1 was driven by a phosphoglycerate kinase gene (pgk) promoter and inserted at an ectopic position, replacing the nonessential genes m02 to m06 (Fig. 8C), virus growth on NIH 3T3 fibroblasts and expression of the late protein gB were restored (Fig. 8D to F). These results indicated that TRS1 can, at least in part, compensate for a lack of m142 or m143. Since TRS1 can block the PKR-dependent antiviral response during an HSV-1 or VV infection (11, 14), the results also support the concept that m142 and m143 are both required to prevent PKR-mediated shutdown of protein synthesis.
FIG. 7.
Alignment of the m142, m143, and TRS1 proteins. Amino acid residues conserved between the three proteins are indicated by black boxes; similar residues are marked with open boxes. The alignment was generated with Multalign software (18). The TRS1 sequence was truncated at both ends, and only the overlapping part is shown. US22 gene family motifs I, III, and IV are marked. Periods represent gaps in gene sequences and nonconserved residues in the consensus sequence.
FIG. 8.
TRS1 rescues m142 and m143 deletion mutants. (A) Multistep growth kinetics of the wild type (wt) and deletion mutants on NIH 3T3 cells stably transduced with a TRS1-expressing retroviral vector. (B) Introduction of the HA-tagged TRS1 gene into the MCMV genome, either by replacing the deleted gene or (C) by insertion at a distant position together with a pgk promoter. (D) Multistep growth curves of the TΔm142 and TΔm143 mutants on NIH 3T3 fibroblasts. (E) Viral-protein expression 24 and (F) 72 h p.i. at an MOI of 0.5 TCID50/cell. The error bars represent the standard errors.
DISCUSSION
Replication of intracellular pathogens, such as viruses, can be inhibited by innate immune defenses of the host cell. One of the best characterized is the interferon-mediated antiviral response, which proceeds in three phases (reviewed in reference 25). First, the cell senses the presence of a virus via toll-like receptors (TLRs) that recognize pathogen-associated molecular patterns (33). TLR signaling triggers the expression and secretion of beta interferon (IFN-β). Binding of IFN-β to type I interferon receptors on the same and on neighboring cells activates the Jak/STAT signaling pathway, which leads to an upregulation of antiviral effector molecules, such as PKR, 2′-5′-oligoadenylate synthetase, and Mx proteins. Viruses, on the other hand, have evolved various mechanisms to counteract the IFN-mediated antiviral response on different levels (reviewed in reference 25).
CMVs already trigger the activation of the IFN response during attachment and entry into the cell. Contact of gB with its receptor activates the IFN-responsive pathway via interferon regulatory factor 3 (3, 4). CMV also activates inflammatory-cytokine production by engaging the pattern recognition receptors TLR2, TLR9, and CD14 (17, 35). However, HCMV counteracts the induction of IFN-β expression via its tegument protein pp65/UL83 (1, 5) and/or via a combined action of the neighboring gene product, pp71/UL82, and the IE2 protein, as suggested by later studies (56, 57). IFN receptor signaling is also inhibited by both HCMV and MCMV. The HCMV IE1 protein forms a complex with the signal transducer and activator of transcription 1 (STAT1) and STAT2 and inhibits the induction of IFN-responsive effector genes (46). In MCMV, the M27 gene product inhibits both type I and type II IFN signaling by selectively down-regulating STAT2 without affecting STAT1 (60).
Like many other viruses, HCMV also encodes proteins that bind dsRNA and inhibit the dsRNA-dependent activation of PKR. This function is fulfilled by the TRS1 and IRS1 proteins of HCMV (11, 14, 24). These two proteins are identical over the N-terminal two-thirds of their amino acid sequences, because they are located in part in the short repeat region of the HCMV genome (12). Each of the two proteins can rescue a VV E3L or an HSV-1 γ134.5 deletion mutant, suggesting at least partly redundant functions. Unfortunately, the importance of these two proteins for the life cycle of HCMV could not yet be analyzed, because a TRS1-IRS1 double-knockout mutant could not be grown in normal fibroblasts, and the generation of reliable complementing cells proved to be difficult (W. Brune, J. E. Adamo, and T. Shenk, Abstr. 27th Int. Herpesvirus Workshop, abstr. 11.13, 2002, and W. Brune, unpublished results).
The present study shows that m142 and m143 are both required to inhibit PKR activation and a shutdown of protein synthesis at late times postinfection. The inability of the Δm142 and Δm143 mutants to prevent PKR activation is by itself not sufficient to prove that the two proteins are directly involved in blocking PKR activation. However, the fact that TRS1, a known inhibitor of PKR activation, can rescue the Δm142 and Δm143 deletion mutants suggests a direct role of m142 and m143 in this process. Hence, this is the first study that demonstrates the importance of the PKR-mediated antiviral response during a cytomegalovirus infection.
The MCMV m142 and m143 proteins show significant sequence similarity to their presumed counterparts in HCMV, TRS1 and IRS1 (Fig. 7). They differ, however, from TRS1 and IRS1 in that each of them is essential for MCMV replication. The HCMV proteins, by contrast, are together essential only for HCMV replication (Brune et al., Abstr. 27th Int. Herpesvirus Workshop), consistent with redundant functions of the two proteins. These findings suggest either that the m142 and m143 proteins block different steps toward PKR activation or that they form an inhibitory complex. The latter possibility is supported by a report which shows that m142 and m143 can bind dsRNA and rescue a VV deficient for the dsRNA-binding protein E3L only when expressed together (13).
Inhibition of the dsRNA-dependent antiviral response is obviously a crucial task for RNA viruses, as their replication inevitably results in the generation of dsRNA molecules. However, a recent study showed that DNA viruses, such as herpesviruses, vaccinia virus, or adenoviruses, also produce dsRNA (59). One possible explanation is that DNA viruses carry genes on both strands of their genomes. This can lead to the formation of dsRNA by hybridization of (partly) complementary mRNAs transcribed from opposing strands. Another possibility is that these viruses need to prevent PKR activation through viral microRNAs. Recent studies have shown that HCMV, like many other herpesviruses, have such microRNAs (19, 23, 49).
The essential roles of m142, m143, and TRS1/IRS1 may not rely exclusively on their abilities to block the PKR-mediated antiviral response. Many herpesvirus proteins are multifunctional, and this may also hold true for the proteins studied here. For instance, previous studies have demonstrated that TRS1 or IRS1 is required for transient complementation of HCMV DNA replication (30, 45). This is consistent with our observation that MCMV DNA is not replicated to high levels in the absence of m142 or m143 (Fig. 4). However, it is possible that MCMV DNA replication is not blocked completely but only inhibited at late times postinfection as an indirect consequence of the antiviral response. In fact, the detection of late-gene transcripts argues for at least limited viral-DNA replication, as γ-gene transcription should occur only after DNA replication.
A recent study has revealed that TRS1 (but not IRS1) is required for efficient assembly of DNA-containing capsids (2), and this function is apparently not secondary to a reduced transcription or translation of viral genes. The molecular mechanism for this additional function of TRS1 has not yet been determined, nor is it known whether m142, m143, or any other MCMV protein serves a similar function.
Considering the multiple functions of the HCMV TRS1 protein, it may appear surprising that TRS1 rescued gB expression and replication in the Δm142 and Δm143 mutants only partly. Insertion of the TRS1 gene into the genomes of the deletion mutants restored virus replication only when TRS1 expression was driven by an extraneous pgk promoter. Since the endogenous promoters of m142 and m143 are known to be weak during the initial phase of infection (26), artificial overexpression of TRS1 appears to be required to compensate for the lack of m142 or m143. This could indicate that these MCMV and HCMV proteins are not fully capable of blocking the antiviral response in cells of a heterologous host species.
The genes of the US22 family probably arose from a common ancestral gene but have diverged in their specific functions. However, it is remarkable that all of the US22 family genes for which a function has been determined are involved in the subversion of innate or adaptive immune functions. At least in some cases, the functions were conserved between the human and the murine cytomegaloviruses: UL36 and M36 inhibit tumor necrosis factor alpha- and Fas-mediated signaling by blocking caspase 8 activation (41, 54), and TRS1/IRS1 and m142/m143 inhibit the IFN-dependent antiviral response by preventing PKR activation (references 11, 13, 14, and 24 and this study). Thus, it should not be surprising if other US22 family genes, whose functions are as yet unknown, are also engaged in immune evasion.
Acknowledgments
We thank Katharina Wolf for valuable help with confocal microscopy and Stipan Jonjic and Lambert Loh for providing antibodies. We also thank Adam Geballe for communicating unpublished results and Michael Nevels for critical reading of the manuscript.
This work was supported by the Emmy Noether Program of the DFG (Br 1730/2-3), SFB 421, and the Georg und Agnes Blumenthal Stiftung.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamo, J. E., J. Schroer, and T. Shenk. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 78:10221-10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune, W., H. Hengel, and U. H. Koszinowski. 1999. A mouse model for cytomegalovirus infection, p. 19.7.1-19.7.13. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 7.Brune, W., C. Ménard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 8.Brune, W., M. Nevels, and T. Shenk. 2003. Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein. J. Virol. 77:11633-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brune, W., M. Wagner, and M. Messerle. 2006. Manipulating cytomegalovirus genomes by BAC mutagenesis: strategies and applications, p. 61-89. In M. J. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Norfolk, United Kingdom.
- 10.Bühler, B., G. M. Keil, F. Weiland, and U. H. Koszinowski. 1990. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J. Virol. 64:1907-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassady, K. A. 2005. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J. Virol. 79:8707-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 13.Child, S. J., L. K. Hanson, C. E. Brown, D. M. Janzen, and A. P. Geballe. 2006. Double-stranded RNA binding by a heterodimeric complex of murine cytomegalovirus m142 and m143 proteins. J. Virol. 80:10173-10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Child, S. J., S. Jarrahian, V. M. Harper, and A. P. Geballe. 2002. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J. Virol. 76:4912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn, W., P. Trang, Q. Zhong, E. Yang, C. van Belle, and F. Liu. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7:1684-1695. [DOI] [PubMed] [Google Scholar]
- 20.Efstathiou, S., G. L. Lawrence, C. M. Brown, and B. G. Barrell. 1992. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J. Gen. Virol. 73:1661-1671. [DOI] [PubMed] [Google Scholar]
- 21.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 22.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakki, M., and A. P. Geballe. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 79:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson, L. K., B. L. Dalton, L. F. Cageao, R. E. Brock, J. S. Slater, J. A. Kerry, and A. E. Campbell. 2005. Characterization and regulation of essential murine cytomegalovirus genes m142 and m143. Virology 334:166-177. [DOI] [PubMed] [Google Scholar]
- 27.Hanson, L. K., B. L. Dalton, Z. Karabekian, H. E. Farrell, W. D. Rawlinson, R. M. Stenberg, and A. E. Campbell. 1999. Transcriptional analysis of the murine cytomegalovirus HindIII-I region: identification of a novel immediate-early gene region. Virology 260:156-164. [DOI] [PubMed] [Google Scholar]
- 28.Hanson, L. K., J. S. Slater, Z. Karabekian, G. Ciocco-Schmitt, and A. E. Campbell. 2001. Products of US22 genes M140 and M141 confer efficient replication of murine cytomegalovirus in macrophages and spleen. J. Virol. 75:6292-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. T. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurak, I., and W. Brune. 2006. Inhibition of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 25:2634-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karabekian, Z., L. K. Hanson, J. S. Slater, N. K. Krishna, L. L. Bolin, J. A. Kerry, and A. E. Campbell. 2005. Complex formation among murine cytomegalovirus US22 proteins encoded by genes M139, M140, and M141. J. Virol. 79:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13:816-825. [DOI] [PubMed] [Google Scholar]
- 34.Keil, G. M., A. Ebeling-Keil, and U. H. Koszinowski. 1987. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J. Virol. 61:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krug, A., A. R. French, W. Barchet, J. A. Fischer, A. Dzionek, J. T. Pingel, M. M. Orihuela, S. Akira, W. M. Yokoyama, and M. Colonna. 2004. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 21:107-119. [DOI] [PubMed] [Google Scholar]
- 36.Li, W., K. Eidman, R. C. Gehrz, and B. Kari. 1995. Identification and molecular characterization of the murine cytomegalovirus homolog of the human cytomegalovirus UL100 gene. Virus Res. 36:163-175. [DOI] [PubMed] [Google Scholar]
- 37.Loh, L. C. 1991. Synthesis and processing of the major envelope glycoprotein of murine cytomegalovirus. Virology 180:239-250. [DOI] [PubMed] [Google Scholar]
- 38.Loh, L. C., N. Balachandran, and W. J. Britt. 1991. Characterization of a membrane-associated phosphoprotein of murine cytomegalovirus (pp50) and its immunological cross-reactivity with a human cytomegalovirus protein. Virology 183:181-194. [DOI] [PubMed] [Google Scholar]
- 39.Mahy, B. W. J., and H. O. Kangro. 1996. Virology methods manual. Academic Press, San Diego, Calif.
- 40.McCormick, A. L., A. Skaletskaya, P. A. Barry, E. S. Mocarski, and V. S. Goldmacher. 2003. Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virology 316:221-233. [DOI] [PubMed] [Google Scholar]
- 41.Ménard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. Campbell, and U. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members for replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 43.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholas, J. 1996. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J. Virol. 70:5975-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. USA 103:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pe'ery, T., and M. B. Mathews. 2000. Viral translational strategies and host defense mechanisms, p. 371-424. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Woodbury, N.Y.
- 48.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 50.Rapp, M., M. Messerle, B. Buhler, M. Tannheimer, G. M. Keil, and U. H. Koszinowski. 1992. Identification of the murine cytomegalovirus glycoprotein B gene and its expression by recombinant vaccinia virus. J. Virol. 66:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 54.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, R. T., and W. A. Bresnahan. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 79:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vink, C., E. Beuken, and C. A. Bruggeman. 2000. Complete DNA sequence of the rat cytomegalovirus genome. J. Virol. 74:7656-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmermann, A., M. Trilling, M. Wagner, M. Wilborn, I. Bubic, S. Jonjic, U. Koszinowski, and H. Hengel. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-γ signaling and antiviral responses. J. Exp. Med. 201:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]