Abstract
Human cytomegalovirus (HCMV) infection results in dysregulation of several cell cycle genes, including inhibition of cyclin A transcription. In this work, we examine the effect of the HCMV infection on expression of the high-mobility group A2 (HMGA2) gene, which encodes an architectural transcription factor that is involved in cyclin A promoter activation. We find that expression of HMGA2 RNA is repressed in infected cells. To determine whether repression of HMGA2 is directly related to the inhibition of cyclin A expression and impacts on the progression of the infection, we constructed an HCMV recombinant that expressed HMGA2. In cells infected with the recombinant virus, cyclin A mRNA and protein are induced, and there is a significant delay in viral early gene expression and DNA replication. To determine the mechanism of HMGA2 repression, we used recombinant viruses that expressed either no IE1 72-kDa protein (CR208) or greatly reduced levels of IE2 86-kDa (IE2 86) protein (IE2 86ΔSX-EGFP). At a high multiplicity of infection, the IE1 deletion mutant is comparable to the wild type with respect to inhibition of HMGA2. In contrast, the IE2 86ΔSX-EGFP mutant does not significantly repress HMGA2 expression, suggesting that IE2 86 is involved in the regulation of this gene. Cyclin A expression is also induced in cells infected with this mutant virus. Since HMGA2 is important for cell proliferation and differentiation, particularly during embryogenesis, it is possible that the repression of HMGA2 expression during fetal development could contribute to the specific birth defects in HCMV-infected neonates.
Human cytomegalovirus (HCMV), a betaherpesvirus, is the major viral cause of birth defects and poses a severe threat to immunodeficient individuals (42a). Infection of permissive cells results in the rapid activation of the immediate-early (IE) genes, whose major products include the IE1 72-kDa (IE1 72; UL123) and IE2 86-kDa (IE2 86; UL122) proteins. A subset of the IE proteins induce the expression of viral early genes. Late genes encode primarily structural proteins and are expressed following viral DNA replication (for a review, see reference 35).
The major IE (MIE) gene is controlled by a strong promoter/enhancer and consists of five exons that are differentially spliced to produce the IE1 72-kDa protein (exons 1 to 4) and the IE2 86-kDa protein (exons 1 to 3 and 5) (for a review, see reference 35). Both of the mRNAs initiate translation in exon 2, and the two proteins share 85 amino acids (aa) at their amino termini; the first 23 aa are encoded by exon 2 and the remaining 62 aa by exon 3. Numerous mutagenesis studies have revealed functional differences between these two proteins. IE1 72 can transactivate the MIE promoter, while IE2 86 is a strong transactivator of both viral and cellular promoters but can repress the MIE promoter (for a review, see reference 35). IE2 86 is essential for growth of the virus (34, 65). In contrast, IE1 72 is dispensable at a high multiplicity of infection (MOI) (23, 24, 36).
HCMV infection alters the expression of many cellular proteins and blocks cell cycle progression in primary human fibroblasts (7, 11, 18, 30, 31, 47, 49, 68, 69). p53 and the retinoblastoma protein (Rb) accumulate in HCMV-infected cells (22, 30, 38). HCMV also induces activation of proto-oncogenes c-jun, c-myc, and c-fos, as well as increased expression of thymidine kinase, DNA polymerase alpha, ornithine decarboxylase, and dihydrofolate reductase (8, 19, 26, 29, 64). While cyclin E and cyclin B protein levels and their associated kinase activities are increased, cyclin A mRNA and protein synthesis is inhibited in HCMV-infected cells (30, 47).
Recently, it has been shown that the architectural transcription factor HMGA2 (high-mobility group AT-hook 2) regulates the transcription of cyclin A (63). HMGA2 is a nuclear protein which, together with HMGA1a and HMGA1b, belongs to the high-mobility group (HMG) family of nonhistone chromatin proteins (for a review, see reference 54). These proteins contain about 100 amino acid residues and possess three copies of a 9-amino-acid motif, the AT-hook, that interacts with the minor groove of AT-rich DNA sequences. HMGA proteins are referred to as architectural transcription factors because of their ability to organize the assembly of nucleoprotein structures (enhanceosomes), resulting in enhancement or repression of transcription. In the case of HMGA2 regulation of cyclin A, HMGA2 was found to form a complex with the transcriptional repressor p120E4F in vitro and in vivo (63). This association interfered with the ability of p120E4F to bind to the cyclic AMP response element in the cyclin A promoter, a site also bound by the transcription factor CREB. HMGA2 also binds to the cyclin A promoter, but only when the gene is transcriptionally activated.
Based on our prior work showing that HCMV infection inhibits the transcription of cyclin A, we were interested in determining whether the mechanism of repression involves HMGA2. In this report, we show that the expression of HMGA2 is specifically repressed during the infection. To determine whether repression of HMGA2 was important for the HCMV infection, we constructed a recombinant virus expressing HMGA2 driven by the MIE promoter. We find that high-multiplicity infection with the HMGA2-expressing virus induces the synthesis of cyclin A mRNA and protein and inhibits virus replication. To further study the mechanism of HMGA2 repression during HCMV infection, we used the HCMV recombinant virus IE2 86ΔSX-EGFP, which has a deletion of aa 136 to 290 in IE2 86 and expresses low levels of the IE2 86 protein (48), and the HCMV recombinant virus CR208, which lacks exon 4 of the MIE gene and thus produces no IE1 72 protein (24, 36). We show that the IE1 deletion mutant virus still inhibits HMGA2 transcription. In contrast, in cells infected with the IE2 86ΔSX-EGFP mutant virus, HMGA2 expression is not significantly affected, suggesting that IE2 is involved in the regulation of the HMGA2 promoter. Cyclin A transcription is also induced, although this effect is slightly delayed relative to HMGA2 expression.
MATERIALS AND METHODS
Cell culture and virus.
Human foreskin fibroblasts (HFF) were obtained from the University of California—San Diego Medical Center and cultured in Earle's minimal essential medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 1.5 μg/ml amphotericin B (Invitrogen), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen). Cells were kept in incubators maintained at 37°C and 7% CO2. The Towne strain of HCMV was obtained from the American Type Culture Collection (VR 977) and propagated as previously described (60). The HCMV recombinant virus that has a deletion of exon 4 of the IE1 72 gene (CR208) and its rescued virus (CR249) were provided by Edward Mocarski (Emory University). IE2 86-EGFP, IE2 86ΔSX-EGFP, and rescued IE2 86ΔSX-EGFP viruses have been previously described (48).
Cell synchronization and infections.
HFF (passage numbers 15 to 20) were synchronized in G0 phase by allowing them to grow to confluence as previously described (47). Three days after growth was contact inhibited, the cells were trypsinized, replated at a lower density to allow progression into the cell cycle, and infected at an MOI of 3 to 5 with HCMV or mock infected with tissue culture supernatants. At various times postinfection (p.i.), cells were washed with phosphate-buffered saline, scraped or trypsinized, and processed as described above.
Bacterial artificial chromosome (BAC) mutagenesis.
A 784-bp fragment containing the Tn7 miniattachment (mini-att) site within the Escherichia coli lacZ alpha fragment was inserted between the US11 and US12 genes of pHB5BAC (10) by homologous recombination mediated by the RecE and RecT recombinases (ET recombination) using the Counter Selection BAC Modification kit (Gene Bridges, Dresden, Germany). The HMGA2 gene, under the control of the HCMV IE promoter, was subsequently inserted between US11 and US12 by Tn7 transposition (25).
To construct a template for ET recombination, a 406-bp AvrII-SalI fragment from the HCMV AD169 genomic fragment EcoRI B (60) was subcloned into the vector pSP72 (Promega). This fragment contained a portion of US11 and US12, as well as the region between the two genes. The resulting plasmid was called pSP:US11-US12. Prior to the first step of ET recombination, the vector pSP:US11-RpsL-Neo-US12 was made by subcloning the RpsL-Neo marker cassette into a SpeI site between US11 and US12 in pSP:US11-US12. This vector was used as the template to amplify the RpsL-Neo marker cassette with arms with 100-bp homology to the region upstream and downstream of the site of insertion. The primers used to amplify the 1.4-kb fragment were 5′-GGATAATTCAGGCATACTACCC-3′ and 5′-TCTGTCGTACTGTCCTTTGTCC-3′. The fragment was electroporated into E. coli DH10B bacteria that had been previously transformed with pHB5BAC and pSC101-BAD-gbaAtet (the expression vector for the RecE/RecT and Redα/Redβ recombinases under the control of an arabinose-inducible and temperature-sensitive promoter from the Gene Bridges kit) and selected with chloramphenicol, tetracycline, and kanamycin. Kanamycin-resistant colonies were screened for the presence of the RpsL-Neo cassette between US11 and US12 in the HB5 BAC by restriction enzyme analysis. The BAC recombinant was referred to as HB5:US11-RpsL-Neo-US12 BAC.
To construct a template for the second step of ET recombination, a 784-bp PmeI fragment containing lacZ/att was excised from the plasmid pMA116 (25) and cloned into an XbaI site located between US11 and US12 to form pSP:US11-lacZ/att-US12. A 1,015-bp fragment containing lacZ/att flanked by arms with 100-bp homology to the US11 and US12 region was amplified from the plasmid using the primers described above. This fragment was then electroporated into DH10B cells harboring HB5:US11-RpsL-Neo-US12 BAC and pSC101-BAD-gbaAtet. Blue colonies were selected on chloramphenicol and streptomycin plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). Recombinant HB5-lacZ/att BACs were confirmed by restriction enzyme analysis and DNA sequencing.
To prepare the plasmid pFB1:IE-HMGA2 used for the Tn7 transposition, a 631-bp BamHI fragment containing the HCMV IE promoter was subcloned into the BamHI site of the vector pFastBac-1 (Invitrogen) to form pFB1:IE. The plasmid pcDNA-HMGA2 (a kind gift from Guidalberto Manfioletti, Trieste, Italy) was digested with HindIII, and the ends of the linearized plasmid were filled in using Klenow enzyme. The fragment was digested with XhoI and subsequently ligated into the StuI-XhoI sites of pFB1:IE, downstream of the promoter, to form pFB1:IE-HMGA2.
To perform the Tn7 transposition, DH10B bacteria containing the HB5-lacZ/att BAC were transformed with pMON7124 (25), which encodes the trans-acting factors needed for transposition. These cells were then transformed with pFB1:IE-HMGA2, and transformants were selected on chloramphenicol, tetracycline, and gentamicin plates which also contained Bluo-Gal (Invitrogen) and IPTG. White colonies, indicative of a transposition event, were screened for the insertion of the IE-HMGA2 cassette by PCR. Positive clones were confirmed by DNA sequencing and detailed restriction enzyme analysis using a Field Inversion Gel Electrophoresis mapper (Bio-Rad). The resulting BAC was named HB5:IE-HMGA2 BAC.
Reconstitution of the HB5:IE-HMGA2 BAC virus.
HFF were electroporated with 6.25 μg of HB5:IE-HMGA2 BAC plus 3.25 μg of pcDNA-pp71tag (a gift from Bodo Plachter, Mainz, Germany). Electroporation was carried out using the BTX ECM-600 electroporator (Genetronics, Inc.) as previously described (37). After electroporation, the cells were seeded into either 6-cm dishes for detection of plaque development or a 12-well plate containing sterile coverslips for immunofluorescence assays. The HMGA2-expressing virus was collected from the cell supernatant at day 14 postelectroporation, after the infection had spread throughout the entire monolayer of cells. This virus was checked by sequencing of the altered region of the viral DNA to confirm that it had not reverted to the wild type (wt). Stocks of HB5-lacZ/att and HB5:IE-HMGA2 viruses were prepared, and their titers were determined by plaque assay as previously described (60).
Preparation of rescued HB5:IE-HMGA2 virus.
A rescued HB5:IE-HMGA2 BAC was made by ET recombination as described above. Briefly, the IE-HMGA2 cassette in the HB5:IE-HMGA2 BAC was replaced with an RpsL-Neo marker cassette to make HB5:US11-RpsL-Neo-US12 BAC. To construct the original HB5-lacZ/att BAC, the RpsL-Neo marker cassette was replaced by a US11-lacZ/att-US12 fragment. After blue colony selection, the identity of the recombinant HB5-lacZ/att BAC was confirmed by restriction enzyme analysis and DNA sequencing. Infectious virus was reconstituted by electroporation of 6.25 μg of rescued HB5:IE-HMGA2 BAC into HFF along with 3.25 μg of pcDNA-pp71tag. Viral stock was prepared from the electroporated cells, and the viral DNA was sequenced in the altered region to verify that the rescued HB5:IE-HMGA2 BAC corresponded to the original BAC.
Quantitative real-time PCR and RT-PCR analysis.
DNA was isolated from the cells infected with HB5-lacZ/att, HB5:IE-HMGA2, and rescued HB5:IE-HMGA2 viruses using a blood minikit (QIAGEN, Valencia, CA). The concentration of each sample was determined by UV spectrophotometry. Quantitative real-time PCR and data analysis were performed as previously described using primers and probes directed against the HCMV UL77 gene (65, 66). The DNA isolated at 1 day p.i. from wild-type virus-infected cells was used to generate a standard curve, allowing comparison of the amount of DNA present in each sample to the amount present in wild-type virus-infected cells at 1 day p.i. Quantitative real-time reverse transcription-PCR (RT-PCR) and data analysis were performed as previously described (65, 66). All reactions were performed using an Applied Biosystems ABI Prism 7700 sequence detection system with the TaqMan One-Step reverse transcription-PCR master mix reagent kit (Applied Biosystems) and oligonucleotide primers and TaqMan dual-labeled (5′-6-carboxyfluorescein-3′ Black Hole quencher) probes (Integrated DNA Technologies, Coralville, Iowa) (Table 1).
TABLE 1.
Quantitative real-time PCR primers and TaqMan probes
| Transcript | Sequence
|
||
|---|---|---|---|
| Forward primer | Reverse primer | TaqMan probe | |
| HMGA2 | AGTCCCTCTAAAGCAGCTCAAAAG | GCCATTTCCTAGGTCTGCCTC | AGAAGCCACTGGAGAAAAACG |
| HMGA1 | ACCAAAGGGAAGCAAAAACAAG | CTTGGTTTCCTTCCTGGAGTTG | CTGCCAAGACCCGGAAAACCACC |
| Cyclin A | CGGTACTGAAGTCCGGGAAC | AACGGTGACATGCTCATCATTT | AGACGAGACGGGTTGCACCCCTT |
| G6PD | TCTACCGCATCGACCACTACC | GCGATGTTGTCCCGGTTC | ATGGTGCTGAGATTTGCCAAACAGGA |
All samples were analyzed with specific primers and probe for the cellular housekeeping gene glucose-6-phosphate dehydrogenase (G6PD) as an internal control for the amount of RNA in each reaction. The RNA isolated at 24 h p.i. from mock-infected cells was used to generate a standard curve for each cellular gene examined. The standard curve was then used to calculate the relative amount of specific RNA in each sample.
Western blotting.
HFF were infected with HB5-lacZ/att, HB5:IE-HMGA2, and rescued HB5:IE-HMGA2 viruses at an MOI of 5 in medium containing 10% fetal bovine serum. Cells were harvested at various time points, lysed in Laemmli reducing sample buffer (50 mM Tris, pH 6.8, 2% sodium dodecyl sulfate [SDS], 10% glycerol, 5% 2-mercaptoethanol, 50 mM leupeptin, 100 mM pepstatin A, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate, 5 mM β-glycerophosphate), and sonicated briefly to shear the DNA. Proteins from an equivalent number of cells were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Membranes were stained with amido black to assess protein loading in each lane. The blots were blocked with 5% nonfat dried milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) and incubated with primary antibodies diluted in blocking solution as follows: monoclonal antibody (MAb) CH16.0 (1:10,000; Goodwin Institute), anti-UL44 MAb (1:5,000; Goodwin Institute), anti-UL57 MAb (1:1,000; Goodwin Institute), anti-pp65 MAb (1:10,000; Goodwin Institute), anti-pp28 MAb (1:6,000; Goodwin Institute), anti-cyclin A sc-751 (1:500; Santa Cruz Biotechnology), β-actin MAb (1:10,000; Sigma-Aldrich), and anti-HMGA2 (1:1,000; a gift from Guidalberto Manfioletti, Trieste, Italy). The β-actin MAb was used as a control for protein loading. Blots were incubated with horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin antibodies, diluted 1:2000 in blocking solution (Calbiochem, San Diego, CA). After the filter was washed in TBST, the proteins were detected using SuperSignal chemiluminescent substrate (Pierce, Rockford, IL) according to the manufacturer's instructions.
Northern blot analysis.
HFF were infected at an MOI of 5 in medium containing 10% fetal bovine serum with IE2 86-EGFP, IE2 86ΔSX-EGFP, or rescued IE2 86ΔSX-EGFP virus and then harvested at either 24 or 96 h p.i. Approximately 1.5 × 107 cells per sample were collected, and mRNA was isolated using the FastTrack 2.0 kit (Invitrogen). Northern blot assays were carried out using the NorthernMax kit according to the manufacturer's protocol (Ambion). One microgram of mRNA was loaded per lane, resolved by agarose gel electrophoresis on a 1% formaldehyde gel, and then transferred to a nylon membrane. Membranes were UV cross-linked, and 32P-labeled probes were synthesized by random priming using the StripEZ DNA kit (Ambion). The HMGA2 probe was generated by PCR using sense primer 5′ ATGAGCGCAAGAGGTGAGG 3′ and antisense primer 5′ GCCATTTCCTAGGTCTGCCTC 3′ (Integrated DNA Technologies, Coralville, Iowa) to amplify a linear fragment that is present in all HMGA2 spliced RNAs. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was synthesized using a 320-bp SacI-XbaI fragment cut from the pTRI-GAPDH plasmid provided in the NorthernMax kit (Ambion). Membranes were hybridized overnight at 42°C and then washed twice in low-stringency wash buffer at 25°C and twice in high-stringency wash buffer at 42°C. Membranes were then exposed to film for autoradiography.
RESULTS
HMGA2 expression is repressed in HCMV-infected cells.
Work in our laboratory and in others has shown that HCMV infection inhibits cell cycle progression and alters the expression of cyclins E, A, and B (7, 11, 18, 30, 31, 47, 49, 68, 69). Since HMGA2 is involved in the regulation of cyclin A transcription, we were interested in whether it might also be involved in the inhibition of cyclin A RNA synthesis during the infection. HFF were synchronized in G0 and subsequently infected with HCMV at an MOI of 5 upon release into G1. Since the HMGA2 antibodies that are available can detect only overexpressed proteins, we assessed the expression of the endogenous HMGA2 by analyzing the levels of its mRNA. Total RNA was isolated from cells at 8, 24, 48, and 72 h p.i., and the levels of the HMGA2 transcript were determined by quantitative real-time RT-PCR. We compared the transcript level in each sample to the amount of HMGA2 RNA in the lysate from 24-h mock-infected cells and calculated the relative difference. Additionally, each sample was normalized with RNA encoding G6PD. The real-time RT-PCR results showed that there was a significant decrease in HMGA2 RNA expression in the HCMV-infected cells relative to the mock-infected cells. The difference was fourfold at 8 h p.i. and approximately 20-fold by 24 h p.i. (Fig. 1A).
FIG. 1.
HMGA2 RNA levels are reduced in infected cells. G0-synchronized cells were released into G1 and infected with HCMV Towne at an MOI of 5 or mock infected. Cells were harvested at 8, 24, 48, and 72 h p.i. Total RNA was isolated and analyzed by quantitative real-time RT-PCR to measure levels of transcripts for HMGA2 (A) and HMGA1 (B). RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. For each time point, two separate experiments were performed and duplicate reactions were analyzed. The mock-infected RNA that was isolated at 24 h p.i. was used to generate a standard curve for each gene. This standard curve was used to calculate the amount of RNA present in each sample. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values.
HMGA1 and HMGA2 proteins both belong to the HMG family and are similar in structure and function (for a review, see reference 54). However, their expression is controlled by different promoters. Therefore, for comparison, we assessed the level of HMGA1 RNA by real-time RT-PCR. In contrast to what was observed for HMGA2 RNA, the level of HMGA1 RNA was reduced less than twofold at 8 and 24 h p.i. and only two- to threefold at later times (Fig. 1B). Thus, the infection has a significantly greater effect on HMGA2 than on HMGA1 RNA expression.
Construction of recombinant HCMVs expressing HMGA2.
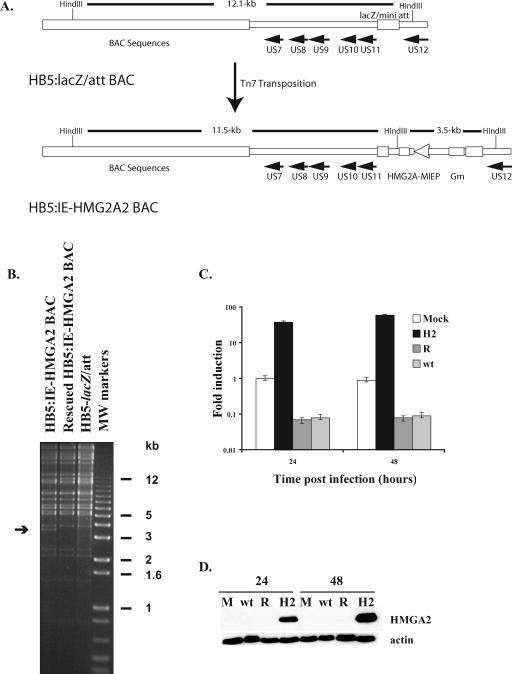
In order to determine whether the inhibition of HMGA2 expression was important for viral replication, we constructed a recombinant virus that expressed HMGA2 (HB5:IE-HMGA2) using the pHB5BAC and Tn7-mediated transposition as described in Materials and Methods. The parental BAC that had the lacZ alpha fragment inserted between the US11 and US12 genes served as a wt control (HB5:lacZ/att). The cDNA encoding HMGA2 was placed under the control of the HCMV MIE promoter in the vector pFastBac-1 (Invitrogen) and inserted between the US11 and US12 genes in the HCMV 12.1-kb HindIII fragment from HB5:lacZ/att (Fig. 2A). We also generated a rescued BAC to ensure that any changes observed in viral replication were not due to mutations that had occurred in other regions of the HCMV genome. The resulting BACs were isolated, and the altered regions were examined for the presence of the correct sequences by DNA sequencing. In addition, the BACs were digested with HindIII restriction endonuclease and subjected to gel electrophoresis to confirm that no large-scale rearrangements of the BAC DNA had occurred during the cloning process (Fig. 2B).
FIG. 2.
Construction and characterization of the HB5:IE-HMGA2 BAC. (A) The DNA fragment containing the cDNA encoding HMGA2 was inserted into the HB5-lacZ/att BAC using Tn7 transposition. Gm, gene for gentamicin resistance. (B) The BAC DNAs for HB5-lacZ/att, HB5:IE-HMGA2, and rescued HB5:IE-HMGA2 were digested with HindIII, and the fragments were separated by agarose gel electrophoresis. The arrow shows the position of the 3.5-kbp fragment in the HB5:IE-HMGA2 BAC DNA that contains the HMGA2 cDNA under the control of the MIE promoter. (C and D) G0-synchronized HFF were released into G1 and mock infected or infected with HB5-lacZ/att (wt), HB5:IE-HMGA2 (H2), or rescued HB5:IE-HMGA2 (R) viruses at an MOI of 5. Total RNA or protein was isolated at 24 and 48 h p.i. and analyzed by quantitative real-time RT-PCR to measure the level of HMGA2 RNA or by Western blotting to assess the amount of HMGA2 protein (C and D, respectively). RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. For each time point, two separate experiments were performed and duplicate reactions were analyzed. The mock-infected RNA that was isolated at 24 h p.i. was used to generate a standard curve for each gene. This standard curve was used to calculate the amount of RNA present in each sample. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values. The y axis of the graph is a log scale. (D) Protein samples from an equivalent number of cells were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with the antibody that detects the HMGA2 protein. Actin was used as a control for equal protein loading. M, mock.
To produce recombinant viruses, we electroporated the BACs into HFF along with a pp71 expression vector and observed the cultures for the development of cytopathic effect. Small plaques could be detected at 6 days postelectroporation in cultures that received any one of the BACs. However, at 10 days postelectroporation we observed a significant delay in the spread of the HB5:IE-HMGA2 virus compared to the HB5:lacZ/att and the rescued viruses. When the entire culture was infected, the viruses were harvested and amplified using fresh HFF.
Viral replication is delayed in cells infected at a high MOI with the HMGA2-expressing virus.
G0-synchronized HFF were infected with HMGA2, HB5-lacZ/att, and rescued viruses at an MOI of 5, and the cells were harvested 24, 48, and 72 h p.i. We first confirmed by quantitative real-time RT-PCR that in the HMGA2-expressing virus-infected cells, there was a high level of HMGA2 RNA that did not decrease as the infection progressed. As shown in Fig. 2C, the amount of mRNA in the cells infected with the HMGA2-expressing virus (H2) was significantly greater than that in the cells infected with the wt and rescued (R) viruses at both 24 and 48 h p.i. The overexpressed HMGA2 protein in the HMGA2-expressing virus-infected cells could also be detected by Western blot analysis with an antibody to HMGA2 (Fig. 2D).
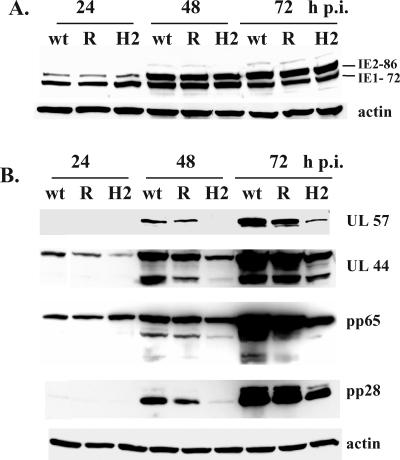
To determine the effect of HMGA2 expression in the infected cells, the infected cell lysates were analyzed by Western blotting with antibodies to specific viral proteins. In cells infected with the HMGA2-expressing virus, the expression of the IE2 86 and IE1 72 proteins at 24 h p.i. was comparable to that observed in the cells infected with the HB5-lacZ/att and rescued viruses (Fig. 3A). However, the levels of other viral proteins showed that the progression of the HCMV infection was significantly delayed by HMGA2 expression. Figure 3B shows that there was a lag in the accumulation of the early-late proteins UL57, UL44, and pp65 and the late protein pp28. Since a decrease in the UL57 and UL44 levels indicated that the HMGA2 expression might have an effect on viral DNA replication, we isolated DNA from the same cultures that were used for Western blot analysis and measured the level of viral DNA at the various time points using quantitative real-time PCR with specific primers and probe to the HCMV UL77 gene as previously described (65, 66). As shown in Fig. 4, significantly less viral DNA was synthesized in the cells infected with the HMGA2-expressing virus than in the cells infected with the wt and rescued viruses. At 48 h p.i., the difference in the amount of viral DNA was 10-fold. Although the level of viral DNA in the HMGA2-expressing virus-infected cells did increase between 48 and 72 h p.i., it was still fivefold lower. These results showed that HMGA2 expression also had a negative effect on viral DNA replication.
FIG. 3.
Viral gene expression is delayed in HFF infected with HMGA2-expressing virus. G0-synchronized cells were released into G1 and infected with HB5-lacZ/att (wt), HB5:IE-HMGA2 (H2), and rescued HB5:IE-HMGA2 (R) viruses at an MOI of 5. Total cell lysates were prepared at 24, 48, and 72 h p.i., and protein samples from an equivalent number of cells were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with the MAb CH16.0, which detects the HCMV IE1 and IE2 proteins (A) or MAbs that detect the UL57, UL44, pp65, and pp28 viral proteins (B). Actin was used as a control for equal protein loading.
FIG. 4.
Viral DNA replication is delayed in cells infected with HMGA2-expressing virus. G0-synchronized cells were released into G1 and infected with HB5-lacZ/att (wt), HB5:IE-HMGA2 (H2), and rescued HB5:IE-HMGA2 (R) viruses at an MOI of 5. DNA was isolated from cells at 24, 48, and 72 h p.i., and the level of viral DNA was measured by quantitative real-time PCR using primers and probe to the HCMV UL77 gene as described in Materials and Methods. DNA levels were normalized to G6PD levels as an internal control for the amount of DNA in each sample. For each time point, two separate experiments were performed and duplicate reactions were analyzed. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values. One of these experiments was the same as that shown in Fig. 3.
HMGA2 induces cyclin A expression in HCMV-infected cells.
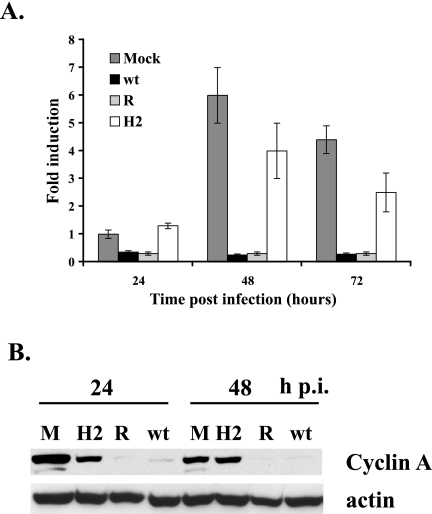
We previously reported that cyclin A expression was inhibited at both the mRNA and the protein level in HCMV-infected cells, but the mechanism of the downregulation was unknown (30, 47). Since HMGA2 plays a role in cyclin A transcription (63), we proceeded to determine if HMGA2 might also be involved in the dysregulation of cyclin A during the infection. HFF that were synchronized in G0 and released into G1 were mock infected or infected at an MOI of 5 with the HMGA2-expressing, wt, and rescued viruses. Cells were harvested at 24, 48, and 72 h p.i., and the levels of cyclin A mRNA were determined by quantitative real-time RT-PCR (Fig. 5A). At all time points, the levels of cyclin A mRNA in the cells infected with the HMGA2-expressing virus were significantly higher than those in the cells infected with the wt and rescued viruses and were comparable to the levels observed in the mock-infected cells. In both the mock-infected and HMGA2-expressing virus-infected cells, the amount of cyclin A mRNA at 48 h p.i. was higher than that at 24 h p.i., which is due to the steady increase in cyclin A mRNA that occurs when cells are released from G0. Lysates obtained at 24 and 48 h p.i. were also analyzed by Western blotting for the level of cyclin A protein (Fig. 5B). The increase in cyclin A mRNA in the mock-infected cells and HMGA2-expressing virus-infected cells was accompanied by a corresponding increase in cyclin A protein expression. The slightly lower level of cyclin A protein in the mock-infected sample at 48 h p.i. than at 24 h p.i. was consistent with the movement of the cells through the cell cycle. These results indicated that there was a strong correlation between HMGA2 expression and cyclin A mRNA and protein levels during the HCMV infection.
FIG. 5.
An HMGA2-expressing virus induces cyclin A expression following high-MOI infection. (A) RNA that was isolated at 24, 48, and 72 h p.i. from the same two infections as shown in Fig. 4 was analyzed by quantitative real-time RT-PCR using primers and probe for the cyclin A transcript. RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. The mock-infected RNA that was isolated at 24 h p.i. was used to generate a standard curve for each gene. This standard curve was used to calculate the amount of RNA present in each sample. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values. (B) The lysates from the cells harvested at 24 and 48 h p.i. in one of these duplicate experiments (the same experiment as that shown in Fig. 3) were analyzed by Western blotting with an antibody to cyclin A. Actin served as a control for protein loading in each lane, and the lysates in the lanes were from an equal number of cells. M, mock infection.
IE2 86 expression is associated with the inhibition of HMGA2 and cyclin A transcription.
HCMV infection leads to the activation and inhibition of a large number of host cell genes, including many genes that are involved in cell cycle control, immunomodulation, and apoptosis (12, 14, 73). Several studies have also suggested that IE2 86, alone or in combination with IE1 72, could have a significant role in the regulation of these processes (17, 32, 33, 53, 55, 61, 74).
To determine if IE2 86 or IE1 72 had any effect on HMGA2 expression, we used the recombinant viruses IE2 86ΔSX-EGFP and CR208. The CR208 virus does not express the IE1 72 protein (24, 36), and there is a significant reduction in the levels of IE2 86 protein, but not IE1 72 protein, in cells infected with the IE2 86ΔSX-EGFP mutant virus (48). The lower levels of IE2 86 protein are not due to a defect in transcription, as the levels of the IE2 86 transcript are comparable in the cells infected with the mutant and wt viruses (48). G0-synchronized HFF were infected at an MOI of 5 with CR208 and its rescued virus CR249 and with the wt IE2 86-EGFP virus and the IE2 86ΔSX-EGFP mutant and rescued viruses. Total RNAs were isolated from cells at 24 and 48 h p.i., and the levels of HMGA2 were analyzed by real-time RT-PCR analysis. We normalized each sample to RNA encoding G6PD. The IE1 72 deletion mutant was similar to the rescued virus with respect to inhibition of HMGA2 expression, although the decrease (n-fold) in the HMGA2 mRNA was slightly less than that in the cells infected with the rescued virus (Fig. 6A). In contrast, the IE2 86ΔSX-EGFP mutant virus did not significantly repress expression of HMGA2 (Fig. 6B). These results were confirmed in a separate experiment by Northern blot analysis of mRNA that was isolated at 24 and 96 h p.i. from cells infected at a high MOI with the wt, IE2 86 mutant, and rescued viruses (Fig. 6C). We also performed Western blot analysis to confirm that IE2 86ΔSX-EGFP mutant showed the previously described decrease in the IE2 86 protein levels (Fig. 6D).
FIG. 6.
Expression of HMGA2 RNA in HCMV-infected HFF depends on IE2 86 but not IE1 72 protein levels. (A) G0-synchronized cells were released into G1 and mock infected or infected with IE1 72 mutant (CR208) virus or rescued (CR249) virus. Cells were harvested at 24 and 48 h p.i. Total RNA was isolated and analyzed by quantitative real-time RT-PCR to measure levels of HMGA2 RNA. RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. For each time point, two separate experiments were performed and duplicate reactions were analyzed. The mock-infected RNA that was isolated at 24 h p.i. was used to generate a standard curve for each gene. This standard curve was used to calculate the amount of RNA present in each sample. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values. (B to D) G0-synchronized cells were released into G1 and mock infected or infected with IE2 86-EGFP (wt), IE2 86ΔSX-EGFP (Δ), and IE2 86ΔSX-EGFP rescued (R) viruses at an MOI of 5. Cells were harvested at 24, 48, and 96 h p.i. (B) Quantitative real-time RT-PCR analysis of 24-h and 48-h samples using specific primers and probe for the HMGA2 transcript. RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. For each time point, three separate experiments were performed and duplicate reactions were analyzed. The graph shows the averages of the three experiments, and the ranges of the highest and lowest values are indicated. (C) Northern blot analysis of mRNA isolated at 24 and 96 h p.i. with an HMGA2-specific probe. The blot was stripped and hybridized to a GAPDH-specific probe as a loading control. M, mock infection. (D) Western blot analysis of the viral IE1 72 and IE2 86 proteins in the samples harvested at 24 and 96 h p.i. using the MAb CH16.0. Levels of cellular actin were used as a protein loading control.
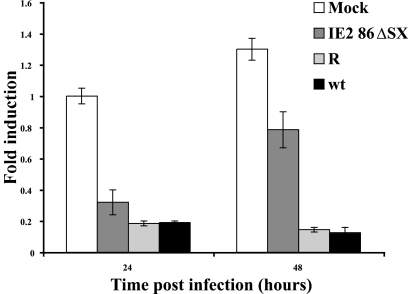
Since the experiment with the HCMV recombinant virus expressing HMGA2 indicated that HMGA2 and cyclin A expression could be linked (Fig. 5), we proceeded to measure the amount of cyclin A RNA in cells infected with the IE2 86ΔSX-EGFP mutant. Figure 7 shows that there was a significant increase in the level of cyclin A RNA in cells infected with the mutant virus, relative to that observed in cells infected with the wt and rescued viruses. Consistent with the requirement for HMGA2 in activation of the cyclin A promoter, this induction of cyclin A was not seen until 48 h p.i., whereas the increase in HMGA2 expression could be seen at 24 h p.i. Taken together, these results suggest that IE2 86 expression is associated with the repression of HMGA2 transcription, which in turn affects the activation of cyclin A transcription.
FIG. 7.
HFF infected with the recombinant mutant virus IE2 86ΔSX-EGFP exhibit elevated cyclin A expression. RNA that was isolated at 24 and 48 h p.i. from two of the three infections shown in Fig. 6B was analyzed by quantitative real-time RT-PCR using primers and probe for the cyclin A transcript. RNA levels were normalized to G6PD levels as an internal control for the amount of RNA in each sample. The mock-infected RNA that was isolated at 24 h p.i. was used to generate a standard curve for each gene. This standard curve was used to calculate the amount of RNA present in each sample. The graph shows the averages of the two experiments, and the range bars indicate the highest and lowest values.
DISCUSSION
HCMV infection blocks the expression of cyclin A mRNA and protein (30, 47). Since the HMGA2 protein has been implicated in the regulation of cyclin A transcription, we examined its expression during HCMV infection. We show that HMGA2 RNA synthesis is also greatly inhibited in HCMV-infected cells. The HMGA1 and HMGA2 proteins have similar structures, and both function as architectural DNA-binding proteins (54). However, the transcription of their genes is controlled by different promoters. In contrast to what was observed for HMGA2, there was only a modest decrease in the levels of HMGA1 RNA, indicating that their transcription is differentially affected by the infection.
To determine whether the repression of HMGA2 was important for viral replication, we constructed a recombinant virus that expressed HMGA2. In cells infected with the recombinant virus at a high MOI there was a significant delay in the progression of the viral infection. Although there were no significant differences in expression of the IE1 and IE2 proteins, there was a decrease in the levels of both early and late proteins. The reduced expression of the viral replication protein UL57, a single-stranded DNA-binding protein, and UL44, a processivity factor for the viral DNA polymerase, was accompanied by a corresponding decrease in viral DNA synthesis (Fig. 3A and B and Fig. 4). Although these results suggest that the presence of HMGA2 has a negative effect on the infection at early times, we cannot exclude the possibility that this was due to the overexpression of HMGA2 in the infected cells.
During HCMV infection, the modulation of factors involved in cellular DNA synthesis, inhibition of apoptosis, and alteration of the host's immune response to the virus are associated with marked dysregulation of the cell cycle and signaling pathways (for a review, see reference 50). HCMV activates many host cell proteins to create an environment that is optimal for viral gene expression and DNA replication. At the same time, however, the virus inhibits selective host cell functions to ensure that its own replication is favored over that of the host. The net effect is that the cell cycle halts in a pseudo-G1 state and host cell DNA synthesis is blocked. These effects occur at all levels of gene expression, including synthesis and posttranscriptional processing of RNAs and posttranslational modification, stabilization, and localization of proteins. For example, inhibition of cyclin A expression and upregulation of cyclin E are both examples of regulation at the transcriptional level (47). Targets of the anaphase-promoting complex E3 ubiquitin ligase, including cyclin B1, geminin, and cdc6, are also prematurely stabilized at early times in the infection (7, 49). The premature accumulation of geminin is particularly important as it is associated with HCMV-mediated inhibition of the licensing of cellular origins of DNA replication (7, 70). Multiple posttranscriptional pathways are also used to maintain high levels of cdk1/cyclin B1 kinase activity (49).
A number of studies suggest that IE2 86 plays a role in the cell cycle modulation. Some have shown that transient expression of IE2 86 alters cell cycle progression, with a block at the G1/S boundary or just after entry into S phase (39, 42, 58, 67, 69). Deletion of aa 451 to 579 abolished the ability of IE2 86 to induce G1 arrest in transient assays in U373 cells, but aa 25 to 85, 544 to 579, and 136 to 290 were not necessary (68). In a recent study, it was shown that a recombinant HCMV with a mutation of aa 548 from Q to R was unable to inhibit host cell DNA synthesis (43). Replication of this virus was impaired, indicating that efficient viral infection requires the inhibition of host cell DNA synthesis. Expression of IE2 86 from a recombinant adenovirus also leads to the activation of E2F-responsive genes, many of which are involved in host cell DNA synthesis (57). In infected cells, p53 is stabilized but the expression of its target gene p21 is repressed, suggesting that the virus impedes p53 function (11, 15, 22, 38). IE2 86 interacts with the C terminus of p53, and in transient-expression assays binding of p53 to target promoters is inhibited (9, 28, 59). There also may be a threshold effect since IE2 86 does not reduce p21 expression in fibroblasts that overexpress p53 (58).
HMGA proteins have been shown to function as architectural factors that can enhance the binding of DNA-interacting proteins and promote the formation of enhanceosomes (45). Therefore, cells in which HMGA proteins are upregulated should have a notable change in the chromatin structure and expression of specific genes. We show here that the HMGA2-expressing virus is able to induce the expression of cyclin A mRNA and protein during the infection (Fig. 5A and B), indicating that HMGA2 repression during HCMV infection is correlated with repression of cyclin A expression. Taken together, these results point to inhibition of HMGA2 expression as another mechanism by which HCMV creates a cellular environment that favors viral replication.
Our studies with the recombinant virus with a mutant IE2 86 gene (IE2 86ΔSX-EGFP) also suggest that IE2 86 plays a role in the repression of HMGA2 and cyclin A expression during the infection. We previously showed that infection with this mutant virus resulted in normal accumulation of IE1 72 and viral early proteins, and viral DNA replication was comparable to that of the wt (48). The most notable molecular defects were that the levels of the IE2 protein (but not mRNA) and the pp65 (UL83) and pp28 (UL99) matrix proteins were significantly reduced. This mutant was also recently used in a study showing that IE2 86 is involved in blocking the expression of cytokines and proinflammatory chemokines during the infection (62). In this paper, we have demonstrated that in the cells infected with the IE2 86ΔSX-EGFP mutant, the lower levels of IE2 86 with a deletion of aa 136 to 290 correlate with higher levels of HMGA2 and cyclin A expression. In contrast, HMGA2 expression was still repressed in cells infected with an IE1 72 deletion mutant.
The mechanism of repression of HMGA2 expression by HCMV is an important question that will require further studies. HMGA2 is an example of a gene that requires histone deacetylase (HDAC) activity for activation of its transcription. It has been shown that treatment of cells with the HDAC inhibitor trichostatin A leads to a decrease in the amount of SP1 and SP3 bound to the HMGA2 promoter as well as a loss of acetylated histones H3 and H4 (21). Several reports have suggested that inhibition of HDAC activity is important for HCMV infection (40, 41, 44, 46). Since IE2 86 can form a complex with HDAC1, and both IE2 86 and IE1 72 can interact with HDAC3, it is possible that repression of HMGA2 by HCMV may be due to direct or indirect interference of IE2 86 with HDAC activity and may require high levels of IE2 86 (41, 71).
An alternative explanation for the inability of IE2 86ΔSX to repress HMGA2 RNA expression is that the domain between aa 136 and 290 is involved in the regulation of HMGA2. Studies from our lab and others have shown that IE2 86 is able to bind many transcriptional regulators in vitro. In our studies with c-Jun, Jun-B, TATA-binding protein, and Rb, we found that the region between aa 136 and 290 was one of three domains that were able to independently mediate these interactions (52, 56). As noted above, IE2 86 has been shown to interact with both HDAC1 and HDAC3 (41, 71), but the domain of IE2 86 mediating this interaction has not yet been reported. IE2 86ΔSX is also missing the sites of sumoylation (aa 175 and 180), as well as the SUMO binding site (aa 200 to 208), which may also be important for the regulation of HMGA2 (1, 27). In addition, it has been shown that mutation of the serine-rich domain (aa 258 to 275) that lies within the deletion has both negative and positive effects on the infection (5). Clearly, IE2 86 is involved in many of the processes responsible for changes in the cell cycle, and the possibility exists that this region is also important for the regulation of HMGA2 expression, either through direct binding or through other, more indirect regulatory mechanisms.
HMGA2 is present at high levels in many tumor cells, and there is considerable evidence that it is involved in cell proliferation and neoplastic transformation (for a review, see reference 54). Transgenic mice expressing a rearranged form of HMGA2 or overexpressing wild-type HMGA2 develop lipomas, natural killer cell lymphomas, or pituitary adenomas (3, 4, 6, 20). Levels of HMGA2 are generally high during embryogenesis and low in the adult, although expression does vary in the different fetal and adult tissues (for a review, see reference 54). During embryogenesis, HMGA2 is involved in spermatogenesis (16) and skeletal muscle differentiation (13). HMGA2-knockout mice display the pygmy phenotype that is characterized by a drastic reduction of body fat content and growth retardation (2, 72). Given the importance of HMGA2 during embryogenesis, it is possible that repression of HMGA2 expression in infected cells could contribute to the birth defects in infants infected with HCMV in utero.
Acknowledgments
We thank Edward Mocarski for providing the HCMV recombinant virus that has a deletion of exon 4 of the IE1 72 gene (CR208) and its rescued virus (CR249), Guidalberto Manfioletti for providing the plasmid pcDNA-HMGA2 and antibody to HMGA2, and Bodo Plachter for providing the plasmid pcDNA-pp71tag. We acknowledge Veronica Sanchez and Elizabeth White for their critical reading of the manuscript.
This work was supported by NIH grants CA73490 and CA034729.
REFERENCES
- 1.Ahn, J. H., Y. Xu, W. J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, A., and K. Chada. 2000. In vivo modulation of Hmgic reduces obesity. Nat. Genet. 24:377-380. [DOI] [PubMed] [Google Scholar]
- 3.Arlotta, P., A. K. Tai, G. Manfioletti, C. Clifford, G. Jay, and S. J. Ono. 2000. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J. Biol. Chem. 275:14394-14400. [DOI] [PubMed] [Google Scholar]
- 4.Baldassarre, G., M. Fedele, S. Battista, A. Vecchione, A. J. Klein-Szanto, M. Santoro, T. A. Waldmann, N. Azimi, C. M. Croce, and A. Fusco. 2001. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc. Natl. Acad. Sci. USA 98:7970-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrasa, M. I., N. Y. Harel, and J. C. Alwine. 2005. The phosphorylation status of the serine-rich region of the human cytomegalovirus 86-kilodalton major immediate-early protein IE2/IEP86 affects temporal viral gene expression. J. Virol. 79:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battista, S., V. Fidanza, M. Fedele, A. J. Klein-Szanto, E. Outwater, H. Brunner, M. Santoro, C. M. Croce, and A. Fusco. 1999. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 59:4793-4797. [PubMed] [Google Scholar]
- 7.Biswas, N., V. Sanchez, and D. H. Spector. 2003. Human cytomegalovirus infection leads to accumulation of geminin and inhibition of the licensing of cellular DNA replication. J. Virol. 77:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boldogh, I., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonin, L. R., and J. K. McDougall. 1997. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J. Virol. 71:5831-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:156-160. [DOI] [PubMed] [Google Scholar]
- 12.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caron, L., F. Bost, M. Prot, P. Hofman, and B. Binetruy. 2005. A new role for the oncogenic high-mobility group A2 transcription factor in myogenesis of embryonic stem cells. Oncogene 24:6281-6291. [DOI] [PubMed] [Google Scholar]
- 14.Challacombe, J. F., A. Rechtsteiner, R. Gottardo, L. M. Rocha, E. P. Browne, T. Shenk, M. R. Altherr, and T. S. Brettin. 2004. Evaluation of the host transcriptional response to human cytomegalovirus infection. Physiol. Genomics 18:51-62. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Z., E. Knutson, A. Kurosky, and T. Albrecht. 2001. Degradation of p21cip1 in cells productively infected with human cytomegalovirus. J. Virol. 75:3613-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chieffi, P., S. Battista, M. Barchi, S. D. Agostino, G. M. Pierantoni, M. Fedele, L. Chiariotti, D. Tramontano, and A. Fusco. 2002. HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene 21:3644-3650. [DOI] [PubMed] [Google Scholar]
- 17.Chiou, S. H., J. H. Liu, W. M. Hsu, S. S. Chen, S. Y. Chang, L. J. Juan, J. C. Lin, Y. T. Yang, W. W. Wong, C. Y. Liu, Y. S. Lin, W. T. Liu, and C. W. Wu. 2001. Up-regulation of Fas ligand expression by human cytomegalovirus immediate-early gene product 2: a novel mechanism in cytomegalovirus-induced apoptosis in human retina. J. Immunol. 167:4098-4103. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estes, J. E., and E.-S. Huang. 1977. Stimulation of cellular thymidine kinases by human cytomegalovirus. J. Virol. 24:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedele, M., S. Battista, L. Kenyon, G. Baldassarre, V. Fidanza, A. J. Klein-Szanto, A. F. Parlow, R. Visone, G. M. Pierantoni, E. Outwater, M. Santoro, C. M. Croce, and A. Fusco. 2002. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 21:3190-3198. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson, M., P. A. Henry, and R. A. Currie. 2003. Histone deacetylase inhibition is associated with transcriptional repression of the Hmga2 gene. Nucleic Acids Res. 31:3123-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn, G., M. Jarosch, J. B. Wang, C. Berbes, and M. A. McVoy. 2003. Tn7-mediated introduction of DNA sequences into bacmid-cloned cytomegalovirus genomes for rapid recombinant virus construction. J. Virol. Methods 107:185-194. [DOI] [PubMed] [Google Scholar]
- 26.Hirai, K., T. Furukawa, and S. A. Plotkin. 1976. Induction of DNA polymerase in WI-38 and guinea pig cells infected with human cytomegalovirus (HCMV). Virology 70:251-255. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 74:2510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, C.-H., M. D. T. Chang, K.-Y. Tai, Y.-T. Yang, P.-S. Wang, C.-J. Chen, Y.-H. Wang, S.-C. Lee, C.-W. Wu, and L.-J. Juan. 2004. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 23:2269-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isom, H. C. 1979. Stimulation of ornithine decarboxylase by human cytomegalovirus. J. Gen. Virol. 42:265-278. [DOI] [PubMed] [Google Scholar]
- 30.Jault, F. M., J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated RB, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukac, D. M., and J. C. Alwine. 1999. Effects of human cytomegalovirus major immediate-early proteins in controlling the cell cycle and inhibiting apoptosis: studies with ts13 cells. J. Virol. 73:2825-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 36.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding ie1 (491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muganda, P., O. Mendoza, J. Hernandez, and Q. Qian. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of the cell cycle. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noris, E., C. Zannetti, A. Demurtas, J. Sinclair, M. De Andrea, M. Gariglio, and S. Landolfo. 2002. Cell cycle arrest by human cytomegalovirus 86-kDa IE2 protein resembles premature senescence. J. Virol. 76:12135-12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42a.Pass, R. F. 2001. Cytomegalovirus, p. 2625-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 43.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 80:3872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 46.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez, V., A. K. McElroy, and D. H. Spector. 2003. Mechanisms governing maintenance of cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J. Virol. 77:13214-13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanchez, V., and D. H. Spector. 2006. Exploitation of host cell cycle regulatory pathways by HCMV, p. 205-230. In M. J. Reddehase (ed.), Cytomegaloviruses. Caister Academic Press, Norwich, Norfolk, United Kingdom.
- 51.Reference deleted.
- 52.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sedger, L. M., D. M. Shows, R. A. Blanton, J. J. Peschon, R. G. Goodwin, D. Cosman, and S. R. Wiley. 1999. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J. Immunol. 163:920-926. [PubMed] [Google Scholar]
- 54.Sgarra, R., A. Rustighi, M. A. Tessari, J. D. Bernardo, S. Altamura, A. Fusco, G. Manfioletti, and V. Giancotti. 2004. Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett. 574:1-8. [DOI] [PubMed] [Google Scholar]
- 55.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, Y.-J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, Y. J., and M. F. Stinski. 2005. Inhibition of cell division by the human cytomegalovirus IE86 protein: role of the p53 pathway or cyclin-dependent kinase 1/cyclin B1. J. Virol. 79:2597-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Speir, E., R. Modali, E.-S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 60.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka, K., J. P. Zou, K. Takeda, V. J. Ferrans, G. R. Sandford, T. M. Johnson, T. Finkel, and S. E. Epstein. 1999. Effects of human cytomegalovirus immediate-early proteins on p53-mediated apoptosis in coronary artery smooth muscle cells. Circulation 99:1656-1659. [DOI] [PubMed] [Google Scholar]
- 62.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tessari, M. A., M. Gostissa, S. Altamura, R. Sgarra, A. Rustighi, C. Salvagno, G. Caretti, C. Imbriano, R. Mantovani, G. D. Sal, V. Giancotti, and G. Manfioletti. 2003. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol. Cell. Biol. 23:9104-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wade, M., T. F. Kowalik, M. Mudryj, E. S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White, E. A., and D. H. Spector. 2005. Exon 3 of the human cytomegalovirus major immediate-early region is required for efficient viral gene expression and for cellular cyclin modulation. J. Virol. 79:7438-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiebusch, L., J. Asmar, R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus immediate-early protein 2 (IE2)-mediated activation of cyclin E is cell-cycle-independent and forces S-phase entry in IE2-arrested cells. J. Gen. Virol. 84:51-60. [DOI] [PubMed] [Google Scholar]
- 68.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiebusch, L., R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 4:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wright, E., M. Bain, L. Teague, J. Murphy, and J. Sinclair. 2005. Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J. Gen. Virol. 86:535-544. [DOI] [PubMed] [Google Scholar]
- 72.Zhou, X., K. F. Benson, H. R. Ashar, and K. Chada. 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376:771-774. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]