Abstract
The human cytomegalovirus major immediate-early protein IE86 is pivotal for coordinated regulation of viral gene expression throughout infection. A relatively promiscuous transactivator of viral early and late gene transcription, IE86 also acts during infection to negatively regulate its own promoter via direct binding to a 14-bp palindromic IE86-binding site, the cis repression sequence (crs), located between the major immediate-early promoter (MIEP) TATA box and the start of transcription. Although such autoregulation does not involve changes in the binding of basal transcription factors to the MIEP in vitro, it does appear to involve selective inhibition of RNA polymerase II recruitment. However, how this occurs is unclear. We show that autorepression by IE86 at late times of infection correlates with changes in chromatin structure around the MIEP during the course of infection and that this is likely to result from physical and functional interactions between IE86 and chromatin remodeling enzymes normally associated with transcriptional repression of cellular promoters. Firstly, we show that IE86-mediated autorepression is inhibited by histone deacetylase inhibitors. We also show that IE86 interacts, in vitro and in vivo, with the histone deacetylase HDAC1 and histone methyltransferases G9a and Suvar(3-9)H1 and that coexpression of these chromatin remodeling enzymes with IE86 increases autorepression of the MIEP. Finally, we show that mutation of the crs in the context of the virus abrogates the transcriptionally repressive chromatin phenotype normally found around the MIEP at late times of infection, suggesting that negative autoregulation by IE86 results, at least in part, from IE86-mediated changes in chromatin structure of the viral MIEP.
Efficient productive infection of cells with human cytomegalovirus (HCMV) is dependent on a temporal cascade of viral gene expression which relies on the finely tuned, coordinated regulation of specific viral gene products at specific stages of the virus infection cycle. Upon infection of permissive cells, the virus undergoes a regulated cascade of gene expression in three broad phases termed immediate early (IE), early, and late, culminating in the release of infectious virions.
The major IE genes are the most abundantly transcribed viral genes at immediate-early times of infection, giving rise to two nuclear phosphoproteins, the 72-kDa (IE72) and 86-kDa (IE86) major IE proteins (59, 62, 69). The expression of IE72 and IE86 is driven by the viral major IE promoter/enhancer (MIEP), and these viral gene products have been implicated in playing a pivotal role in the coordinated regulation of viral gene expression. IE86 acts as a very strong but relatively nonspecific transcriptional activator of viral early and late gene expression; its effect is synergized by the viral IE72 protein. The activation of viral promoters by IE86 is likely to be mediated by direct DNA binding of IE86 to specific DNA-binding sites as well as protein-protein interactions with general transcription factors and promoter-specific transcription factors (10, 11, 15, 18, 25, 29, 34, 57, 58). In contrast, IE72 and IE86 have an established role in MIEP autoregulation: IE72 positively regulates its own promoter through a mechanism likely to involve NF-κB (13, 52, 60), and IE86 negatively autoregulates through, so far, undefined mechanisms. This IE86-mediated repression of the IE86 promoter occurs through direct binding of IE86 to a short DNA sequence, the cis repression sequence (crs), whose function is orientation independent but site dependent; the crs must lie between the TATA box of the promoter and the start of transcription (1, 12, 24, 30-32, 43, 56, 60, 67, 72). While other IE86-binding sites besides the crs have been reported, these appear only to augment the level of repression mediated by the crs (22). Although IE86-mediated repression does not involve the inhibition of binding of basal transcription factors to the MIEP (24), it has been shown in vitro that IE86 binding to the crs does result in an inhibition of recruitment of RNA polymerase II to the promoter (72). However, despite these observations, the exact mechanism by which IE86 represses its own promoter is, so far, unknown.
It has become increasingly clear that transcriptional regulation of cellular genes is profoundly linked to chromatin structure and that the higher-order chromatin structure of specific genes is often subject to extensive remodeling in order for them to become more or less accessible to the cellular transcription machinery (4, 55, 68). Such chromatin remodeling is presumed to be dynamic with a balance occurring between histone acetylation (leading to transcriptional activation) and histone deacetylation with subsequent histone methylation (leading to transcriptional silencing). These observations have formed the basis of the so-called histone code hypothesis, which stipulates that the transcriptional activity of cellular promoters is based on the quality and quantity of posttranslational modification of nucleosomes around promoter DNA (55, 68); there is good evidence that this hypothesis extends to the regulation of viral promoters as well (23, 26, 27, 33, 37, 40, 41, 49).
In the case of HCMV, expression from the viral MIEP is known to be regulated by its chromatin structure and that, when active, the viral MIEP is associated with acetylated histones, indicative of an open transcriptionally active chromatin conformation. In contrast, in nonpermissive cells or cells from healthy seropositive individuals carrying latent viral genomes, the viral MIEP is associated with nonacetylated, methylated histones and silencer proteins, such as heterochromatin protein 1 (HP1) (40, 49), both of which are markers of a transcriptionally inactive chromatin state (3, 28, 54, 61).
Such changes in chromatinization of the viral MIEP in different cell types is likely to result, at least in part, from the differential binding of cellular transcription factors to DNA sequences in the MIEP; such factors are then able to bind and recruit chromatin remodeling enzymes, such as histone deacetylases (HDACs) and histone methyltransferases (HMTs), which result in a more compact transcriptionally inactive chromatin template (71). On this basis, we asked whether the ability of IE86 to repress its own promoter is also mediated by such chromatin remodeling and, specifically, whether IE86 is able to physically and functionally interact with cellular HDACs and HMTs which, when recruited to the MIEP via the crs, result in transcriptional repression.
Our results show that IE86 is indeed able to physically and functionally interact with the histone deacetylase HDAC1 and the histone methyltransferases G9a and Suvar(3-9)H1 and that these interactions result in repression of the viral MIEP. Consistent with this, we also observed changes in the chromatin structure of the MIEP during the course of productive infection such that at late times, the MIEP is profoundly associated with chromatin markers of transcriptional repression. However, mutation of the crs in the context of the virus genome abrogates such IE86-mediated repression and the chromatin remodeling of the viral MIEP which is normally observed at late times of infection, and this is correlated with unregulated high levels of IE72 and IE86 expression at late times of infection.
MATERIALS AND METHODS
Cell culture and virus infection.
Human foreskin fibroblasts (HFFs) and U20S [an osteosarcoma cell line that expresses the retinoblastoma (RB) protein; designated RB(+ve)] were maintained in Eagle's minimal essential medium supplemented with 10% fetal calf serum (FCS). SAOS2 cells [an osteosarcoma cell line that does not express RB protein; designated RB(−ve)] were maintained in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% FCS. All cells were incubated at 37°C in 5% CO2. Cells were infected with approximately 5 PFU per cell as previously described (46). All the cell lines were fully permissive for HCMV infection as defined by expression of viral immediate-early, early, and late gene products, although the efficiencies of infections differed between cell types; HFFs were fully permissive for HCMV infection, and SAOS2 and U2OS cells were also fully permissive for infection (as determined by expression of the true late protein pp28), but the efficiencies of infections were about 50 to 60% for SAOS2 and 10 to 20% for U2OS cells, compared to 100% for HFFs (data not shown).
For analyses with trichostatin A (TSA) treatment, cells were incubated with TSA (330 nM) or ethanol as a solvent control for 18 h prior to transfection or infection.
A recombinant HCMV with a mutated crs (CRS217) was constructed by insertion of the crs HindIII site-change MIEP plasmid (see below), coupled with a gpt insert, in the viral unique long region open reading frames 127 to 129. The gpt insert was subsequently removed (and unique long region open reading frames 127 to 129) by plasmid rescue and selection using 6-thioguanine in Lesch-Nyhan fibroblasts, as previously described (17).
Plasmids.
pRG224 and pRG222 are MIEP reporter constructs driving expression of a luciferase reporter. They include the EcoRV-SacII fragment of HCMV MIEP (positions −929 to +77 with respect to the start of major IE transcription) cloned into pUHC13-3, which contains a luciferase reporter (16). pRG224 carries a wild-type crs, whereas pRG222 contains a change from CGTTTAGTGAACCG (wild type) to CGTTTAGaagcttG (lowercase letters indicate changes); this mutation also carries a HindIII site for easy recognition (crs HindIII site change).
pGex3XIE72 and pGex3XIE86 plasmids for the expression of glutathione S-transferase (GST)-IE72 and GST-IE86 fusion proteins, respectively, as well as vectors for the expression of GST-IE86 deletion mutants, have all been previously described (11).
Expression vectors for GST fused to either HDAC1 residues 1 to 382 [HDAC1(N)] or residues 332 to 482 [HDAC1(C)] have been previously described (8).
pcDNA3IE72 was made by cloning the EcoRI/XhoI fragment from pBSIE1 (11), containing the HCMV major IE72 cDNA, into pcDNA3 (Invitrogen). Similarly, pcDNA3IE86 was constructed by cloning the HCMV IE86 cDNA from pGex3XIE2 (11) into pcDNA3 by using BamHI/EcoRI.
cDNASuvar(3-9)H1-HA, for expressing hemagglutinin-tagged Suvar(3-9), was constructed by cloning a cDNA of Suvar(3-9)H1 into the XbaI/NotI sites of pcDNA3 and was a gift of A. Bannister. cDNA3HDAC1 was constructed by inserting the BamHI/XbaI fragment from PHK3HDAC1 (8), containing a full-length cDNA of HDAC1, into BamHI/XbaI-digested pcDNA3. cDNA3G9a has been described previously (63) and was constructed by cloning a full-length cDNA of G9a into pcDNA3.
pcDNA3IE86(1-290), encoding only amino acids 1 to 290 of IE86, was constructed by deleting an XhoI fragment from pcDNA3IE86.
Transient transfection assays.
For transient transfection, approximately 5 × 106 cells were transfected with 5 μg of luciferase reporter construct together with 10 μg of cotransfected plasmid by calcium phosphate precipitation. The amount of cotransfected plasmid was kept constant throughout by the addition of empty pcDNA3 vector. Cells were assayed 48 h posttransfection for luciferase activity, as described previously (21). The results are averages from three independent experiments.
For experiments using TSA, 5 × 106 U2OS or SAOS2 cells were transfected with 2.5 μg of luciferase reporter and 5 μg of cDNA3 or cDNA3IE86 by calcium phosphate coprecipitation. Twenty-four hours posttransfection, cells were treated with 333 ng/ml TSA for 24 h. This concentration of TSA gave optimum histone H4 acetylation with little corresponding cell toxicity (data not shown). Cells were then harvested and analyzed for luciferase activity.
GST fusion interaction assays.
GST fusion protein {GST, GST-IE72, GST fused with amino acids 1 to 579 of IE86 [GST-IE86(1-579)], and GST-IE86(1-290)} expression, purification, and interaction with [35S]methionine-labeled proteins were carried out exactly as described previously (11). Vectors for the generation of [35S]methionine-labeled gelsolin by coupled in vitro transcription/translation (Promega) have also been described previously (11). cDNA3HDAC1, cDNA3Suvar(3-9)H1, and cDNA3G9a vectors were also used directly to generate [35S]methionine-labeled HDAC1, Suvar(3-9)H1, and G9a, respectively, by virtue of the T7 polymerase site contained in pcDNA3.
Plasmids for the expression of [35S]methionine-labeled full-length IE86 and the N terminus of IE86 (amino acids 1 to 290) have been described elsewhere (11, 19).
Deacetylase assay.
Deacetylase assays were carried out essentially as described previously (65). Lyophilized peptide corresponding to the first 24 residues of bovine histone H4 (trifluoroacetic acid salt; Affiniti Research Products) was chemically acetylated with [3H]sodium acetate (5.3 Ci [197.3 GBq] mmol−1; 10 mCi [370 MBq] ml−1 in ethyl alcohol [EtOH]; New England Nuclear). GST fusion protein (1 μg) attached to Sepharose beads was rotated with 200 μl EBC buffer (0.05 M Tris-Hcl [pH 8.0], 0.12 M Nacl, 0.5% Nonidet P-40, 0.1 M NaF, 0.2 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride), supplemented with bovine serum albumin (40 μg ml−1) for 10 min at room temperature. Nuclear extracts of primary human fibroblasts cells (1 ml) were added and tubes rotated for 2 h at room temperature. Samples were microcentrifuged (5 min at 6,000 rpm), and the GST pellet was washed four times with 1 ml NETN (20 mM Tris [pH 8.0], 100 mM Nacl, 1 mM EDTA, 0.5% NP-40). Beads were resuspended in 200 μl EBC (without NP-40 and phenylmethylsulfonyl fluoride), to which 5 × 104 cpm of acetylated H4 peptide was added before the tubes were incubated at 37°C for 2 h. The reaction was stopped by the addition of 65 μl 1 M HCl-0.16 M acetic acid mix. Released [3H]acetate was extracted with ethyl acetate, the supernatant was transferred to a scintillation vial containing 1 ml scintillation cocktail (PerkinElmer), and the amount of free 3H released from the peptide was counted. It is important to note that all reaction mixes contained equal amounts of GST fusion protein, Sepharose beads, and peptide.
IP/Western blot analysis.
Where stated, 2 × 107 SAOS2 or U2OS cells were transiently transfected by calcium phosphate precipitation with 10 μg of pcDNA3, 10 μg of cDNA3IE86(1-579), or 10 μg of pcDNA3IE86(1-290) together with 10 μg of cDNA3HDAC1 or 10 μg of cDNA3Suvar(3-9)H1 or 10 μg of cDNA3G9a. After 36 h, total cell extracts were prepared and analyzed by immunoprecipitation (IP) followed by Western blot analysis as previously described (9). Briefly, extracts were immunoprecipitated for 1 h at 4°C with E13 (1:1,000 dilution; Chemicon), which recognizes HCMV IE72 and IE86 [both IE86(1-579) and IE86(1-290)] proteins, with Suvar(3-9)H1 antibody (1:200 dilution; Santa Cruz Biotechnology), which detects Suvar(3-9)H1, or with anti-HDAC1 antibody (Upstate). Equivalent concentrations of isotype-matched antibody controls were also present in all experiments. Samples were then incubated at 4°C for 90 min with protein A-Sepharose (Pharmacia) and analyzed by Western blot hybridization with anti-G9a antibody (1:1,000; Abcam) (after E13 immunoprecipitation) or with E13 antibody at a 1:1,000 dilution (after HDAC1 or Suvar immunoprecipitation) by using an ECL kit (Amersham).
Because the IE86(1-290) polypeptide would comigrate with the immunoglobulin G (IgG) heavy chain (approximately 45 kDa) of the antibody used for immunoprecipitation upon any subsequent Western blot analysis using E13 antibody, the ExactaCruz system (Santa Cruz Biotechnology) was employed as detailed by the manufacturer in the Western blot analysis after immunoprecipitations with anti-HDAC1 or anti-Suvar(3-9)H1. This specifically prevents the detection of any IgG heavy chain contamination in the immunoprecipitate upon the subsequent Western blot assay.
ChIP.
Chromatin immunoprecipitations (ChIPs) were carried out essentially as described previously (35, 40, 49). Briefly, 105 control primary human fibroblasts or fibroblasts infected with HCMV 24 to 96 h postinfection were fixed with 1% formaldehyde for 10 min and then lysed. DNA associated with histones was immunoprecipitated with control serum (Sigma-Aldrich, Dorset, United Kingdom), anti-acetyl histone H4 antiserum (ChIP grade, 1:200 dilution; Upstate, Charlottesville, VA), anti-dimethyl K9-H3 antiserum (ChIP grade, 1:200 dilution; Upstate, Charlottesville, VA), or anti-HP-1 antiserum (1:200 dilution; Serotec, Oxford, United Kingdom) as previously described (40).
For detection of the MIEP of HCMV, DNA from disrupted nucleosomes was precipitated and amplified by PCR with a sense primer (5′-TGG GAC TTT CCT ACT TGG-3′) and antisense primer (5′-CCA GGC GAT CTG ACG GTT-3′) complementary to positions −272 and +13 relative to the MIEP start site. PCR products were transferred to nitrocellulose and hybridized to an MIEP-specific 32P (Amersham, Woking, United Kingdom)-radiolabeled probe. The probe fragment was generated by PCR of HCMV DNA using a sense primer (5′-ATT ACC ATG GTG ATG CGG TT-3′) and antisense primer (5′-GGC GGA GTT GTT ACG ACA T-3′). All amplifications by PCR were performed using AmpliTaq Gold (Applied Biosystems, Foster City, CA) with the addition of 2 × MasterAmp PCR Enhancer (Cambio, Cambridge, United Kingdom). The cycle parameters for amplification by PCR were 95°C for 5 min and then 25 cycles at 94°C (40 s), 50°C (40 s), and 72°C (90 s).
Amplification of the human γ-globin promoter by PCR was performed using a sense primer (5′-GCC TTG ACC AAT AGC CTT GAC A-3′) and antisense primer (5′-GAA ATG ACC CAT GGC GTC TG-3′) which have been used in previous analyses of this region (5).
IE72/IE86 expression analyses.
HFFs infected with Towne or CRS217 deletion mutant virus were harvested at 24 to 96 h postinfection to determine the levels of IE RNA and protein expression. Total RNA was isolated using TRIzol reagent as described by the manufacturer, and then 10 μg of total RNA was resolved on a 0.8% agarose gel. Following transfer to a nitrocellulose membrane, the filter was then incubated with a 32P-radiolabeled exon-5-specific probe to detect the p86, p60, and p40 transcripts.
Hybridoma supernatants of mouse monoclonal antibodies p63-27 and SMX, which recognize IE72 and IE86, respectively, were gifts of Bodo Plachter (45). For Western blot analysis, total proteins from 6 × 103 cells were separated on sodium dodecyl sulfate (SDS)-8.5% polyacrylamide gels and transferred to nitrocellulose filters. Protein transfer was confirmed by Ponceau S staining of filters. Following a blocking, filters were then incubated for 3 h with primary antibody diluted in 10% FCS in phosphate-buffered saline (PBS-FCS). After three 5-min washes in PBS-0.5% Nonidet P-40, the filters were rinsed in PBS and incubated for 1 h with rabbit anti-mouse IgG-horseradish peroxidase conjugate (DAKO) or rabbit anti-human IgG-horseradish peroxidase conjugate (DAKO), each diluted 1/1,000 in PBS-FCS and detected by use of an ECL kit (Amersham).
RESULTS
IE86 interacts with HDAC1 in vitro and in vivo.
In an attempt to better understand the mechanisms by which IE86 might mediate its well-established role in autorepression, we screened a human lung fibroblast cDNA expression library for IE86-binding proteins; one cDNA identified in this manner was that of the histone deacetylase HDAC1 (J. Fairley et al., Abstr. 20th Int. Herpesvirus Workshop, abstr. 171, 1995), a member of a family of enzymes able to remove highly charged acetyl groups from core histones (14, 20). Such deacetylated histones are known to be targets for other chromatin remodeling enzymes, such as HMTs; such methylated histones (particularly those methylated at lysine 9 or lysine 27), in turn, become targets for recruitment of silencing proteins, such as HP1 (3, 28, 39, 54, 61). Recruitment of HDAC1 to the MIEP via IE86 binding to the crs could, therefore, be a mechanism to help explain IE86-mediated autorepression.
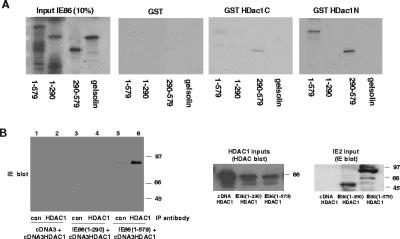
To confirm the physical interaction between IE86 and HDAC1, we embarked on a series of experiments analyzing this interaction in vitro and in vivo. Firstly, we analyzed the interaction between [35S]methionine-labeled IE86 and domains of HDAC1 fused to GST in standard GST fusion pull-down assays (Fig. 1A). It was difficult for us, as it was for others, to generate a full-length GST-HDAC1 fusion (data not shown). Consequently, we used GST fused to N- and C-terminal domains of HDAC1 as used previously by others (8). Such analyses showed that full-length IE86 was specifically able to interact with the N-terminal domain of HDAC1. We observed little binding of IE86 to the HDAC1 C-terminal domain. Binding to GST-HDAC1(N) was not observed with an IE86 deletion leaving only amino acids 1 to 290. In contrast, binding to GST-HDAC1(N) was also observed with an IE86 deletion leaving only amino acids 291 to 579, a region of IE86 that contains domains that have been implicated in binding to a number of other cellular proteins (2, 11, 18, 70). As we and others have observed before with the binding to a number of cellular proteins, full-length IE86(1-579) appeared to bind less efficiently to GST-HDAC1(N) than this short domain of IE86(291-579). This may be due to N-terminal regions of IE86 interfering with the interaction domains of the C terminus of IE86 (11, 58). An irrelevant [35S]methionine-labeled protein, gelsolin, showed no binding to either GST-HDAC1(N) or GST-HDAC1(C), and we observed no such interactions between IE86 and GST control protein.
FIG. 1.
IE86 interacts with HDAC1 in vitro and in vivo. (A) GST or GST fused to the C-terminal (GST HDac1C) or N-terminal (GST HDac1N) end of HDAC1 on glutathione beads was used as a target for binding to [35S]methionine-labeled full-length IE86 (1-579), a domain of IE86 containing amino acids 1 to 290 (1-290), a domain of IE86 containing amino acids 291 to 579 (291-579), or control gelsolin. Input proteins (1/10) are also shown, and molecular mass markers are in kDa. (B) SAOS2 cells were transfected with pcDNA3 (tracks 1 and 2), IE86(1-290) (tracks 3 and 4), or IE86(1-579) (tracks 5 and 6) together with pcDNA3HDAC1 expression vectors. Cell extracts were immunoprecipitated with control (con) antibody or anti-HDAC1 antibody, and immunoprecipitated complexes were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blot analysis using an anti-IE86 antibody. Extracts were also analyzed directly for levels of expression of HDAC1 (HDAC1 inputs) by HDAC1 Western blot analysis or for expression of IE86 (IE86 inputs) by IE Western blot analysis.
We also confirmed the interaction between IE86 and HDAC1 in vivo in mammalian cells (Fig. 1B). Unfortunately, RB is a well-known independent interaction partner of both HDAC1 (8, 36) and IE86 (18, 58). Consequently, it is possible that the interaction of IE86 with HDAC1 could be mediated by RB “bridging.” In order to rule this out, we used an RB(−ve) cell line, SAOS2 cells, for this analysis. We coexpressed IE86 and HDAC1 by transfection in SAOS2 cells and analyzed the interaction by IP/Western blotting. As SAOS2 cells contain no endogenous RB protein, any observed interaction between IE86 and HDAC1 cannot be due to bridging by RB. Figure 1B, track 6, clearly shows that immunoprecipitation of HDAC1 in cells coexpressing IE86 and HDAC1 resulted in coimmunoprecipitation of IE86. We observed no such coimmunoprecipitation of IE86 by using a nonspecific antibody control instead of HDAC1-specific antibody (Fig. 1B, track 5). Similarly, we saw no IE86 coimmunoprecipitated in cells transfected with HDAC1 alone, as expected (Fig. 1B, track 2). Consistent with our in vitro and in vivo analyses, IE86(1-290) also showed no binding to HDAC1 (track 4). Finally, we observed identical results when we, alternatively, immunoprecipitated with anti-HDAC1 antibody and then used E13 antibody for Western blot analysis or if IE86 and HDAC1 were coexpressed in RB(+ve) U2OS cells (data not shown).
IE86 has functional histone deacetylase-binding activity.
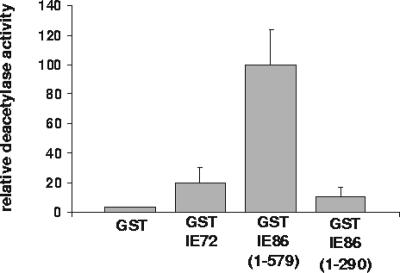
If IE86 is functioning to repress its own promoter by the recruitment of histone deacetylase, we would predict that IE86 should be able to sequester functional histone deacetylase activity from cellular extracts. Figure 2 shows that, as predicted, full-length GST-IE86 protein does bind high levels of functional histone deacetylase enzymes from nuclear extracts of MRC5 human fetal lung fibroblasts. In contrast, control GST protein showed no such interaction. Although IE72 shares the first 85 amino acids of IE86, GST-IE72, in our hands, has only low levels of deacetylase-binding activity. Consistent with the GST fusion pull-down and IP/Western blot analyses, a deletion mutant of IE86 devoid of amino acids 291 to 597, with only amino acids 1 to 290 of IE86, was also unable to bind appreciable levels of deacetylase.
FIG. 2.
IE86 has functional histone deacetylase-binding activity. GST, GST-IE72, GST-IE86(1-579), or GST-IE86(1-290) on glutathione beads was used as bait for nuclear extracts from primary human fibroblasts cells. Bound histone deacetylase activity was then assayed. The results shown are the means from triplicate samples of one experiment; the activity level of histone deacetylase binding to GST control protein was set at an arbitrary value of 1.
IE86 and HDAC1 act together to autorepress the viral MIEP in cotransfection assays.
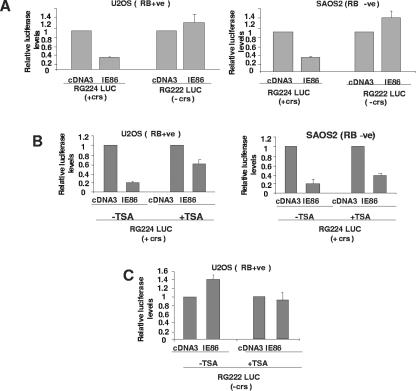
If IE86-mediated autorepression occurred via recruitment of HDACs, it would be predicted that this repression should be, at least in part, abrogated by inhibitors of histone deacetylase activity. To test this hypothesis, we used cotransfection assays to analyze IE86-mediated repression of the viral MIEP in the presence or absence of TSA, a global inhibitor of histone deacetylase activity. Firstly, we wanted to analyze the effect of IE86 on the viral MIEP in both the absence and presence of RB, as RB is an established interaction partner of a number of corepressors (7) and also IE86 (18, 58). Consequently, we carried out this assay with U2OS cells [RB(+ve)] and SAOS2 cells [RB(−ve)]. Figure 3A shows that, as expected, a luciferase expression vector based on the viral MIEP bearing the crs site (RG224) was reproducibly repressed up to fivefold upon coexpression of IE86. This repression requires the crs site, as a luciferase vector based on the MIEP with a mutant crs site (RG222) was not repressed by IE86. Also, as this IE86-mediated repression occurred in both RB(−ve) and RB(+ve) cell lines, repression of the MIEP by IE86 appears to be RB independent.
FIG. 3.
IE86-mediated repression of the MIEP is crs-dependent and inhibited by TSA. (A) U2OS [RB(+ve)] or SAOS2 [RB(−ve)] cells were transfected with an MIEP luciferase vector (5 μg) carrying the crs (RG224 LUC) or with a crs mutation (RG222 LUC) in the presence of 10 μg of control empty vector (cDNA3) vector or 5 μg of cDNA3IE86 (IE86) plus 5 μg of cDNA3. Cell extracts were then analyzed for luciferase activity. The results shown are the means from at least three independent experiments. (B) U2OS and SAOS2 cells were transfected and assayed as described for panel A with an MIEP luciferase vector (2.5 μg) carrying the crs (RG224 LUC) with pcDNA3 (5 μg) or pcDNA3IE86 (5 μg), but in addition, these transfections were carried out in the absence (−) or presence (+) of TSA. The results shown are the means from two independent experiments. (C) U2OS and SAOS2 cells were transfected and assayed as described for panel A but with an MIEP luciferase vector (2.5 μg) with no crs (RG222 LUC) and with pcDNA3 (5 μg) or pcDNA3IE86 (5 μg), but in addition, these transfections were carried out in the absence (−) or presence (+) of TSA. The results shown are the means from two independent experiments.
We next tested the ability of IE86 to mediate repression of the MIEP in the presence of TSA. As TSA can be toxic, the concentrations of TSA used were based on titrations which showed maximum acetylation of histone H4 by Western blot analysis with corresponding minimum cell death (data not shown). Although TSA treatment never fully reversed IE86-mediated repression, inhibition of histone deacetylase activity by TSA routinely results in the alleviation of IE86-mediated autorepression of RG224 (Fig. 3B), consistent with the belief that IE86-mediated repression of the MIEP is, at least in part, mediated by histone deacetylases. This ability of TSA to abrogate IE86-mediated repression of the MIEP was specific for vectors carrying the crs, as TSA had little effect on a promoter with a crs deletion in the presence of IE86 (see Fig. 3C). Alleviation of IE86-mediated repression by TSA was routinely greater in RB(+ve) U2OS cells (threefold) than in RB(−ve) SAOS2 cells (twofold). This also suggests that repression of the MIEP by IE86 has histone deacetylase-mediated components that are both RB dependent and RB independent; we also cannot rule out the possibility that other RB family members may be involved in IE86-mediated repression of the MIEP.
We next asked whether cotransfection of IE86 with HDAC1 led to any increased autorepression of the MIEP compared to use of IE86 alone. Transfection assays with RB(+ve) U2OS cells (Fig. 4A) or RB(−ve) SAOS2 cells (Fig. 4B) showed that both IE86 and HDAC1 independently repressed MIEP vectors carrying the crs (pRG224). However, cotransfection of IE86 and HDAC1 together was only, at best, additive with respect to repression of the MIEP carrying the crs (pRG224).
FIG. 4.
IE86 and HDAC1 act together to repress the MIEP. U2OS [RB(+ve)] (A) or SAOS2 [RB(−ve)] (B) cells were transfected with an MIEP luciferase vector carrying the crs (RG224 LUC) in the presence of 10 μg of control empty vector (cDNA3) vector, 5 μg of cDNA3IE86 (IE86) plus 5 μg of cDNA3, 5 μg of cDNA3HDAC1 (HDAC1) plus 5 μg cDNA3, or 5 μg of cDNA3IE86 plus 5 μg of cDNA3HDAC1 together. Cell extracts were then analyzed for luciferase activity. The results show the means from at least three independent experiments. The same transfections, with the exception that MIEP luciferase vector with a crs deletion (pRG222 LUC) was used, were also carried out with U2OS cells (C) and SAOS2 cells (D).
The same analysis with the pRG222, which carries a crs mutation (Fig. 4C and D), showed that, as expected, IE86 did not repress this promoter but that, as opposed to that of the MIEP carrying the crs, the low level of repression of the promoter with a crs mutation mediated by HDAC1 was now abrogated by IE86 coexpression. We have observed a similar abrogation of HDAC-mediated repression of cellular promoters by IE86 and is consistent with the previous observations that the IE72 and IE86 proteins can bind and sequester chromatin remodeling enzymes to promote viral or cellular gene expression (41; J. Murphy and J. Sinclair, unpublished observations) (see Discussion).
IE86 interacts with the HMT G9a in vitro and in vivo.
Chromatin-mediated repression is now well established to result from deacetylation of histones mediated by histone deacetylases. However, it has also become clear that such hypoacetylated histones are themselves targets for further modification, which includes the methylation of specific lysine residues which results in transcriptional repression (53). Methylation of lysine 9 of histone H3 is such a modification clearly associated with transcriptional repression (54, 61) and is carried out in the cell by a number of specific HMTs, two of which are G9a (63, 64) and Suvar(3-9)H1 (39, 48). Unlike the case with histone acetylation, lysine residues in histones can be multiply methylated by specific HMTs. G9a is believed to be important for mono- and dimethylation of histone H3, whereas Suvar(3-9)H1 is predominantly involved in the trimethylation of lysine residues in histone H3 (50). Methylated histone H3, particularly trimethylated H3, is a known target for recruitment of silencing proteins such as HP1 and is believed to be the foundation for heterochromatic silencing (28, 50, 54, 61), although an involvement with euchromatin silencing is increasingly likely (42, 66). On this basis, we asked whether the mechanism for IE86-mediated autorepression also involved the recruitment HMTs.
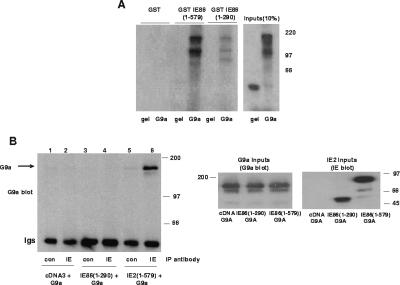
We started by analyzing G9a. Figure 5A shows that full-length IE86(1-579) and G9a interact specifically in GST fusion pull-down assays. No such interaction was observed between IE86 and GST control protein. Similarly, we observed little interaction between G9a and a GST-IE86 fusion containing only amino acids 1 to 290 of IE86. Gelsolin, a sticky negative-control protein confirmed that the interactions between full-length IE86 and G9a were specific.
FIG. 5.
IE86 interacts with G9a in vitro and in vivo. (A) GST, GST fused to full-length IE86 (1-579), or GST fused to amino acids 1 to 290 of IE86 (1-290) on glutathione beads was used as a target for binding to [35S]methionine-labeled full-length G9a or control gelsolin (gel). Input proteins (1/10) are also shown, and molecular mass markers are in kDa. (B) SAOS2 cells were transfected with pcDNA3 (tracks 1 and 2), IE86(1-290) (tracks 3 and 4), or IE86(1-579) (tracks 5 and 6) together with G9a expression vectors. Cell extracts were immunoprecipitated with control antibody or anti-IE antibody, and immunoprecipitated complexes were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blot analysis using a G9a antibody. Extracts were also analyzed directly for levels of expression of G9a (G9a inputs) by G9a Western blot analysis or IE86 (IE86 inputs) by IE Western blot analysis.
We next confirmed that the interaction we had observed in vitro also occurs in vivo. Fig. 5B shows an IP/Western blot analysis of cells coexpressing IE86 and G9a by cotransfection. Immunoprecipitation of IE86 with specific antibody from RB(−ve) SAOS2 cells cotransfected with IE86 and G9a resulted in coprecipitation of G9a as detected by Western blot analysis (track 6). We observed no such coimmunoprecipitation of IE86 and G9a when nonspecific antibodies were used for the IP (track 5) or in the absence of IE86 coexpression (track 2). Similarly, we saw no interaction between IE86(1-290) and G9a (track 4).
As expected, we also observed identical results when IE86 and G9a were coexpressed in RB(+ve) U2OS cells (data not shown). These results suggest that full-length IE86 and G9a are able to interact in vivo in either the presence or absence of RB.
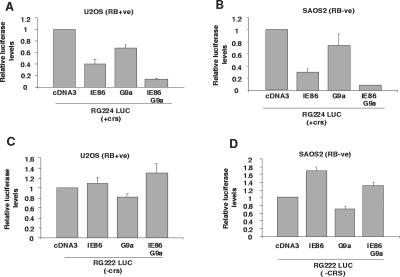
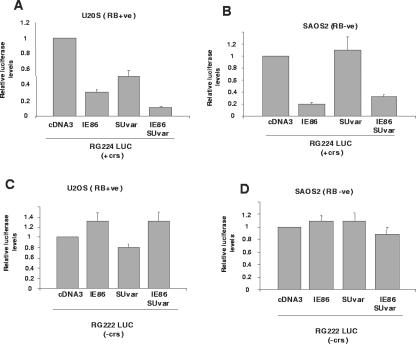
IE86 and G9a act in concert to repress the viral MIEP.
On the basis of IE86 and G9a physically interacting with each other, we tested whether IE86 and G9a functionally interact to repress MIEP activity. Figure 6 shows that IE86, as expected, routinely repressed MIEP activity; this IE86-mediated inhibition occurred in both RB(+ve) U2OS cells (Fig. 6A) and RB(−ve) SAOS2 cells (Fig. 6B) and, as in previous cases, this repression was not observed following the transfection of pRG222 with IE86 (Fig. 6C and D). The transfection of G9a alone with pRG224 resulted in a low level of inhibition of the MIEP. In contrast, the cotransfection of IE86 and G9a together resulted in MIEP inhibition to levels that were more than just independently additive in the pRG224-transfected cells. This effect was observed for both RB(+ve) U2OS cells and RB(−ve) SAOS2 cells, again suggesting that the functional interaction between IE86 and G9a resulting in increased repression of the MIEP does not require interactions with RB. Consistent with this repression being mediated via the crs sequence, the cotransfection of G9a and IE86 with pRG222 resulted in no increase in the level of repression compared to that with G9a alone (Fig. 6C and D).
FIG. 6.
IE86 and G9a act together to repress the MIEP. U2OS [RB(+ve)] (A) or SAOS2 [RB(−ve)] (B) cells were transfected with an MIEP luciferase vector carrying the crs (RG224 LUC) in the presence of 10 μg of control (con) empty vector (cDNA3) vector, 5 μg of cDNA3IE86 (IE86) plus 5 μg of cDNA3, 5 μg of cDNA3G9a (G9a) plus 5 μg of cDNA3, or 5 μg of cDNA3IE86 plus 5 μg of cDNA3G9a together. Cell extracts were then analyzed for luciferase activity. The results shown are the means from at least three independent experiments. The same transfections, with the exception that the MIEP luciferase vector with a crs deletion (pRG222 LUC) was used, were also carried out with U2OS cells (C) and SAOS2 cells (D).
IE86 and Suvar(3-9)H1 physically interact in vitro and in vivo.
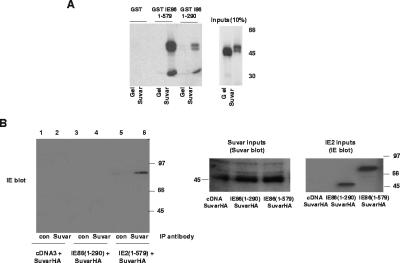
Although histone methylation by Suvar(3-9)H1 is generally associated with constitutive heterochromatin, it has also been shown to be involved in the repression of euchromatic genes, in which it can be recruited to promoters via RB (3, 42). Consequently, we next sought to determine if IE86 also interacts with this HMT. Figure 7A shows, from GST fusion pull-down assays, that IE86 interacts strongly with Suvar(3-9)H1. We observed a much weaker interaction, in comparison, of Suvar(3-9)H1 and a GST-IE86(1-290) expression construct (with amino acids 291 to 579 deleted).
FIG. 7.
IE86 interacts with Suvar(3-9)H1 in vitro and in vivo. (A) GST, GST fused to full-length IE86 (1-579) or GST fused to amino acids 1 to 290 of IE86 (1-290) on glutathione beads was used as a target for binding to [35S]methionine-labeled full-length Suvar(3-9)H1 (Suvar) or control gelsolin (gel). Input proteins (1/10) are also shown, and molecular mass markers are in kDa. (B) U2OS cells were transfected with pcDNA3 (tracks 1 and 2), IE86(1-290) (tracks 3 and 4), or IE86(1-579) (tracks 5 and 6) together with Suvar(3-9)H1 expression vectors. Cell extracts were immunoprecipitated with control antibody or anti-Suvar antibody, and immunoprecipitated complexes were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blot analysis using an E13 antibody. Extracts were also analyzed directly for levels of expression of Suvar(3-9)H1 (Suvar inputs) or IE86 (IE86 inputs) by direct Western blotting of cell extracts.
We confirmed this physical interaction in vivo using an IP/Western blot analysis of U20S cells cotransfected with IE86 and hemagglutinin-tagged Suvar(3-9)H1 (Fig. 7B). Immunoprecipitation of Suvar(3-9)H1 from RB(+ve) U2OS cells cotransfected with IE86 and Suvar(3-9)H1 resulted in coprecipitation of IE86 as detected by Western blot analysis (track 6). We observed no such coimmunoprecipitation of IE86 when nonspecific antibodies were used for the IP (track 5) or in the absence of IE86 coexpression (track 2). Similarly, we saw no interaction between IE86(1-290) and Suvar(3-9)H1 (track 4). These results suggest that full-length IE86 and Suvar(3-9)H1 are able to interact in vivo in RB(+ve) U2OS cells.
We saw, in contrast to our observations with G9a, only interactions between IE86 and Suvar(3-9)H1 in RB(+ve) U2OS cells. Despite multiple attempts, we have been unable to show this interaction in SAOS2 cells (data not shown). This suggests that the physical interaction between IE86 and Suvar(3-9)H1 requires the presence of RB, as has been suggested for other systems (3).
IE86 and Suvar(3-9)H1 functionally interact to repress the viral MIEP in RB(+ve) cells.
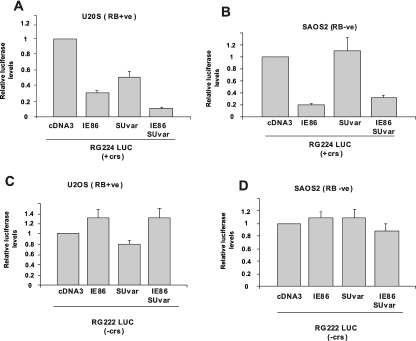
We next tested whether the physical interaction we had observed between IE86 and Suvar(3-9)H1, at least in the presence of RB, was similarly reflected in a functional interaction with respect to repression of the MIEP (Fig. 8). Once again, we cotransfected IE86 with or without Suvar(3-9)H1 into RB(+ve) U2OS cells (Fig. 8A) or RB(−ve) SAOS2 cells (Fig. 8B) and analyzed the effects on luciferase expression driven from the MIEP. As expected, IE86 routinely repressed the MIEP in U2OS cells (Fig. 8A). This IE86-mediated repression was increased by cotransfection with Suvar(3-9)H1 (Fig. 8A).
FIG. 8.
IE86 and Suvar(3-9)H1 act together to repress the MIEP, but only in the presence of RB. SAOS2 [RB(−ve)] (A) or U2OS [RB(+ve)] (B) cells were transfected with an MIEP luciferase vector carrying the crs (RG224 LUC) in the presence of 10 μg of control empty vector (cDNA3) vector, 5 μg of cDNA3IE86 (IE86) plus 5 μg of cDNA3, 5 μg of cDNA3Suvar(3-9)H1-HA plus 5 μg of cDNA3 (Suvar), or 5 μg of cDNA3IE86 plus 5 μg of cDNA3Suvar(3-9)H1-HA (IE86 Suvar) together. Cell extracts were then analyzed for luciferase activity. The results show the means from at least three independent experiments. The same transfections, with the exception that the MIEP luciferase vector with a crs deletion (pRG222 LUC) was used, were also carried out with U2OS cells (C) and SAOS2 cells (D).
In contrast, a similar analysis in RB(−ve) SAOS2 cells (Fig. 8B) showed that in the absence of RB, there is no corepression by IE86 and SuVar(3-9)H1, suggesting that any ability of IE86 and Suvar(3-9)H1 together to corepress the MIEP interaction is RB dependent. Again, also consistent with the corepression of the MIEP in U2OS and SAOS2 cells by IE86 and Suvar(3-9)H1 being mediated via the crs, the pRG222 luciferase reporter with a crs deletion showed no such corepression by IE86 and Suvar(3-9)H1 together in U2OS and SAOS2 cells (Fig. 8C and D).
The viral MIEP becomes associated with repressive chromatin markers at late times of infection.
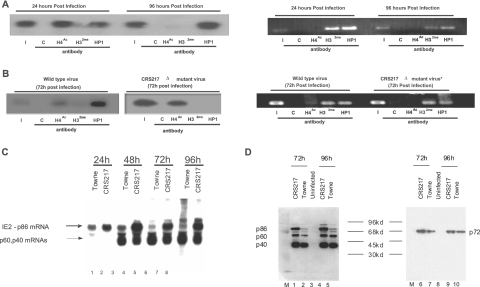
If autorepression of the viral MIEP at late times of infection is, indeed, a result of the recruitment of functional HDACs and HMTs to the MIEP, then we would predict that the state of chromatinization of the viral MIEP should change during the course of virus infection. Therefore, we used ChIP assays to analyze the state of histone modification around the viral MIEP at both early and late times of virus infection, when the MIEP is highly active and when IE86 should be mediating autorepression, respectively. Figure 9A shows that in cells infected with HCMV at a multiplicity of infection of 0.5 for 24 h, a substantial amount of the viral MIEP is associated with acetylated histone H4, consistent with the known high levels of major IE transcription which occur at this time during infection. A smaller amount of the viral MIEP appears to be in a transcriptionally repressive chromatin structure as shown by the association of the MIEP with the silencer protein HP1. This likely reflects intrinsic repression of the viral MIEP observed at very early times of infection by factors such as ND10 (see Discussion). However, as virus infection progresses, the MIEP in viral genomes becomes substantially more associated with the silencing protein HP1 such that at 96 h postinfection, the MIEP is almost totally associated with the silencing protein HP1 and it shows no association with acetylated histone H4.
FIG. 9.
The viral MIEP undergoes chromatin remodeling at late times of infection, and it is dependent on the crs. (A) Fibroblasts were infected with HCMV and analyzed by ChIPs at 24 h or 96 h postinfection using a control antibody (C), an acetylated histone H4-specific antibody (H4Ac), a dimethylated histone H3-specific antibody (H32me), or an antibody specific for HP-1. Immunocomplexes were analyzed by PCR to detect the viral MIEP. Input material (I) is also shown. The same samples were also analyzed by a PCR specific for the human γ-globin promoter (right panel). (B) Fibroblasts were infected with wild-type HCMV or HCMV with a mutated crs (CRS217 Δ) and analyzed by ChIPs at 72 h postinfection as described for panel A above. The same samples were also analyzed by a PCR specific for the human γ-globin promoter (right panel). (C) Total RNA from fibroblasts infected with Towne or CRS217 virus was isolated at 24 to 96 h postinfection, and 10 μg of the RNA was analyzed by Northern blotting. The filter was incubated with a 32P-radiolabeled exon-5-specific probe and hybridization detected by autoradiography. (D) Total protein isolated from 6 × 103 fibroblasts infected with Towne or CRS217 virus between 72 and 96 h postinfection was analyzed by Western blot analysis for IE72 expression using an exon-4-specific antibody, and then the filter was reprobed with an anti-exon-5 antibody to show levels of IE86/p60/p40 expression.
The right panel of Fig. 9A shows a ChIP analysis of the human γ-globin gene promoter, a promoter which is silenced in differentiated cell types (5). This promoter is not affected by CMV infection and remains in the same transcriptionally repressed state. As expected, it remained associated with dimethylated histone H3 and HP1 and was not subject to chromatin remodeling, confirming that the results shown with the viral MIEP-specific PCR do not result from differential immunoprecipitation of chromatin samples.
These observations would be entirely consistent with a progressive switch from high levels of transcriptional activity of the MIEP at immediate-early/early times of infection to a general state of repression of the MIEP at late times of infection. We would also predict that this progression from a transcriptionally active open chromatin configuration to a more compacted transcriptionally silenced chromatin configuration is dependent on autorepression by IE86, specifically, in a crs-dependent way. Consequently, we tested whether a virus genome with crs deleted would still be subject to the same sequence of changes in chromatin structure around the MIEP which are associated with progression of infection to late times of the lytic cycle in wild-type virus with a functional crs. Figure 9B shows that, at 72 h postinfection with wild-type virus (a multiplicity of infection of 2), the MIEP is substantially associated with HP1, as expected from the previous experiment. The observed association of a proportion of the MIEPs with acetylated H4, even at 72 h postinfection, probably reflects asynchronous infection of the cell population: activation and subsequent IE86-mediated repression of the MIEP may well be out of sync in the total cell population (see Discussion). In contrast, the same analysis with a virus with a crs mutation showed the viral MIEP to be associated almost exclusively with acetylated histone H4 with no association with HP1 at this time point of infection. The data in Fig. 9C confirm that the infections with CRS217 and wild-type viruses were temporally at comparable stages, since the expression of the p40/p60 late RNAs, whose expression is independent of the MIEP, were at equivalent levels, suggesting that the infections were proceeding at relatively equivalent rates. The right panel of Fig. 9B also shows control ChIP assays for the same samples with the human γ-globin gene promoter, once again confirming that the results shown with the MIEP-specific PCR do not result from differential immunoprecipitation of chromatin samples. This strongly argues that the recruitment of HP1 and the formation of a transcriptionally silenced chromatin structure of the viral MIEP apparently mediated by IE86, at late times of infection, absolutely require a functional crs.
Having observed that the deletion of the crs region from HCMV resulted in a dramatic decrease in the methylation (and concomitant increase in acetylation) of histones bound to the MIEP, we wanted to confirm that this correlated with increased major IE gene expression at later times of infection, times when the MIEP is normally silenced. Figure 9C shows a Northern blot analysis of IE86, IE60, and IE40 expression in both wild-type (Fig. 9C, lanes 1, 3, 5, and 7) and CRS217 mutant (Fig. 9C, lanes 2, 4, 6, and 8) virus-infected cells at 24 to 96 h postinfection. At 24 h postinfection, similar levels of IE86 expression are observed in cells infected with wild-type (Fig. 9C, lane 1) and CRS217 (Fig. 9C, lane 2) viruses. However, in the cells infected with wild-type virus, the level of IE86 RNA expression decreased, which is consistent with transcriptional silencing of the MIEP. In contrast, in cells infected with CRS217 virus, the level of IE86 RNA did not decrease between 24 and 96 h postinfection. This difference was specific to IE86, as both the p40 and p60 transcripts, which are expressed at later times during infection (44), showed similar levels of abundance at later times of infection. A similar analysis of IE72 and IE86 by Western blotting (Fig. 9D) shows that the transcriptional activity of the MIEP directly correlates with protein expression as well. The expression of IE86 (Fig. 9D, lanes 1 to 5) was more abundant in the CRS217-infected cells (lanes 1 and 4) than in wild-type HCMV-infected cells (lanes 2 and 5) at 72 to 96 h postinfection. Similarly, the IE72 protein was also more abundant in CRS217-infected cells (lanes 6 and 9) than in wild-type HCMV-infected cells (lanes 7 and 10). However, this difference was specific to the major IE genes, as no difference in p40 or p60 late gene expression was observed in the same cells.
DISCUSSION
Coordinated regulation of specific viral gene products at specific stages of the virus infection cycle is a well-documented phenomenon during herpesvirus infection. As well as IE gene expression positively acting to ensure that viral genes are expressed in a temporally correct fashion, many other herpesviruses infections also result in negative autoregulation of viral IE gene expression, presumably so as not to waste valuable resources in expressing genes which are no longer needed during later stages of the virus infection cycle. Autoregulation of the HCMV MIEP by IE86 is well established, but how IE86 mediated such regulation was previously unclear. Our results now show that autorepression by IE86 at late stages of viral infection is correlated to changes in chromatin structure around the MIEP during the course of infection and that this is likely to result from physical and functional interactions between IE86 and chromatin remodeling enzymes which are normally associated with transcriptional repression of cellular promoters.
As expected (1, 12, 24, 30-32, 43, 56, 60, 67, 72), IE86 routinely repressed the HCMV MIEP, and this repression was absolutely dependent on the crs present in the MIEP. However, this autorepression by IE86 was also substantially inhibited by the histone deacetylase inhibitor TSA, suggesting that IE86-mediated autorepression, at least in part, occurs through mechanisms involving histone deacetylases.
Consistent with this, our results also showed that IE86 interacted, in vitro and in vivo, with the histone deacetylase HDAC1; we observed a specific and reproducible interaction between full-length IE86 and HDAC1. This interaction also occurred in RB(−ve) cells, ruling out the possibility that IE86 and HDAC1 interact indirectly by bridging through a common interaction partner of both proteins, RB. Consistent with this, IE86 was also able to bind functional histone deacetylase. Recently, Nevels et al. (41) suggested that another major immediate-early protein of HCMV, IE72, is able to physically interact with another HDAC family member, HDAC3. However, we observed only low levels of interaction between IE72 and functional histone deacetylase enzyme activity in our analyses.
Although we clearly observed physical interaction between IE86 and HDAC1 both in vitro and in vivo, cotransfection of IE86 and HDAC1 did not result in any cooperative or synergistic effects on repression of the viral MIEP greater than the observed effects of IE86 or HDAC1 alone. In many experimental systems, cotransfection of a repressor and corepressors, whose effects are mediated through physical interactions, often results in repression levels that are cooperative or synergistic in such assays. It is possible that the cell types used for transfections here already have substantial levels of HDAC1; HDAC1 is never limiting, so the expression of additional levels of HDAC1 has no obvious effects on the level of repression mediated by IE86 alone in these cells.
Deacetylated histones are, themselves, known to be targets for further modifications which are associated with chromatin-mediated repression; deacetylated histones are subsequent targets for methylation by HMTs. Consequently, we also analyzed the ability of IE86 to recruit such HMTs. We observed a strong interaction between IE86 and G9a, an HMT which is known to mono- and dimethylate histone H3 on specific lysine residues associated with transcriptional repression. This interaction occurred in vitro and in vivo and did not require the presence of RB. In contrast to HDAC1, cotransfection of IE86 with G9a routinely resulted in levels of repression of the MIEP which were more than additive compared to levels of repression of the MIEP by IE86 or G9a alone. As with the physical interaction between IE86 and G9a, this functional interaction between these two proteins was also RB independent.
Mono- and dimethylated H3 histones are subsequent targets for trimethylation by HMTs such as Suvar(3-9)H1, and following trimethylation, these histones act to recruit silencing proteins such as HP1, and this is believed to be the foundation for heterochromatic silencing (28, 50, 54, 61). However, HP1 has also been shown to be involved in repression of euchromatic genes, in which it can be recruited to promoters via RB (3, 42). On this basis, we asked whether IE86 is also able to recruit HMTs involved in trimethylation of H3 as part of its mechanism for autorepression. Once again, we observed a good interaction between IE86 and Suvar(3-9)H1 both in vitro and in vivo. However, we observed the in vivo interaction only in RB(+ve) cell lines, arguing that the interaction between IE86 and Suvar(3-9)H1 is likely to be bridged by RB. Consistent with this, cotransfection of IE86 with Suvar(3-9)H1 routinely resulted in corepression of the MIEP; levels of repression of the MIEP were increased when IE86 and Suvar(3-9)H1 were coexpressed compared to expression of IE86 or Suvar(3-9)H1 alone. However, this was only observed in RB(+ve) cell lines.
These results suggest that negative autoregulation by IE86 at late times of infection could result, at least in part, from IE86-mediated recruitment of such chromatin remodeling enzymes. Whether or not IE86 acts as a nucleation center for all three chromatin remodeling factors simultaneously, with Suvar(3-9)H1 recruitment also involving a further interaction with RB, as has been suggested for other transcriptional repressors (6, 61, 66), is, as yet, unclear. We do know that IE86 can bind HDAC1 and RB simultaneously (J. Sinclair and L. Teague, unpublished observations), but whether or not recruitment of a large repressor complex containing all these factors by IE86 occurs or whether repressors are recruited sequentially and independently by IE86, our observations suggested that changes in chromatin structure of the viral MIEP should occur as HCMV infection progresses to late times of the virus life cycle.
These data do not preclude the possibility that there are other mechanisms important for the regulation of IE gene expression during other stages of infection. Previous studies have shown that steady-state levels of IE72 and IE86 RNA are decreased as early as 8 h postinfection (38, 47), a time at which we do not observe the recruitment of repressive chromatin to the MIEP. It is possible that this repression at IE times of infection does not involve chromatin remodeling but rather involves a direct, IE86-mediated steric hindrance of RNA polymerase II binding to the MIEP, as has been suggested from in vitro analyses. However, it is clear that, at late times of infection, the viral MIEP is associated with chromatin markers indicative of transcriptional repression. Therefore, it is not unlikely that HCMV employs multiple mechanisms during infection to regulate major immediate-early gene expression.
At immediate-early times of infection, the viral MIEP is predominantly associated with markers of transcriptionally active chromatin, consistent with the concomitant high levels of major IE transcription which are known to occur at this time of infection. However, as the virus infection progressed, the MIEP in viral genomes became totally associated with the silencing protein HP1, a defined marker of transcriptionally repressed chromatin. These analyses, however, also showed that at 24 h postinfection, a proportion of viral MIEPs were already associated with HP1, suggesting that chromatin-mediated repression of the viral MIEP by IE86 may be occurring as early as 24 h postinfection. Alternatively, the ChIP assay will depict an analysis of the whole infected population of cells, some of which may not be fully supportive of IE gene expression immediately upon infection; cells in G2/M are not permissive for IE gene expression, for, so far, undefined reasons (51). As these cells will initiate IE gene expression only after cells have cycled back into G1, activation and subsequent IE86-mediated repression of the MIEP may well be out of sync in the total cell population. This is likely why some MIEPs show an association with acetylated H4 histone at apparently later times of infection: the infection in some cells is delayed until the cell returns to an environment conducive for a lytic infection. Similarly, it is now becoming clear that proteins with intrinsic antiviral functions, such as cellular ND10, may mediate their repressive action very early on in infection via chromatinization of the viral MIEP (D. L. Woodhall, M. B. Reeves, I. J. Groves, G. W. G. Wilkinson, and J. H. Sinclair, submitted for publication). This may alternatively explain why a proportion of the viral MIEPs are in a repressive chromatin structure at very early times of infection.
Notwithstanding this, it is clear that as the infection proceeds to late stages, there is a progressive switch from high levels of transcriptional activity of the MIEP at immediate-early/early times of infection to a general state of repression of the MIEP at late times of infection, and this is concomitant with an almost total association of viral MIEPs with HP1 protein at 96 h postinfection. Confirmation that such chromatin remodeling of the viral MIEP during the time course of HCMV infection was dependent on IE86 binding to the viral crs came from the observation that removal of the crs in the context of the virus resulted in a total abrogation of the recruitment of the transcriptionally repressive chromatin protein HP1 normally found around the viral MIEP at late times of infection and, consistent with this, increased IE72 and IE86 transcription and protein expression at later times of infection when the MIEP is normally transcriptionally silent.
Taken together, our observations strongly suggest that autorepression of the HCMV major immediate-early promoter/enhancer at later times of infection results from the IE86-mediated recruitment of chromatin remodeling enzymes to the MIEP resulting in modification of the MIEP to a repressive chromatin phenotype at late times of infection. Recently, Nevels et al. (41) suggested that the ability of IE72 and IE86 to interact with HDAC3 is key to IE-mediated activation of viral early gene expression. We have also observed previously that IE86 can activate cellular promoters essentially by preventing their HDAC-mediated repression (J. Murphy, M. Bain, and J. Sinclair, unpublished observations). We believe that the differential ability of IE86 to repress its own promoter but activate cellular and other viral promoters through physical interaction with HDACs may ultimately depend on whether IE86 binds and recruits HDACs to promoters (as in the case of binding to the crs in the MIEP) or whether it sequesters HDACs away from other specific promoter elements. Experiments to address this specifically are in progress.
Acknowledgments
We thank Joan Baillie and Linda Teague for excellent technical assistance. We are indebted to Tony Kouzarides and Andy Bannister for reagents.
This work was supported by grants from the Medical Research Council (United Kingdom) and The Wellcome Trust.
REFERENCES
- 1.Arlt, H., D. Lang, S. Gebert, and T. Stamminger. 1994. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J. Virol. 68:4117-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asmar, J., L. Wiebusch, M. Truss, and C. Hagemeier. 2004. The putative zinc finger of the human cytomegalovirus IE2 86-kilodalton protein is dispensable for DNA binding and autorepression, thereby demarcating a concise core domain in the C terminus of the protein. J. Virol. 78:11853-11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 5.Bottardi, S., A. Aumont, F. Grosveld, and E. Milot. 2003. Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood 102:3989-3997. [DOI] [PubMed] [Google Scholar]
- 6.Boulias, K., and I. Talianidis. 2004. Functional role of G9a-induced histone methylation in small heterodimer partner-mediated transcriptional repression. Nucleic Acids Res. 32:6096-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm, A., and T. Kouzarides. 1999. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 24:142-145. [DOI] [PubMed] [Google Scholar]
- 8.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597-601. [DOI] [PubMed] [Google Scholar]
- 9.Bryant, L. A., P. Mixon, M. Davidson, A. J. Bannister, T. Kouzarides, and J. H. Sinclair. 2000. The human cytomegalovirus 86-kilodalton major immediate-early protein interacts physically and functionally with histone acetyltransferase P/CAF. J. Virol. 74:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caswell, R., L. Bryant, and J. Sinclair. 1996. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J. Virol. 70:4028-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caswell, R., C. Hagemeier, C. J. Chiou, G. Hayward, T. Kouzarides, and J. Sinclair. 1993. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J. Gen. Virol. 74:2691-2698. [DOI] [PubMed] [Google Scholar]
- 12.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari, B. A., E. Poma, T. F. Kowalik, S. M. Huong, and E. S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 67:4981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves, R. F., J. M. Brown, J. Vieira, and E. S. Mocarski. 1995. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J. Gen. Virol. 76:2151-2160. [DOI] [PubMed] [Google Scholar]
- 18.Hagemeier, C., R. Caswell, G. Hayhurst, J. Sinclair, and T. Kouzarides. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier, C., S. Walker, R. Caswell, T. Kouzarides, and J. Sinclair. 1992. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J. Virol. 66:4452-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassig, C. A., and S. L. Schreiber. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300-308. [DOI] [PubMed] [Google Scholar]
- 21.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C. H., and J. Y. Chen. 2002. Identification of additional IE2-p86-responsive cis-repressive sequences within the human cytomegalovirus major immediate early gene promoter. J. Biomed. Sci. 9:460-470. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins, P. J., U. K. Binne, and P. J. Farrell. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jupp, R., S. Hoffmann, A. Depto, R. M. Stenberg, P. Ghazal, and J. A. Nelson. 1993. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J. Virol. 67:5595-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 29.Lang, D., S. Gebert, H. Arlt, and T. Stamminger. 1995. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J. Virol. 69:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 22:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lashmit, P. E., M. F. Stinski, E. A. Murphy, and G. C. Bullock. 1998. A cis repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection. J. Virol. 72:9575-9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, G., J. Wu, P. Luu, P. Ghazal, and O. Flores. 1996. Inhibition of the association of RNA polymerase II with the preinitiation complex by a viral transcriptional repressor. Proc. Natl. Acad. Sci. USA 93:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 36.Magnaghi-Jaulin, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601-605. [DOI] [PubMed] [Google Scholar]
- 37.Meier, J. L. 2001. Reactivation of the human cytomegalovirus major immediate-early regulatory region and viral replication in embryonal NTera2 cells: role of trichostatin A, retinoic acid, and deletion of the 21-base-pair repeats and modulator. J. Virol. 75:1581-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meier, J. L., and M. F. Stinski. 1997. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J. Virol. 71:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melcher, M., M. Schmid, L. Aagaard, P. Selenko, G. Laible, and T. Jenuwein. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20:3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 43.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 46.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puchtler, E., and T. Stamminger. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 49.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. USA 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12:1591-1598. [DOI] [PubMed] [Google Scholar]
- 51.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schotta, G., M. Lachner, A. H. Peters, and T. Jenuwein. 2004. The indexing potential of histone lysine methylation. Novartis Found. Symp. 259:22-47, 163-169. [PubMed] [Google Scholar]
- 54.Schotta, G., M. Lachner, K. Sarma, A. Ebert, R. Sengupta, G. Reuter, D. Reinberg, and T. Jenuwein. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreiber, S. L., and B. E. Bernstein. 2002. Signaling network model of chromatin. Cell 111:771-778. [DOI] [PubMed] [Google Scholar]
- 56.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stenberg, R. M. 1996. The human cytomegalovirus major immediate-early gene. Intervirology 39:343-349. [DOI] [PubMed] [Google Scholar]
- 60.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stewart, M. D., J. Li, and J. Wong. 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stinski, M. F., D. R. Thomsen, R. M. Stenberg, and L. C. Goldstein. 1983. Organization and expression of the immediate early genes of human cytomegalovirus. J. Virol. 46:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 64.Tachibana, M., K. Sugimoto, M. Nozaki, J. Ueda, T. Ohta, M. Ohki, M. Fukuda, N. Takeda, H. Niida, H. Kato, and Y. Shinkai. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16:1779-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 66.Vandel, L., E. Nicolas, O. Vaute, R. Ferreira, S. Ait-Si-Ali, and D. Trouche. 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21:6484-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waheed, I., C. J. Chiou, J. H. Ahn, and G. S. Hayward. 1998. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology 252:235-257. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Y., W. Fischle, W. Cheung, S. Jacobs, S. Khorasanizadeh, and C. D. Allis. 2004. Beyond the double helix: writing and reading the histone code. Novartis Found. Symp. 259:3-21, 163-169. [PubMed] [Google Scholar]
- 69.Wathen, M. W., and M. F. Stinski. 1982. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J. Virol. 41:462-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wright, E., M. Bain, L. Teague, J. Murphy, and J. Sinclair. 2005. Ets-2 repressor factor recruits histone deacetylase to silence human cytomegalovirus immediate-early gene expression in non-permissive cells. J. Gen. Virol. 86:535-544. [DOI] [PubMed] [Google Scholar]
- 72.Wu, J., R. Jupp, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J. Virol. 67:7547-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]