Abstract
The ubiquitous human polyomavirus JC (JCV) is a small double-stranded DNA virus that establishes a persistent infection, and it is often transmitted from parents to children. There are at least 14 subtypes of the virus associated with different human populations. Because of its presumed codivergence with humans, JCV has been used as a genetic marker for human evolution and migration. Codivergence has also been used as a basis for estimating the rate of nucleotide substitution in JCV. We tested the hypothesis of host-virus codivergence by (i) performing a reconciliation analysis of phylogenetic trees of human and JCV populations and (ii) providing the first estimate of the evolutionary rate of JCV that is independent from the assumption of codivergence. Strikingly, our comparisons of JCV and human phylogenies provided no evidence for codivergence, suggesting that this virus should not be used as a marker for human population history. Further, while the estimated nucleotide substitution rate of JCV has large confidence intervals due to limited sampling, our analysis suggests that this virus may evolve nearly two orders of magnitude faster than predicted under the codivergence hypothesis.
Polyomaviruses, with genomes of approximately 5 kb, are the smallest known double-stranded DNA viruses and infect a variety of avian and mammalian species, including humans. The persistent human polyomavirus JC (JCV) has an estimated seroprevalence of 70 to 90% (26). While 20 to 80% of adults continuously excrete JCV in their urine (8), almost all infections are benign, only causing the demylating neurological disease progressive multifocal leukoencephalopathy in immunocompromised patients (1, 38). JCV has attracted considerable attention, as it can be divided into a number of subtypes (n > 14) that are associated with different human population groups. For example, types 3 and 6 are found in Africans, type 7A in southeast Asians, and types 1 and 4 in Europeans (11). This, together with evidence that the virus is often transmitted from parent to child postnatally but in a quasi-vertical manner (22, 41), has led to the theory that the virus has codiverged with human populations over thousands of years (2, 31). This, in turn, has resulted in the widespread use of JCV as a marker for human evolution and migration (28, 32, 40). Likewise, the estimated rate of substitution in JCV, at ∼4 × 10−7 synonymous substitutions per site per year (subs/site/year), is based on the assumption of codivergence with human populations, with host divergence times used to calibrate those of the virus (17, 32).
Although long-term codivergence and consequently low rates of nucleotide substitution have been supported in some DNA viruses, specifically herpesviruses and papillomaviruses (5, 24, 25), the extent of codivergence between JCV and human populations has not been rigorously tested. However, a previous study of genetic variation noted differences in the demographic histories of JCV and human populations, implying that factors besides human population structure have shaped viral diversity (39). Similarly, an evolutionary rate for JCV that is independent of calibration through codivergence has not been obtained, making it difficult to ascertain if rates derived so far are valid. Indeed, if the polyomaviruses do evolve as slowly as estimated under a codivergence hypothesis, then an independent estimate, based on sequence variation observed over a short time period (as described in reference 15), should be impossible, as substitutions would accumulate too slowly to measure. Further, the assumption that all DNA viruses evolve orders of magnitude more slowly than RNA viruses has recently been challenged. In particular, an interhost rate of approximately 10−4 subs/site/year, close to that of many RNA viruses, has been observed in the small, 5-kb single-stranded DNA parvoviruses (30), and an intrahost rate of roughly 10−5 subs/site/year was estimated for the human polyomavirus BK (BKV) (10). Herein we provide a systematic study of JCV-human codivergence.
MATERIALS AND METHODS
Sequences.
Three hundred thirty-three JCV sequences, 4,798 bp in length, were compiled from GenBank (the “total data set”). These sequences represented complete viral genomes, excluding the 276-bp 5′ noncoding region and two internal fragments, approximately 9 and 47 bp in length, which contained several unalignable regions. GenBank accession numbers, host ethnicity (as given by the publishing author), isolate name, and isolation dates are given in Table S1 in the supplemental material. Sequences were aligned manually using the Se-Al program (http://evolve.zps.ox.ac.uk/). We obtained sampling dates, through direct communication with authors or from published material, for 231 of these isolates and 11 additional isolates for which host ethnicity was not known. In cases where sequences assigned the same subtype were also isolated from the same population at the same time point by the same author, two of these sequences were randomly selected (http://www.randomizer.org/). This resulted in a data set of 158 dated sequences, termed the “dated data set,” highlighted in boldface in Table S1 in the supplemental material.
To compare the population dynamics (including population growth rates) of JCV and their human hosts, we compiled a corresponding data set of 158 human mitochondrial DNA (mtDNA) sequences from the mitochondrial database mtDB (http://www.genpat.uu.se/mtDB/). This data set reflected, as far as possible, the populations from which the viral isolates were sampled. Entire mtDNA genomes, with the exception of the 1,120-bp noncoding “D-loop,” which evolves at a higher rate than the rest of the mitochondrial genome (19), were aligned manually. Accession numbers and mitochondrial host populations are given in Table S2 in the supplemental material.
Phylogenetic analyses.
To determine the phylogenetic relationships of all JCV strains, we used maximum likelihood (ML), neighbor-joining (NJ), and Bayesian Markov chain Monte Carlo (MCMC) approaches to infer three individual trees. ML and NJ phylogenies were estimated with the GTR+I+Γ4 model of nucleotide substitution, available in PAUP* (33). ML trees used SPR (subtree pruning regrafting) and TBR (tree bisection-reconnection) branch swapping, with 100,000 and 200,000 rearrangements, respectively. All parameter values were estimated from the data, and bootstrap values were calculated using 1,000 replicate NJ trees on the ML substitution model. Bayesian trees were estimated with the program MrBayes 3, with the HKY85+I+Γ4 model of nucleotide substitution (29). The MCMC chain was run for 9 million generations (with a burn-in of 850,000 generations), with sampling every 1,000 generations. The tree with the highest posterior probability, i.e., the MAP (maximum a posteriori) tree, was found, and posterior probabilities for nodes were calculated from a consensus tree derived from the same MCMC chain, sampling every 500 generations after a burn-in of 850,000 generations. All trees were midpoint rooted, as no suitable outgroup is known. (The most closely related virus, BKV, is ∼22% divergent from the JCV strains in our data set. As the maximum diversity of the JCV sequences is only 2.7%, BKV is not sufficiently similar to constitute a reliable outgroup.)
These three trees were used as a basis for constructing the JCV phylogeny that was used as input for the TreeMap analysis (see below). Because TreeMap does not allow viruses to have multiple hosts, JCV subtypes which both shared a host population and were located on sister or neighboring branches were combined onto a single branch. The ML phylogeny gave a polytomy comprising the three clades 7A, 7C1/C2, and 7B1/B2, yet the NJ tree indicated an initial divergence of 7A and the MAP tree indicated an initial divergence of 7B1/B2. To ensure all possible topologies were explored, concise trees were constructed from both resolutions (labeled a and b, respectively). Subtypes 2B, 2E, and 2D3 were excluded because of either their unresolved positions or limited sampling.
The phylogenetic tree of human populations, labeled i, was constructed in accordance with those proposed by Cavalli-Sforza and Feldman (6) and Cavalli-Sforza et al. (7). The variant human tree ii was constructed by placing the Caucasoid branch in the position proposed by Ayub et al. (3) and Uinuk-ool et al. (36). Finally, tree iii shows the Indian population branching separately because of controversy regarding the affinity of Indian populations to Asians and Europeans (4) (see Fig. S3 in the supplemental material).
Cophylogenetic reconciliation and significance testing.
To determine the degree of JCV and human phylogenetic congruence, we used the program TreeMapv2.0 (http://taxonomy.zoology.gla.ac.uk/rod/treemap.html) (9, 20). A “tanglegram” was created by matching each subtype with the host population in which it is predominantly found. From this, a graph (a “jungle”) was created which includes all optimal mappings of viral tree nodes onto host tree nodes. The potentially optimal solutions, or POpt, for each of the tanglegrams were determined by weighing the noncoevolutionary events (NCEs) required to reconcile the host and virus trees (see Table S3 in the supplemental material). NCEs include viral duplication, host population transfer, and the loss of a virus by a host population. When determining the POpt, an upper limit was put on NCEs at the point where NCE + 1 fails to result in reconciliations with a greater number of codivergence events. Those maps in the jungle that were optimal with respect to these evolutionary events (i.e., maps that infer the maximum number of codivergences with the minimum number of NCEs to explain the phylogenetic congruencies and incongruencies) were analyzed (20). To test the null hypothesis that the JCV tree is no more congruent with the host tree than a random tree would be, 100 viral phylogenies in which the branches were randomized were mapped onto host phylogenies. We then determined which proportion of these reconciliations showed the same or more codivergence events, or the same or fewer NCEs, as the “optimal” trees. Using the same analysis, the consensus tree of the 158 human mtDNA genome data set was also compared to the host phylogenies described above.
Nucleotide substitution rates and population dynamics.
To estimate rates of nucleotide substitution in JCV and to compare the population dynamics of JCV and human mtDNA, we used a Bayesian MCMC approach (the BEASTv1.3 package; http://evolve.zoo.ox.ac.uk). This method considers differences in branch lengths among viruses sampled at different times and explores evolutionary models whose parameters include tree topology, substitution rate, and population size changes. Bayesian skyline plots, with 10 population groups of unique sizes, were used to infer demographic history (10 grouped intervals were used) (16). Phylogenies were evaluated using a chain length of 40 million states under the HKY85+Γ4 substitution model and with uncertainty in the data reflected in the 95% high-probability density (HPD) intervals. An uncorrelated lognormal relaxed molecular clock model (14) was employed for JCV genome analyses, while a strict clock and a fixed (known) substitution rate were used for the analysis of human mtDNA, as these sequences have been shown to evolve in a roughly clock-like manner (19). Population growth curves were estimated over a 22-million-state chain with a fixed substitution rate parameter.
RESULTS
JCV subtype and host ethnicity determination.
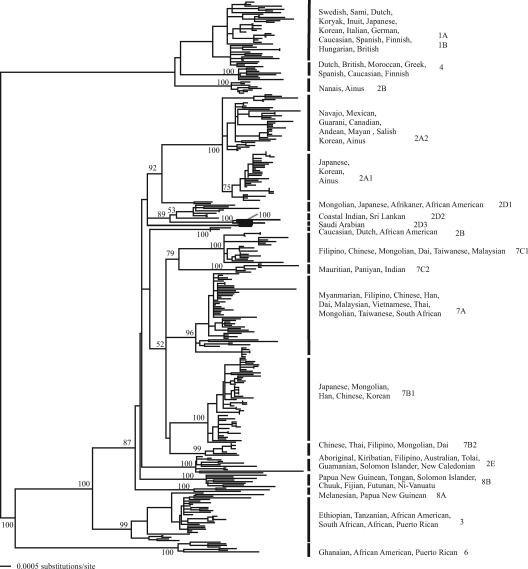
Phylogenetic trees of 333 JCV genomic sequences were inferred using ML (Fig. 1), NJ, and Bayesian MCMC approaches (see Fig. S1a and S1b in the supplemental material). The midpoint-rooted ML, NJ, and Bayesian MCMC MAP trees exhibited very similar topologies, as did trees estimated with TBR and SPR branch swapping. The major subtypes of JCV largely form distinct clades, generally associated with geographical regions. However, it is also clear that the correspondence between subtypes and specific host populations is not as strict as previously suggested. Indeed, a number of clear exceptions can be found, the most striking of which is the basal divergence of the JCV clade containing subtypes 1 and 4, which infect European populations, from all other subtypes. While in human phylogenies the African sequences are the most divergent, this does not appear to be the case in JCV. Furthermore, within the European clade there are multiple JCV strains isolated from the Ainus, Nanais, Koryak, and Japanese populations. Other exceptions to standard interpretations of human population history follow: (i) the JCV subtypes from the Middle East and India are separated from the European JCV subtypes; (ii) the Caucasoid clade does not form a sister group with the northeast Asia/Native American clade; (iii) the presence of two phylogenetically distinct Indian subtypes (7C2 and 2D2); (iv) the presence of two distinct clades of European viruses (subtypes 1 and 4), both containing viruses sampled from Britain, Finland, The Netherlands, and Spain; (v) the presence of multiple Korean isolates within the Native American subtype (2A2); and (vi) the lack of monophyly for those viruses from Native Americans (present in subtypes 1a and 2A2).
FIG. 1.
Phylogenetic tree of 333 JCV genomes inferred using a maximum likelihood approach. The tree is midpoint rooted, and clades are labeled with the range of host ethnicities found in the clade (designations are those submitted by the publishing author) followed by the subtype designation. Bootstrap values, calculated using 1,000 replicate NJ trees, are labeled for relevant nodes with >50% support.
Host-virus codivergence analyses.
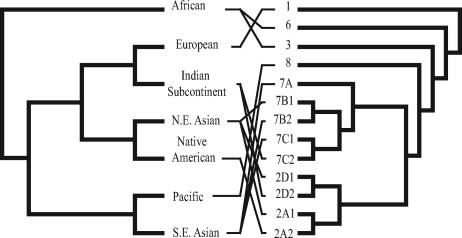
To address the extent of correspondence between viral subtypes and host populations more rigorously, we constructed two summary viral trees (identical except for the resolution of the polytomy comprising the three clades 7A, 7C1/C2, and 7B1/B2). The thirteen well-supported subtype clades, which are predominantly associated with a specific host population, were each represented by a single branch.
To compare the evolutionary histories of virus and host, the consensus JCV trees were each mapped onto all three possible human population phylogenies (i, ii, and iii), creating “tanglegrams” (Fig. 2; see Fig. S3 in the supplemental material; Table 1). Although some mismatch is to be expected given human population admixture, it is striking that none of the six cophylogenetic solution sets had reconciliations with more than five lineage codivergences (i.e., 10 codivergence events). A significance test demonstrated that, given 100 random viral topologies, more than 60 can be mapped onto each host tree and still give five or more codivergences (P ≥ 0.61 ± 0.05). Similarly, nonsignificant P values were obtained when testing the minimal number of NCEs (P ≥ 0.58 ± 0.05). In sum, there does not appear to be any significant support for codivergence, as the observed JCV trees are no more congruent with human trees than random viral trees would be. In contrast, the consensus tree of the 158 globally sampled mtDNA genomes, which clearly have codiverged with that of the human population, shows a significant level of congruence with one of the three consensus human phylogenies (P = 0.02 ± 0.014), confirming the robustness of the TreeMap approach employed here.
FIG. 2.
Tanglegrams a to i of JCV (right) and human population (left) phylogenies (see Table 1; see Fig. S3 in the supplemental material).
TABLE 1.
JCV-human phylogenetic reconciliation analysis
| Virus-host topology | Maximum codivergence events in POpt | Minimum NCEs in POpt | Significance of maximum codivergence (P) | Significance of minimum NCEs (P) |
|---|---|---|---|---|
| a-i | 10 | 25 | 0.68 | 0.63 |
| b-i | 10 | 25 | ||
| a-ii | 6 | 26 | 0.76 | 0.84 |
| b-ii | 10 | 26 | ||
| a-iii | 10 | 25 | 0.61 | 0.58 |
| b-iii | 10 | 25 |
Comparison of viral and host population dynamics.
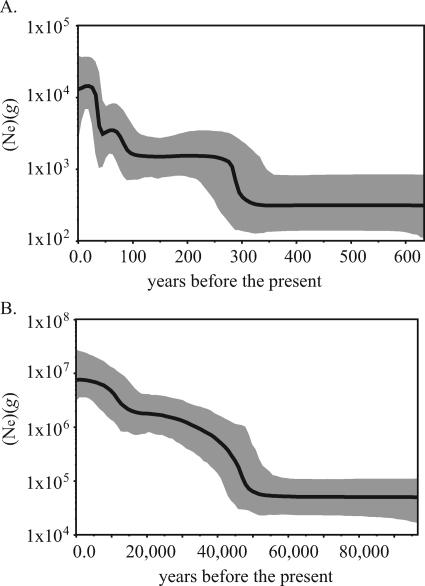
To estimate the population dynamics of JCV, we compiled 158 diverse viral sequences for which dates of isolation, ranging from 1970 to 2003, were available. The ML phylogeny (see Fig. S2 in the supplemental material) showed the same topology, with respect to host geography, as the phylogeny of 333 undated sequences. Using a Bayesian MCMC approach with a relaxed molecular clock (14), we coestimated the rate of JCV molecular evolution and the demographic history of the virus using a “skyline plot” (16). A corresponding data set of 158 mtDNA sequences was compiled and the same analysis was performed, except that the rate of mtDNA evolution was set to 1.7 × 10−8 subs/site/year, a clock rate previously determined for complete mtDNA sequences, excluding the D-loop (19).
The predicted age of the human mtDNA phylogeny was consistent with accepted estimates of a most recent common ancestor (MRCA) 100,000 to 200,000 years ago (ya) (27) and an increase in population size corresponding to major cultural changes, beginning approximately 50,000 ya (21) (Table 2; Fig. 3B). Furthermore, the posterior estimates of the 10 Θ parameters (equivalent to Ne × g, where g is generation length and Ne is the effective population size) ranged from 5.3 × 104 to 9.2 × 106. Given that the human lineage has had a long-term harmonic mean Ne of ≥10,000 (34, 37) and a g of approximately 20 years, the estimated Θ's are also consistent with human population history. In contrast, the posterior population parameters estimated for JCV did not correspond to those of its human host, as would be predicted under a model of codivergence. The upper 95% confidence interval for the inferred MRCA of JCV did not exceed 3,100 ya (Table 2), while the values estimated for JCV Θ's only ranged from 5.6 × 102 to 3.4 × 104. Given the prevalence of JCV in the human population, if the viral g was truly equivalent to that of humans (i.e., transmitted essentially vertically), these Θ's should be the same order of magnitude as those estimated for the human population.
TABLE 2.
Bayesian MCMC estimates of JCV and human mtDNA evolutionary dynamics
| Genetic material analyzed | ML data set factors
|
||||||
|---|---|---|---|---|---|---|---|
| Clock | Sequence length (bp) | Isolation date range | Mean rate (subs/site/yr) | HPD of rate (subs/site/yr) | Mean MRCA (yr) | HPD of MRCA (yr) | |
| 158 JCV coding regions | Relaxed | 4,798 | 1970-2003 | 1.7 × 10−5 | 2.1 × 10−6, 3.1 × 10−5 | 1.3 × 103 | 3.1 × 102, 3.1 × 103 |
| 158 human mtDNA genomes (no D-loop) | Strict | 15,454 | NA | Set to 1.7 × 10−8 | NA | 1.4 × 105 | 1.0 × 105, 1.8 × 105 |
FIG. 3.
Skyline plots estimated from Bayesian MCMC analyses of (A) 158 JCV genomes found in geographically diverse populations and (B) 158 human mtDNA sequences with a similar geographical distribution. A priori nucleotide substitution rates of 1.7 × 10−5 and 1.7 × 10−8 subs/site/year were specified, respectively, while all other parameters were allowed to vary. The black line shows the median estimate of Θ (Ne × g) throughout the given time period. The gray area gives the 95% HPD interval of these estimates.
An independent estimate of the rate of nucleotide substitution in JCV.
The mean substitution rate for JCV estimated using a relaxed molecular clock was 1.7 × 10−5 subs/site/year (Table 2). However, the HPD spanned an order of magnitude, from 2.1 × 10−6 to 3.1 × 10−5 subs/site/year. Thus, while we are unable to arrive at a precise estimate of the evolutionary rate of JCV, our analysis suggests that the substitution rate is significantly more rapid than that of 10−7 subs/site/year, as would be predicted under a model of virus-host codivergence. To reconstruct virus population growth dynamics under the assumption that the mean posterior rate of the initial analysis approximates the true rate, we conducted an additional relaxed-clock Bayesian skyline analysis, during which a JCV evolutionary rate of 1.7 × 10−5 subs/site/year was specified a priori. The resulting population growth curve was then compared to that of human mitochondrial genomes, evolving at a mean rate of 1.7 × 10−8 subs/site/year (Fig. 3). Like the mitochondrial population dynamics described above, JCV shows a period of constant population size, followed by population expansion. However, the population expansion in JCV only began in the last 350 years, far more recently than the beginning of human population expansion, and most likely reflects the increased size and mobility of the human host population in the recent past.
DISCUSSION
If JCV had been transmitted primarily vertically and had codiverged with human populations, we would expect to see a strong geographical grouping of the viral isolates matching the structure of human populations, with any mismatch caused by recent human admixture. However, phylogenies of JCV and humans are no more similar than would be expected by chance, in contrast to the pattern seen in human mtDNA. Furthermore, had vertical transmission and codivergence been the norm, both JCV and human mtDNA should exhibit broadly similar population dynamics. Yet, based on our estimates for the age of human and viral genetic diversity as well as the patterns of population growth, this does not appear to be the case. Possibly the greatest discrepancy between the predicted human and JCV histories is that, while modern humans originated in Africa and populations subsequently diverged (19), the JCV strains associated with African populations are not basal in JCV phylogenies. This seems to indicate that either a basal African strain of JCV has gone extinct or that JCV did not infect humans before their migration from Africa. This discrepancy cannot be resolved by simply changing the position of the root on the JCV tree; if the African subtype 3 is chosen as the outgroup, then the European subtypes and the African subtype 6 form a sister group that is basal to every other ethnicity, while if the tree is rooted with subtype 6, then the European subtypes are basal to the African subtype 3. Neither of these phylogenies match the history of human populations.
Thus, our analysis suggests that JCV has not strictly codiverged with human populations. While specific strains do exist predominantly within certain populations and some parts of the JCV phylogeny hint at codivergence, such as the close association of subtypes 2A1 (east Asians) and 2A2 (Native Americans), geographical association does not provide adequate evidence for long-term codivergence. As such, we caution against using this virus to make extensive inferences about the evolution of human populations. That factors other than codivergence could account for the similar geographical distribution of humans and JCV was proposed by Wooding (39) after finding differences in demographic history. Both JCV and human subpopulations exhibit distinctive genetic features likely caused by population isolation and genetic drift, but human population structure cannot explain many of the observed phylogenetic patterns in JCV, such as the relative similarity of viruses sampled in subpopulations from Africa and Asia and the genetic diversity of European strains. In contrast, despite their limitations, sequence data derived from human mitochondria and Y chromosomes may be far better suited for deducing the details of human population migration.
Under an assumption of JCV-human codivergence, the rate of viral nucleotide substitution estimated by calibrating the viral phylogeny with host divergence times should approximately match the rate obtained in a host-independent analysis of substitution rate. While host-calibrated viral clocks have resulted in estimates of the synonymous substitution rate at 4 × 10−7 subs/site/year (17), no host-independent rate had been estimated, either to confirm this rate or to test assumptions of vicariance upon which it is based. Here, we attempted to obtain such an estimate, based solely on the extent of sequence variation observed in JCV isolates over a period of 34 years. Our analyses suggest a significantly more rapid rate of evolution than that obtained under a model of codivergence. Although limited long-term viral sampling resulted in fairly large confidence intervals for the rate of JCV evolution (indicating that all estimations of the substitution rate must be made with caution and that further data are required), these rapid rates, together with the lack of evidence for codivergence, suggest that human phylogenetic history does not provide suitable calibration points for JCV.
A high rate of evolutionary change in JCV is also compatible with analyses of intrahost diversity in populations of BKV, the only other known human polyomavirus. In particular, a 52-year-old patient was found to harbor BK viruses that differed by 0.55% (10). Assuming a clonal infection ∼50 years before, this level of diversity would indicate a rate of 5 × 10−5 subs/site/year, similar to the estimates for JCV we find here. Likewise, the 0.15% diversity in a patient infected for approximately 40 years yields a rate of 2 × 10−5 subs/site/year (10). A separate study of healthy transplant recipients found less intrahost diversity, yet the phylogenetically grouped BKV populations in four out of six patients still showed nucleotide differences (35). Performing the equivalent conservative calculation as that described above suggests that if the patients contracted the virus as infants, rates of evolution are between 4.0 × 10−6 and 7.0 × 10−6 subs/site/year. If, on the other hand, the virus was contracted at the time of the kidney transplant, evolutionary rates would range from 1.8 × 10−4 to 7.8 × 10−4 subs/site/year. In either of these scenarios, the rate appears to be closer to the rate derived independently from codivergence assumptions than to the codivergence-based rate estimates. Finally, employing the covarion model of nucleotide substitution, which has been proposed as a means to reconcile virus and host divergence times (18), failed to extend the divergence times of JCV; although the covarion model gave a significantly better fit than a noncovarion model, the total length of the phylogenetic tree was reduced in the former (analysis performed using MrBayes; methods and results are available from the authors on request).
In the case of some DNA viruses, notably herpesviruses and papillomaviruses, it has been possible to estimate substitution rates using well-established patterns of host-virus codivergence (5, 24, 25). The rates inferred in these viruses are generally low, in the range of 10−7 to 10−9 subs/site/year (5, 23). In the case of the herpesviruses (which range from 150 to 230 kb in length), these rates are also compatible with the notion that there is a universal rate of mutation in DNA microbes which is proportional to genome size (estimated at ∼0.003 mutations/genome/replication [12, 13]). While the rapid rates of evolution recently observed in the 5-kb autonomous carnivore parvoviruses of ∼10−4 to 10−5 subs/site/year (30) would seem to support this notion, the low rates found in the 8-kb papillomaviruses imply that genome size is not the only factor influencing substitution rates. It is evident that viral generation times (replication rates) will strongly influence substitution rates per unit of time, although accurate measurements of generation times are lacking for most viruses. Our study suggests that it is necessary to reexamine many previously held suppositions regarding substitution rates in DNA viruses and to more accurately determine the similarities and differences between long- and short-term as well as intra- and interhost substitution rates.
Supplementary Material
Acknowledgments
We are grateful to Michael A. Charleston and Andrew P. Jackson for assistance with TreeMapv2.0.
This work was supported by a Howard Hughes Medical Institute fellowship to L.A.S.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agostini, H. T., C. Ryschkewitsch, and G. L. Stoner. 1996. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J. Clin. Microbiol. 34:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agostini, H. T., R. Yanagihara, V. Davis, C. F. Ryschkewitsch, and G. L. Stoner. 1997. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: markers of viral evolution and human migration. Proc. Natl. Acad. Sci. USA 94:14542-14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayub, Q., A. Mansoor, M. Ismail, S. Khaliq, A. Mohyuddin, A. Hameed, K. Mazhar, S. Rehman, S. Siddiqi, M. Papaioannou, A. Piazza, L. L. Cavalli-Sforza, and S. Q. Mehdi. 2003. Reconstruction of human evolutionary tree using polymorphic autosomal microsatellites. Am. J. Phys. Anthropol. 122:259-268. [DOI] [PubMed] [Google Scholar]
- 4.Bamshad, M., T. Kivisild, W. S. Watkins, M. E. Dixon, C. E. Ricker, B. B. Rao, J. M. Naidu, B. V. R. Prasad, P. G. Reddy, A. Rasanayagam, S. S. Papiha, R. Villems, A. J. Redd, M. F. Hammer, S. V. Nguyen, M. L. Carroll, M. A. Batzer, and L. B. Jorde. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res. 11:994-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, H.-U. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2:140-143. [DOI] [PubMed] [Google Scholar]
- 6.Cavalli-Sforza, L. L., and M. W. Feldman. 2003. The application of molecular genetic approaches to the study of human evolution. Nature 33:266-275. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli-Sforza, L. L., P. Menozzi, and A. Piazza. 1994. The history and geography of human genes. Princeton University Press, Princeton, N.J.
- 8.Chang, H., M. Wang, R. T. Tsai, H. S. Lin, J. S. Huan, W. C. Wang, and D. Chang. 2002. High incidence of JC viruria in JC-seropositive older individuals. J. Neurovirol. 8:447-451. [DOI] [PubMed] [Google Scholar]
- 9.Charleston, M. A. 1998. Jungles: a new solution to the host/parasite phylogeny reconciliation problem. Math. Biosci. 149:191-223. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y., P. M. Sharp, M. Fowkes, O. Kocher, J. T. Joseph, and I. J. Koralnik. 2004. Analysis of 15 novel full-length BK virus sequences from three individuals: evidence of a high intra-strain genetic diversity. J. Gen. Virol. 85:2651-2663. [DOI] [PubMed] [Google Scholar]
- 11.Cui, X., J. C. Wang, A. Deckhut, B. C. Joseph, P. Eberwein, C. L. Cubitt, C. F. Ryschkewitsch, H. T. Agostini, and G. L. Stoner. 2004. Chinese strains (type 7) of JC virus are afro-asiatic in origin but are phylogenetically distinct from the Mongolian and Indian strains (type 2D) and the Korean and Japanese strains (type 2A). J. Mol. Evol. 58:568-583. [DOI] [PubMed] [Google Scholar]
- 12.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake, J. W., and C. B. C. Hwang. 2005. On the mutation rate of herpes simplex virus type 1. Genetics 170:969-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond, A. J., S. Y. W. Ho, M. J. Phillips, and A. Rambaut. 2006. Relaxed molecular clocks and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond, A. J., O. G. Pybus, A. Rambaut, R. Forsberg, and A. G. Rodrigo. 2003. Measurably evolving populations. Trends Ecol. Evol. 18:481-488. [Google Scholar]
- 16.Drummond, A. J., A. Rambaut, B. Shapiro, and O. G. Pybus. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22:1185-1192. [DOI] [PubMed] [Google Scholar]
- 17.Hatwell, J. N., and P. M. Sharp. 2000. Evolution of human polyomavirus JC. J. Gen. Virol. 81:1191-1200. [DOI] [PubMed] [Google Scholar]
- 18.Holmes, E. C. 2003. Molecular clocks and the puzzle of RNA virus origins. J. Virol. 77:3893-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingman, M., H. Kaessmann, S. Pääbo, and U. Gyllensten. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408:708-713. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, A. P., and M. A. Charleston. 2004. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 21:45-57. [DOI] [PubMed] [Google Scholar]
- 21.Kaessmann, H., V. Wiebe, G. Weiss, and S. Pääbo. 2001. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat. Genet. 27:155-156. [DOI] [PubMed] [Google Scholar]
- 22.Kunitake, T., T. Kitamura, J. Guo, F. Taguchi, K. Kawabe, and Y. Yogo. 1995. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J. Clin. Microbiol. 33:1448-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGeoch, D. J., and S. Cook. 1994. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 238:9-22. [DOI] [PubMed] [Google Scholar]
- 24.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong, C.-K., S.-Y. Chan, M. S. Campo, K. Fujinaga, P. Mavromara-Nazos, V. Labropoulou, H. Pfister, S.-K. Tay, J. TerMeulen, L. L. Villa, and H.-U. Bernard. 1993. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J. Virol. 67:6424-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padgett, B. L., and D. L. Walker. 1973. Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. J. Infect. Dis. 127:467-470. [DOI] [PubMed] [Google Scholar]
- 27.Pakendorf, B., and M. Stoneking. 2005. Mitochondrial DNA and human evolution. Annu. Rev. Genomics Hum. Genet. 6:165-183. [DOI] [PubMed] [Google Scholar]
- 28.Pavesi, A. 2005. Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times. J. Gen. Virol. 86:1315-1326. [DOI] [PubMed] [Google Scholar]
- 29.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 30.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugimoto, C., T. Kitamura, J. Guo, M. N. Al-Ahdal, S. N. Shchelkunov, B. Otova, P. Ondrejka, J. Y. Chollet, S. El-Safi, M. Ettayebi, G. Gresenguet, T. Kocagoz, S. Chaiyarasamee, K. Z. Thant, S. Thein, K. Moe, N. Kobayashi, F. Taguchi, and Y. Yogo. 1997. Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proc. Natl. Acad. Sci. USA 94:9191-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimoto, C., M. Hasegawa, A. Kato, H.-Y. Zheng, H. Ebihara, F. Taguchi, T. Kitamura, and Y. Yogo. 2002. Evolution of human polyomavirus JC: implications for the population history of humans. J. Mol. Evol. 54:285-297. [DOI] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer, Sunderland, Mass.
- 34.Takahata, N. 1993. Allelic genealogy and human evolution. Mol. Biol. Evol. 10:2-22. [DOI] [PubMed] [Google Scholar]
- 35.Takasaka, T., N. Goya, H. Ishida, K. Tanabe, H. Toma, T. Fujioka, S. Omori, H. Y. Zheng, Q. Chen, S. Nukuzuma, T. Kitamura, and Y. Yogo. 2006. Stability of the BK polyomavirus genome in renal-transplant patients without nephropathy. J. Gen. Virol. 87:303-306. [DOI] [PubMed] [Google Scholar]
- 36.Uinuk-ool, T. S., N. Takezaki, O. A. Derbeneva, N. V. Volodko, and R. I. Sukernik. 2004. Variation of HLA class II genes in the Ngasan and Ket, two aboriginal Siberian populations. Eur. J. Immunogenet. 31:43-51. [DOI] [PubMed] [Google Scholar]
- 37.Wall, J. D. 2003. Estimating ancestral population sizes and divergence times. Genetics 163:395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber, T., and E. O. Major. 1997. Progressive multifocal leukoencephalopathy: molecular biology, pathogenesis and clinical impact. Intervirology 40:98-111. [DOI] [PubMed] [Google Scholar]
- 39.Wooding, S. 2001. Do human and JC virus genes show evidence of host-parasite codemography? Infect. Genet. Evol. 1:3-12. [DOI] [PubMed] [Google Scholar]
- 40.Yogo, Y., C. Sugimoto, H.-Y. Zheng, H. Ikegaya, T. Takasaka, and T. Kitamura. 2004. JC virus genotyping offers a new paradigm in the study of human populations. Rev. Med. Virol. 14:179-191. [DOI] [PubMed] [Google Scholar]
- 41.Zheng, H. Y., T. Kitamura, T. Takasaka, Q. Chen, and Y. Yogo. 2004. Unambiguous identification of JC polyomavirus strains transmitted from parents to children. Arch. Virol. 149:261-273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.