Abstract
Background
Laser scanning microscopy is a powerful tool for analyzing the structure and function of biological specimens. Although numerous commercial laser scanning microscopes exist, some of the more interesting and challenging applications demand custom design. A major impediment to custom design is the difficulty of building custom data acquisition hardware and writing the complex software required to run the laser scanning microscope.
Results
We describe a simple, software-based approach to operating a laser scanning microscope without the need for custom data acquisition hardware. Data acquisition and control of laser scanning are achieved through standard data acquisition boards. The entire burden of signal integration and image processing is placed on the CPU of the computer. We quantitate the effectiveness of our data acquisition and signal conditioning algorithm under a variety of conditions. We implement our approach in an open source software package (ScanImage) and describe its functionality.
Conclusions
We present ScanImage, software to run a flexible laser scanning microscope that allows easy custom design.
Background
Laser scanning microscopies (LSM) include some of the most important imaging modalities in biology. For example, confocal laser scanning microscopy (CLSM) is the tool of choice for high-resolution fluorescence microscopy in cultured and fixed tissue preparations [1-3]. Two-photon excitation laser scanning microscopy (2PLSM) is ideal for fluorescence microscopy in scattering media, such as living skin and brain tissue [4,5].
Historically, new developments and applications of laser scanning microscopy have been limited primarily by the availability of new hardware. The development of suitable laser light sources and powerful data acquisition systems running on desktop computers brought CLSM to fruition [3]. In the case of 2PLSM, the development of mode-locked pulsed light sources operating in the red and near IR spectral range [6] made it practical [5].
Several lines of commercial CLSMs and 2PLSMs are available. A manufacturer typically will offer one principal model and allow the customer to choose between different versions and options. Fundamentally, in terms of optics and software, a single general purpose design is offered. However, exciting applications demand specialized design. Often these applications include some of the most promising experimental developments. Currently, most hardware components of CLSMs and 2PLSMs (lasers, optics, scanning mirrors, detectors, and computers) are mature and readily available. The development of cutting edge applications using custom design is often limited by the availability of suitable software to control laser scanning microscopes.
In our laboratory we have written flexible software to control LSMs (ScanImage). In a LSM, a signal (typically fluorescence) is detected as a focused laser beam is scanned across a sample. The signal emitted from within a number of tiny, contiguous regions (called pixels) [3,7] is collected over short time intervals (the pixel time, Tp). The integrated signal is converted to a digital pixel value and the collection of all the pixels from the scanned region defines the image. The location of the pixel in the image is determined by the location of the laser at the time of acquisition. Synchronization of the position of the laser beam and the signal collection is therefore essential for imaging.
In order to time scanning and acquisition precisely and to achieve high performance in terms of signal-to-noise ratio, on-line averaging, and real-time image refresh, many commercially available laser-scanning microscopes (e.g. Olympus Fluoview™, Olympus America Inc., Melville, NY; Bio-Rad Radiance 2100™, Bio-Rad Laboratories, Hercules, CA) use proprietary signal conditioning electronics and data acquisition boards. Commercial systems are suitable for many applications and have also been customized for specific experimental needs [4,8-10]. However, complete custom design is often more advantageous and cost effective [11-13]. Two major impediments to custom design are the specialized software and hardware required for laser scanning microscopy.
Here we present a software package called ScanImage for collection of laser-scanned images that functions at a high level without the need for elaborate custom hardware [14,15]. The key idea is to use fast data acquisition boards and CPU-based numeric computations to perform most of the tasks that are accomplished by DSP boards and analog integrators in typical commercial systems. Thus ScanImage simplifies the design and construction of custom microscopes. An additional advantage is that ScanImage is written in MATLAB (version 6.1 or later; MathWorks Inc., Natick, MA), and thus already fully integrated into a high-level image analysis environment.
Results and Discussion
Description of ScanImage
ScanImage controls a laser-scanning microscope (Figure 1A). It is written entirely in MATLAB and makes use of standard multifunction boards (e.g. National Instruments, Austin, TX) for data acquisition and control of scanning. The software generates the analog voltage waveforms to drive the scan mirrors, acquires the raw data from the photomultiplier tubes (PMTs), and processes these signals to produce images. ScanImage controls three input channels (12-bits each) simultaneously, and the software is written to be easily expandable to the maximum number of channels the data acquisition (DAQ) board supports and that the CPU can process efficiently. The computer bus speed dictates the number of samples that can be acquired before an overflow of the input buffer occurs, while the CPU speed and bus speed combine to determine the rate of data processing and ultimately the refresh rate of images on the screen. Virtually no customized data acquisition hardware is required for either scan mirror motion or data acquisition.
Figure 1.
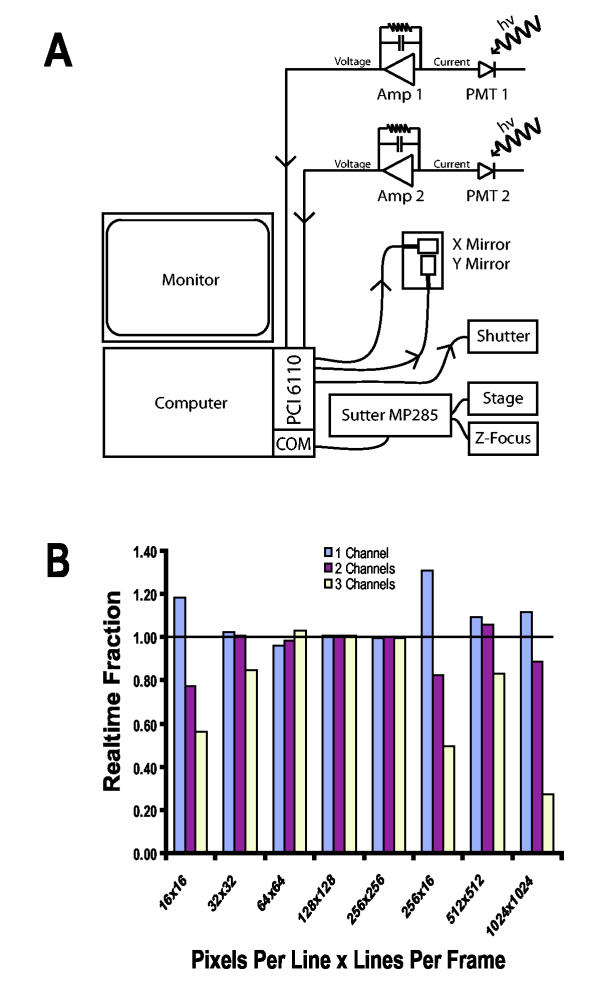
Wiring Diagram and Performance Benchmarking for ScanImage A) Typical wiring diagram for a microscope running ScanImage. Scanning, data acquisition, and laser shuttering are accomplished by a single data acquisition board (PCI 6110, National Instruments). Stage and Z-focus motors are controlled through a Sutter 3-axis motor controller (Sutter MP285) that is programmed through a serial communications (COM) port. B) Performance of ScanImage. We computed the realtime fraction as the average time it took ScanImage to process and display the acquired data (i.e. a stripe; see Figure 2) normalized to the acquisition time for that portion of data. Values were averaged over 20 frames acquired at each configuration. The realtime fraction was computed using an 800 MHz Pentium III computer with 512 MB of RAM. A realtime fraction above one indicates the software was able to run in realtime (i.e. ScanImage had completed processing and displaying all of the previously collected data before the next portion of data was fully acquired). For realtime fractions less than one, the display lags the acquisition but no data is lost. The low realtime fraction when acquiring 16 lines per frame on multiple channels indicates that we are approaching the fundamental speed limit for graphics updates.
Typical pixel times in biological laser scanning microscopy are > 2 μs (the pixel time depends on the pixels per line and the time per line; see Figure 2 for more information on the image construction algorithm). We have benchmarked ScanImage while acquiring three channels simultaneously with ~3 μs pixel times (Figure 1B). Under these demanding conditions, ScanImage updates and saves the images with minimal delay from the time they are actually obtained (Figure 1B). The actual size of the image (in microns) depends on the microscope optics and the angles navigated by the scan mirrors.
Figure 2.
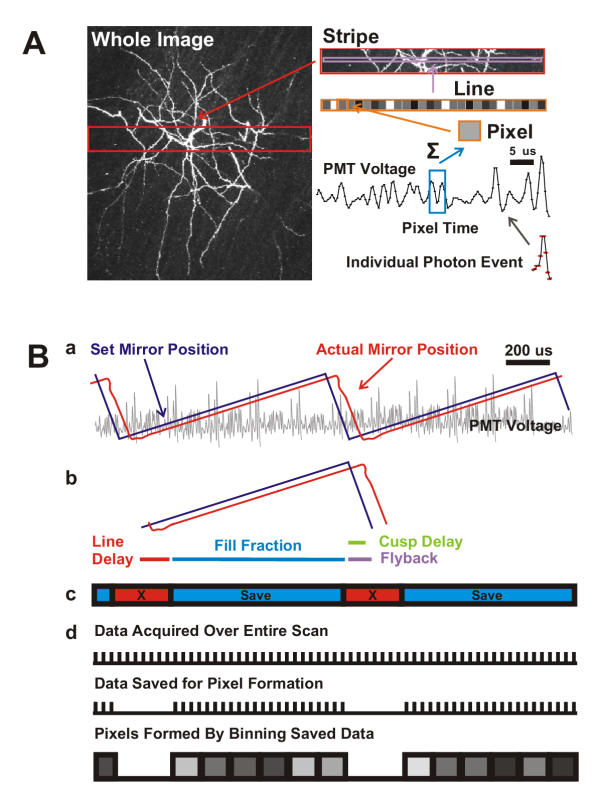
Image Construction and Signal Processing in ScanImage A) Schematic depicting the algorithm of image creation in ScanImage. Pixels are constructed by summing the PMT voltage over the pixel time Tp. To achieve fast (i.e. realtime) refresh rates we break the acquisition of an image down into a set of stripes acquired and displayed in succession. The stripes are concatenated to form the entire image. B) Further details of image construction in ScanImage. a) Mirror position signal delivered by ScanImage (blue trace) and the actual mirror position (red trace) for two lines of acquisition (PMT voltage shown in gray). b) Definitions of acquisition parameters. The flyback (purple) and line delay (red) regions of the acquired PMT signal are discarded from final image data, which keeps only the fill fraction (blue) of the PMT voltage for image construction. Because the scan mirrors have inertia, their actual position lags the set position by ~7% of the scan period, which is corrected by the cusp delay (green). c) ScanImage retains the data acquired during the linear portions of the scan (blue) and discards the intervening data (red). d) Samples are summed to form the pixels in the final image.
We have chosen MATLAB as our development environment for its powerful numerical manipulation abilities and data acquisition (DAQ) engine. The DAQ engine directly communicates with the acquisition board and presents a flexible, object-oriented environment in which to construct DAQ software. In many scientific and engineering communities, MATLAB has become the dominant language for desktop numerical computing [16]. Integration of custom written analysis functions and incorporation of additional software is much more tractable for novice programmers in MATLAB, simplifying much of the experimental design process.
Data Acquisition in ScanImage
The signal from the detector (typically a PMT) is a current that needs to be converted to a voltage before acquisition. This conversion can be done with a standard amplifier [17] (see Stanford Research Systems, Sunnyvale, CA for examples) equipped with a low pass filter. The roll-off frequency of the filter has to be <0.5 fs, where fs is the rate at which the detector signal is sampled. In ScanImage we compromise between the degradation in the signal-to-noise ratio due to limited sampling of the signal and the increasing computational load placed on the CPU with higher sample rates.
With current computer hardware we operate ScanImage with fs = 1.25 MHz. This sampling frequency allows nearly ideal imaging (in terms of apparent quantum efficiency and bleedthrough between pixels, discussed below) for pixel times as short as 3 μs. An appropriate roll-off frequency would be ~300–500 kHz. In the future, faster computer architectures will allow more rapid sampling and number crunching and hence shorter pixel times. The sampled data is streamed to memory. An overview of the image construction algorithm used in ScanImage is shown in Figure 2A.
Sampled data are summed in real-time over the pixel times. Once the data is collected, it is compared with the output signals to the scan mirrors. The position of the scan mirrors dictates where the pixels lie in the final image. The mirror driver electronics (see Cambridge Technology, Cambridge, MA for examples) and the command voltage output to the drivers from the DAQ board determine the mirror position. During a cycle of the mirror motion useful data are collected only when the mirrors are moving at constant velocity (i.e. linear voltage versus time regime; see Figure 2B). Data acquired when the mirrors are turning around or flying back to their starting positions (flyback) are removed (Figure 2B; figure caption for details).
The timing for image formation requires synchronization of the mirror scan forms and the data acquisition engine. This is achieved by triggering data acquisition and data output using a TTL trigger generated by the DAQ board (i.e. under the control of a software panel). This mechanism for triggering also allows ScanImage to be externally triggered by a TTL from another application or hardware device. Appropriate sampling rates and output rates are hard-coded in ScanImage, while many other acquisition parameters can be chosen based on the demands of particular applications.
Quality of the Images Collected by ScanImage
We evaluated the performance of our signal conditioning method (Figures 2 &3). We compared the variability of the integral of a simulated PMT signal to the variability of the signal sum computed at a fixed sample rate as is done in ScanImage (see Figure 2B for details). In simulations the PMT voltage was modeled as a Poisson process with mean photon rate of 0.5 photons per μs. Each single-photon pulse was Gaussian with unit amplitude and 2.35 μs full-width-at-half-maximum (FWHM) [18]. Each simulated pulse was constructed at 500 MHz. A 20 ms waveform was created by summing Poisson-distributed pulses and dividing them into 2000 pixel intervals (duration 10 μs). Each of these intervals constituted a trial in the analysis of variability described below. Trials were sampled at 200 kHz to 5 MHz in 100 kHz steps. A typical trial sampled at different rates is shown in the inset of Figure 3A.
Figure 3.
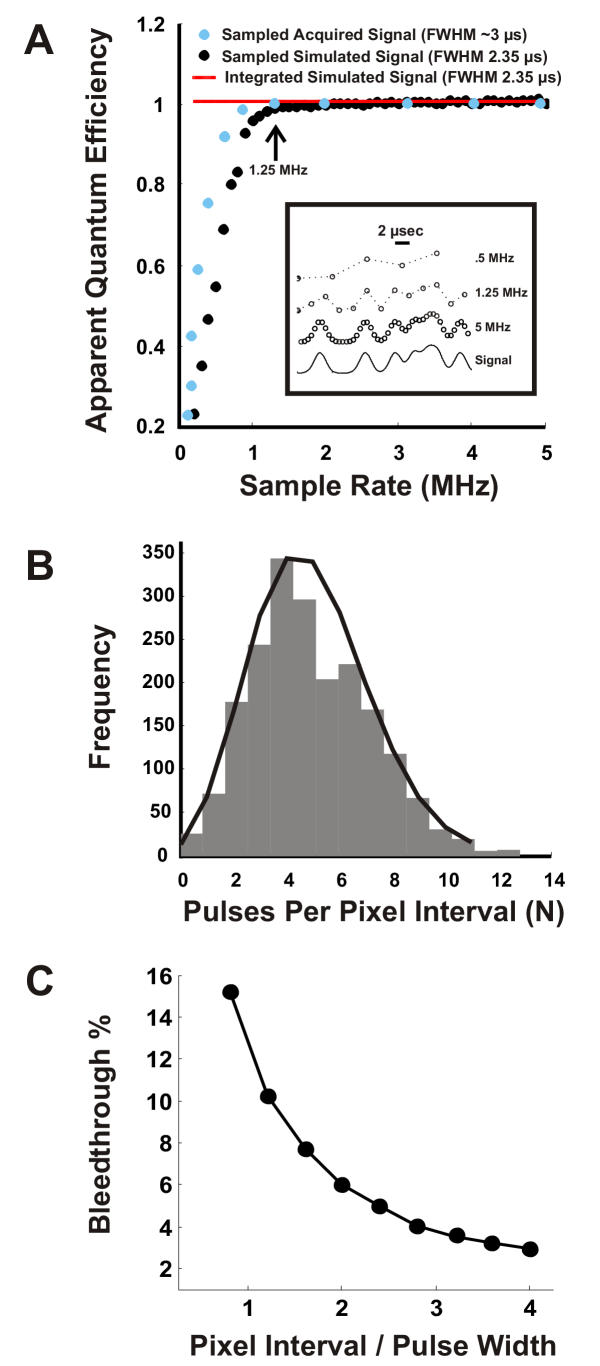
Image Quality in ScanImage A) Apparent quantum efficiency as a function of sampling rate in ScanImage for a single-photon-pulse with a 2.35 μs FWHM and unit amplitude. The red trace shows the apparent quantum efficiency (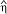 ) of the signal (see inset) determined by digital integration. The black trace shows
) of the signal (see inset) determined by digital integration. The black trace shows 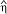 sampled at 200 kHz to 5 MHz.
sampled at 200 kHz to 5 MHz. 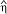 approaches 1 as the sample rate approaches 1 MHz. The blue points are the normalized apparent quantum efficiencies from an actual PMT illuminated by a constant light source (data pre-filtered at 330 kHz). ScanImage uses a sample rate of 1.25 MHz (arrow), which yields nearly ideal signal-to-noise
approaches 1 as the sample rate approaches 1 MHz. The blue points are the normalized apparent quantum efficiencies from an actual PMT illuminated by a constant light source (data pre-filtered at 330 kHz). ScanImage uses a sample rate of 1.25 MHz (arrow), which yields nearly ideal signal-to-noise 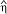 ≈ 1. This analysis is only valid for pulses that are al least 2.35 μs in duration. Inset: 20-μs pulse interval comprised of 2.35-μs events sampled at different rates. B) Histogram of the events in the pixel interval (bars) and Poisson fit (line). C) Model of bleedthrough of photons from adjacent pixels. A single-photon-pulse with a 2.5 μs FWHM was placed randomly in pixel intervals of widths 2 to 10 μs. The number of trials at each pixel interval was 10,000. The percent bleedthrough equaled 100*(1 - IPP/IEP) where IEP is the integral of the entire pulse and IPP is the integral of the region of the pulse that fell in the pixel interval. Shown is the mean value of the percent bleedthrough for each pixel time normalized with respect to the pulse width (Pixel Interval/Pulse Width). The error bars (SEM) were smaller than the marker points and were omitted.
≈ 1. This analysis is only valid for pulses that are al least 2.35 μs in duration. Inset: 20-μs pulse interval comprised of 2.35-μs events sampled at different rates. B) Histogram of the events in the pixel interval (bars) and Poisson fit (line). C) Model of bleedthrough of photons from adjacent pixels. A single-photon-pulse with a 2.5 μs FWHM was placed randomly in pixel intervals of widths 2 to 10 μs. The number of trials at each pixel interval was 10,000. The percent bleedthrough equaled 100*(1 - IPP/IEP) where IEP is the integral of the entire pulse and IPP is the integral of the region of the pulse that fell in the pixel interval. Shown is the mean value of the percent bleedthrough for each pixel time normalized with respect to the pulse width (Pixel Interval/Pulse Width). The error bars (SEM) were smaller than the marker points and were omitted.
How does the signal to noise ratio vary with sampling frequency? A useful measure of the quality of the signal is the coefficient of variance (CVy). CVy is the ratio of the standard deviation (σy) to the mean (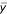 ) of the signal summed over a pixel interval (y). CVy is related to the apparent quantum efficiency (
) of the signal summed over a pixel interval (y). CVy is related to the apparent quantum efficiency (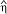 ) of the acquisition system and the mean number of pulses per pixel interval (
) of the acquisition system and the mean number of pulses per pixel interval (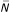 ) by Equation 1:
) by Equation 1:
![]()
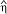 is inversely related to the noise introduced by the signal conditioning in addition to the inherent shot noise of the light source and multiplicative noise due to the variable pulse-amplitude distribution [19]. For optimal sampling with uniform pulse amplitudes,
is inversely related to the noise introduced by the signal conditioning in addition to the inherent shot noise of the light source and multiplicative noise due to the variable pulse-amplitude distribution [19]. For optimal sampling with uniform pulse amplitudes, 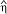 will be 1 since
will be 1 since 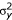 =
= 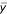 =
= 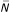 (see Equation 1); this can be accomplished by direct integration of the signal (Figure 3A, red line). Thus we can interpret any reduction in
(see Equation 1); this can be accomplished by direct integration of the signal (Figure 3A, red line). Thus we can interpret any reduction in 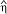 from 1 as a decrease of the signal-to-noise ratio (SNR) of the system.
from 1 as a decrease of the signal-to-noise ratio (SNR) of the system.
We verified our simulation by fitting the histogram of the number of events (N) to a Poisson model (Figure 3B). This analysis yielded a mean value for 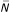 = 4.96 ± .09 (p < .05) photons per pixel interval (10 μs duration) as expected. We then analyzed the trials sampled at different frequencies to determine the effects of finite sample frequency on
= 4.96 ± .09 (p < .05) photons per pixel interval (10 μs duration) as expected. We then analyzed the trials sampled at different frequencies to determine the effects of finite sample frequency on 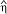 . Variation of the pulse amplitudes can be modeled directly by convolving the pulse amplitude distribution with our model, but does not alter the optimal signaling criteria; it merely reduces the maximum value of
. Variation of the pulse amplitudes can be modeled directly by convolving the pulse amplitude distribution with our model, but does not alter the optimal signaling criteria; it merely reduces the maximum value of 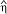 to a value lower than 1 [18].
to a value lower than 1 [18].
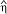 was computed at each sample rate from the simulated pulses (Figure 3A). Note that the digital integrator shows nearly ideal behavior (
was computed at each sample rate from the simulated pulses (Figure 3A). Note that the digital integrator shows nearly ideal behavior (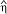 ≈ 1). As expected, for low sampling frequencies
≈ 1). As expected, for low sampling frequencies 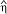 is small, reflecting the fact that data (single photon events) are missed by the sampling scheme.
is small, reflecting the fact that data (single photon events) are missed by the sampling scheme. 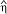 increases with sample rate until it approaches 1 around the Nyquist limit of 1 MHz (i.e. since we simulated the PMT voltage with 2.35 μs pulses). We repeated this analysis with data coming from an actual PMT filtered at ~330 KHz (blue points in Figure 3A). The lower filtering frequency broadens the pulses (3–4 μs FWHM) and hence increases the performance of our algorithm at lower sample rates. At 1.25 MHz, the image has an essentially ideal
increases with sample rate until it approaches 1 around the Nyquist limit of 1 MHz (i.e. since we simulated the PMT voltage with 2.35 μs pulses). We repeated this analysis with data coming from an actual PMT filtered at ~330 KHz (blue points in Figure 3A). The lower filtering frequency broadens the pulses (3–4 μs FWHM) and hence increases the performance of our algorithm at lower sample rates. At 1.25 MHz, the image has an essentially ideal 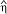 .
.
We also modeled the expected bleedthrough of photons from adjacent pixels (Figure 3C). A simulated test pulse with a FWHM of 2.5 μs was placed randomly in pixel intervals of widths 2 to 10 μs. The percent bleedthrough was calculated as the percentage of the area of the pulse that fell outside of the pixel interval in which it was placed. We see that the bleedthrough for the shortest pixel times recommended (~3.0 μs) is ~10%. Longer pixel times under normal imaging conditions (Pixel Interval/Pulse Width ~2) yield a bleedthrough of only ~6%.
Currently ScanImage performs well with a 1.25 MHz acquisition rate and a 330 kHz filter frequency (blue points and arrow in Figure 3A) depending on the application. Using sampling rates above 1.25 MHz (together with higher filter frequencies) is currently an issue of bus and CPU speed as mentioned. The sample rate can be adjusted manually in the software to a multiple of 1.25 MHz (e.g. 2.5 MHz) without any corruption of image quality. This yields an effective range of pixel times of 2–3-μs. Pixel times as low as 2 μs are feasible with a 500 kHz filter acquiring at 1 ms per line.
Conclusions
We have shown that our algorithm creates digital images with near-ideal noise properties and realtime refresh rates.
ScanImage Features
We now briefly describe the software that we have created to turn the data acquisition algorithm into a full image acquisition program. The features presented here can be easily extended through the MATLAB interface. The wiring diagram for a LSM using ScanImage is given in Figure 1A.
Multi-Channel Acquisition
Many applications, including quantitative calcium imaging, can involve the use of two dyes with different spectral properties (e.g. green calcium sensitive, Fluo-4, and red calcium insensitive, Alexa Fluor-594) [15]. Optics can be easily designed to separate and simultaneously collect both of the emitted wavelengths in separate channels. We have integrated support for processing 3 separate fluorescence channels independently in ScanImage. The data are collected, processed, and displayed simultaneously in real time (Figure 1B).
3-Dimensional Image Acquisition
The microscope focus is under the control of ScanImage allowing automated collection of stacks of images from different focal planes. These stacks can be set and collected easily in ScanImage using any configuration. Kalman filtering can be implemented per focal plane for noise reduction.
Structure of Acquired Data
ScanImage stores data in TIF files for easy portability and compatibility. TIF files can include multiple frames for storage of three-dimensional, multi-channel, and time series data in the same file. The pixel intensities are saved as 16-bit unsigned integers, exploiting the full dynamic range of the detector and DAQ boards. Different acquisition channels are interleaved. The experimental details are written into a header string that is placed into the ImageDescription field of the generic TIF header, and can be accessed via the standard header reading modules included in all image-processing programs, including MATLAB.
Automation of Stage Controls
LSM often involves finding the region (or regions) of interest (ROIs) in a complex and large specimen. The ability to select and save ROIs can help in the design of complex imaging experiments. ScanImage allows the user to define an arbitrary number of ROIs for imaging. ScanImage uses its control of the stage and focus to automate collection of focal stacks of data as well as for saving and retrieving ROIs. Through a system of experimental cycles it is possible to select multiple ROIs and image them in succession at a selected repetition rate any number of times.
Focus Interface
Before data acquisition the structure to be imaged needs to be located. For this purpose a rapid acquisition mode is necessary where the user can observe the image in realtime as the specimen and focal positions move. ScanImage includes a focus mode to accomplish this. The image seen in focus mode is identical to the one that will be acquired. Multiple channels can be focused simultaneously in realtime. Image zooming and rotation are accomplished in the focus mode by changing the signal output to the mirrors.
Modular Design of ScanImage: User Functions
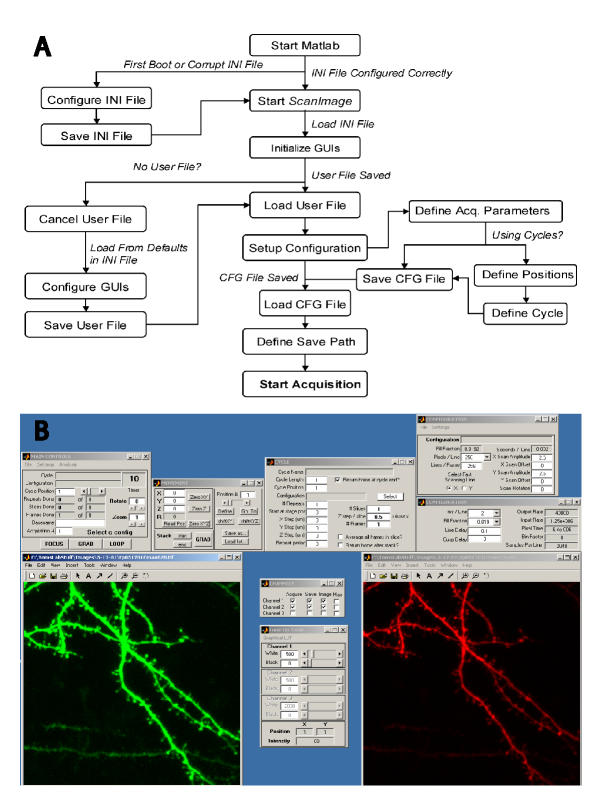
One of the key advantages of ScanImage is the user-friendly development environment, MATLAB. Users can easily develop and test data analysis functions off-line and incorporate them into the software without fear of interfering with the data acquisition. A flowchart of ScanImage from a programming perspective is given in Figure 4A and a screenshot of the software is shown in Figure 4B.
Figure 4.
Flowchart and Screenshot of ScanImage A) ScanImage uses a series of text files to remember configurations for different microscopes, users, and experimental configurations. Each aspect of the software and the associated files is detailed in the ScanImage manual. B) A screenshot of ScanImage acquiring data from 2 channels simultaneously.
Any MATLAB function can be automatically executed after an image is acquired to aid in analysis of the experiment while it is being conducted (see ScanImage manual for details). In addition to MATLAB functions, programmers can compile C and Fortran code and run it with MATLAB calls in the same user function.
Modular Design of ScanImage: Controlling Additional Devices
From a programming perspective, the code allows the generation of custom graphical interfaces that can be incorporated easily into ScanImage (see Figure 4A). As an example, additional serial ports may need to be integrated to run other optical equipment. A GUI can be constructed with its own initialization file and run in parallel to ScanImage (see the ScanImage manual for details). This GUI can use the same convenient tools for handling GUIs that ScanImage uses while offering the programmer flexibility in both design and development.
Availability and Distribution
ScanImage is freely available as OSI Certified open source software. The license can be found in the ScanImage folder in the ScanImageLicense.txt file. All terms of the license, including details on referencing ScanImage in published reports, must be followed when using ScanImage.
The software and documentation on installation are available from: http://svobodalab.cshl.edu/software_main.html.
Please email any questions or suggestions about the software to scanimage@cshl.edu
Methods
Programming
All programming and benchmarking was done in MATLAB 6.1 running on a Windows 2000 Professional PC with an 800 MHz Pentium III processor and 512 MB of RAM.
List of Abbreviations
2PLSM two-photon excitation laser scanning microscopy
CLSM confocal laser scanning microscopy
DAQ data acquisition
CPU central processing unit
TTL transistor-transistor logic
PMT photo-multiplier tube
GUI graphical user interface
SNR signal-to-noise-ratio
LUT look-up table
DSP digital signal processing
COM communications port
FWHM full-width-at-half-maximum
SEM standard error of the mean
RAM random access memory
Author's Contributions
BLS and KS conceived and planned this project. TAP and BLS wrote the majority of the code, with contributions from KS. Simulations and experiments were performed by TAP with KS. TAP and KS drafted the manuscript.
Acknowledgments
Acknowledgements
We thank Brian Chen, Karen Zito, and Josh Trachtenberg for beta-testing early versions of ScanImage. We thank Ryohei Yasuda for helpful comments on the analysis. We thank Gordon Shepherd, Karen Zito, and Thomas Oertner for helpful comments on the manuscript.
This work was funded in part by a grant from the National Institutes of Health (Grant# R01 EB01464-01).
Contributor Information
Thomas A Pologruto, Email: pologrut@cshl.org.
Bernardo L Sabatini, Email: bernardo_sabatini@hms.harvard.edu.
Karel Svoboda, Email: svoboda@cshl.org.
References
- White EL, Keller A. Intrinsic circuitry involving the local axon collaterals of corticothalamic projection cells in mouse Sm1 cortex. J Comp Neurol. 1987;1987:13–26. doi: 10.1002/cne.902620103. [DOI] [PubMed] [Google Scholar]
- Goldstein SR, Hubin T, Smith TG. An improved, no moving parts video rate confocal microscope. Micron and Microscopica Acta. 1992;23:437–442. doi: 10.1016/0739-6260(92)90019-A. [DOI] [Google Scholar]
- Pawley JB, ed . Plenum Press: New York. 3 1995. Handbook of Biological Confocal Microscopy. [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Denk W, Svoboda K. Photon upmanship: why multiphoton imaging is more than a gimmick. Neuron. 1997;18:351–357. doi: 10.1016/S0896-6273(00)81237-4. [DOI] [PubMed] [Google Scholar]
- Gosnell TR, Taylor AJ, eds Selected Papers on Ultrafast Laser Technology. Milestone Series SPIE Press: Bellingham, WA. 1991.
- Pawley JBaCV. Practical laser-scanning confocal light microscopy: Obtaining optimal performance from your instrument. In: Celis JE, editor. in Cell Biology: A Laboratory handbook. Academic Press: New York; 1994. pp. 44–64. [Google Scholar]
- Majewska A, Yiu G, Yuste R. A custom-made two-photon microscope and deconvolution system. Pflugers Arch. 2000;441:398–408. doi: 10.1007/s004240000435. [DOI] [PubMed] [Google Scholar]
- Lechleiter JD, LDT. Sieneart I. Multi-photon laser scanning microscopy using an acoustic optical deflector. Biophysical Journal. 2002;83:2292–2299. doi: 10.1016/S0006-3495(02)73989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, et al. Anatomical and functional imaging of neurons using 2-photon laser scanning microscopy. J Neurosci Meth. 1994;54:151–162. doi: 10.1016/0165-0270(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Sunderland WJ, Wilkinson RS. High-resolution imaging of synaptic structure with a simple confocal microscope. New Biol. 1989;1:75–82. [PubMed] [Google Scholar]
- Tsai PS, et al. Principles, design and construction of a two photon scanning microscope for in vitro and in vivo studies. In: Frostig R, editor. In Methods for In Vivo Optical Imaging. CRC Press; 2002. pp. 113–171. [Google Scholar]
- Mainen ZF, et al. Two-photon imaging in living brain slices. Methods. 1999;18:231–239. doi: 10.1006/meth.1999.0776. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–94. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Oertner TG, et al. Facilitation at single synapses probed with optical quantal analysis. Nat Neurosci. 2002;5:657–664. doi: 10.1038/nn867. [DOI] [PubMed] [Google Scholar]
- Trefethen E, et al. MultiMatlab: MATLAB on multiple processors. Cornell Theory Center. 1996. pp. 96–239.
- Horowitz P, Hill W. Cambridge: Cambridge University Press. 2. 1989. The Art of Electronics; p. 1125. [Google Scholar]
- Jones R, OIiver CJ, Pike ER. Experimental and theoretical comparison of photon-counting and current measurements of light intensity. Appl Opt. 1970;10:1673–1680. doi: 10.1364/AO.10.001673. [DOI] [PubMed] [Google Scholar]
- Tan YP, et al. Fast scanning and efficient photodetection in a simple two-photon microscope. J Neurosci Methods. 1999;92:123–35. doi: 10.1016/S0165-0270(99)00103-X. [DOI] [PubMed] [Google Scholar]