Abstract
Background
The bioavailability of iron is quite low since it is usually present as insoluble complexes. To solve the bioavailability problem microorganisms have developed highly efficient iron-scavenging systems based on the synthesis of siderophores that have high iron affinity. The systems of iron assimilation in microorganisms are strictly regulated to control the intracellular iron levels since at high concentrations iron is toxic for cells. Streptomyces pilosus synthesizes the siderofore desferrioxamine B. The first step in desferrioxamine biosynthesis is decarboxylation of L-lysine to form cadaverine, a desferrioxamine B precursor. This reaction is catalyzed by the lysine decarboxylase, an enzyme encoded by the desA gene that is repressed by iron.
Results
The binding of the DmdR (acronym for divalent metal dependent repressor) to the desA promoter in presence of Fe2+ or other divalent ions has been characterized. A 51 bp DNA fragment of the desA promoter containing the 9 bp inverted repeat was sufficient for binding of the DmdR repressor, as observed by the electrophoretic mobility shift assay. The desA mobility shift was prevented by neutralizing DmdR with anti-DmdR antibodies or by chelating the divalent metal in the binding reaction with 2,2'-dipyridyl. Binding to the desA promoter was observed with purified DmdR repressors of Streptomyces coelicolor or Rhodococcus fascians suggesting that there is a common mechanism of iron-regulation in actinomycetes. The complete desA promoter region was coupled using transcriptional fusions to the amy reporter gene (encoding α-amylase) in low copy or multicopy Streptomyces vectors. The iron-regulated desA promoter was induced by addition of the iron chelating agent 2,2'-dipyridyl resulting in a strong expression of the reporter gene.
Conclusions
The iron-regulated desA promoter can be used for inducible expression of genes in Streptomyces species, as shown by de-repression of the promoter when coupled to a reporter gene.
Background
Iron is an important nutrient for microorganisms since it plays an essential role for cell growth. Iron acts as a cofactor of a large number of enzymes, forms part of cytochromes and is required for nitrogen fixation and DNA synthesis [1-3]. Despite its abundance in soil, the bioavailability of iron is quite low since it is usually present as insoluble complexes [3,4]. To solve the bioavailability problem microorganisms have developed highly efficient iron-scavenging systems based on the synthesis of siderophores that have high iron affinity [5-7]. The systems of iron assimilation in microorganisms are strictly regulated to control the intracellular iron levels since at high concentrations iron is toxic for cells [8-15]. Siderophores form a six-coordinated octaedral complex with ferric iron with extremely high affinity (stability constants) ranging from 1023 to 1052.
Streptomyces pilosus synthesizes the siderofore desferrioxamine B [16]. Desferrioxamine B is used clinically to treat disorders related to iron overload and pathological iron deposition in man [17]. The first step in desferrioxamine biosynthesis is decarboxylation of L-lysine to form cadaverine, a desferrioxamine B precursor (Fig. 1). This reaction is catalyzed by the lysine decarboxylase, an enzyme encoded by the desA gene that is repressed by iron [18,19].
Figure 1.
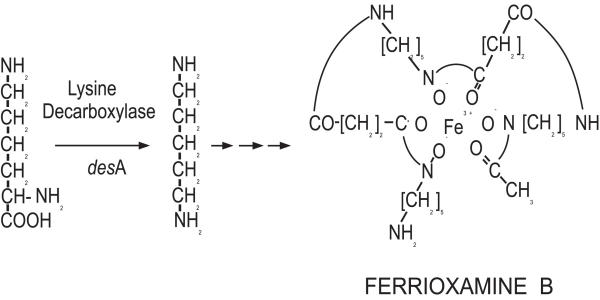
Biosynthesis of desferrioxamine B from lysine. The desferrioxamine B component binds Fe3+ to form ferrioxamine B. Lysine decarboxylase encoded by the desA gene catalyzes the first step of the biosynthetic pathway.
The desA promoter possess a region involved in iron regulation containing a symmetrical 19 bp sequence that overlaps with the -10 box and the transcription initiation site of the desA gene [20]. This palindromic sequence is very similar to the iron box recognized by the DtxR iron regulator of Corynebacterium diphtheriae [21] and the dmdR gene of Corynebacterium (formerly Brevibacterium) lactofermentum [22]. A DmdR protein similar to DtxR occurs in several Streptomyces strains [23] (Flores FJ and Martín JF, unpublished results).
The availability of iron-regulated promoters may provide a very useful biological sensor to test the presence of iron in different environments. In addition, an iron-regulated promoter provides an interesting tool for controlled expression of genes at specific times during the fermentation by addition of either iron or iron-chelating agents.
It was, therefore, of great interest to characterize the binding of the DmdR repressor to the desA promoter and to study the derepression of the desA gene of S. pilosus following addition of iron chelators.
We report in this article that DmdR repressors from different actinomycetes bind to the desA promoter in presence of iron making this system an excellent regulated promoter for inducible or repressible expression of genes in different actinomycetes.
Results
Characterization of the desA promoter
A detailed analysis of the promoter region of the desA gene of S. pilosus was performed to define the regulatory region involved in iron-regulation and to study if the desA promoter could be regulated by the DmdR repressor of different actinomycetes. As shown in Fig. 2, the desA promoter region contains a 19 bp palindromic region overlapping with the transcription initiation site defined by Günter et al. [20]. To ascertain that this putative repressor binding sequence was involved in iron regulation a DNA fragment of 51 bp extending from -31 to +21 was synthesized, labeled and used as a probe in electrophoretic mobility shift assays. DmdR proteins purified from Streptomyces coelicolor and Rhodococcus fascians were used for the binding assays in the electrophoretic mobility shift assays.
Figure 2.
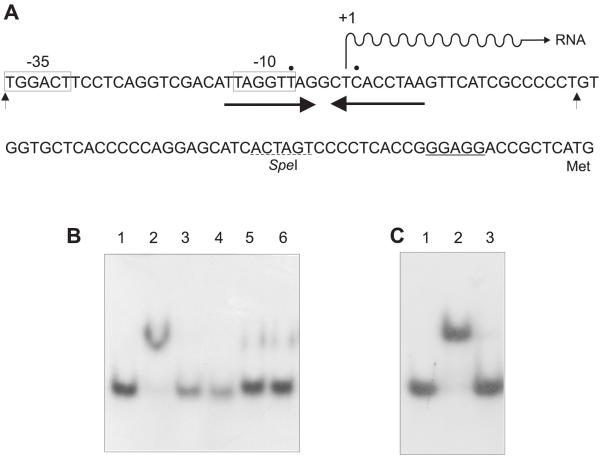
A Nucleotide sequence of the promoter operator region of the desA gene containing the -10 and -35 boxes; the transcription start point is indicated as +1. The mRNA is indicated by a wavy line and the putative ribosome binding site GGAGG upstream of the first translated codon ATG is underlined. The convergent thick arrows indicate the palindromic sequence of binding of the DmdR protein (operator). The two vertical arrows indicate the DNA fragment used for the gel mobility shift experiments. B Electrophoretic mobility shift assay of the [32P]labeled desA probe and DmdR protein from S. coelicolor. Lanes 1, labeled free desA probe; 2, labeled desA, mixed with DmdR (note the band shift); 3, labeled desA with excess unlabeled desA; 4, labeled desA with DmdR and anti-DmdR antibodies; 5, labeled desA with DmdR and 2,2'-dipyridyl (200 mM) without Mn2+; 6, labeled desA with DmdR and 2,2'-dipyridyl (200 mM). C Same as in B but with DmdR purified from R. fascians. Lanes 1, labeled desA; 2, labeled desA with DmdR; 3, labeled desA with DmdR and excess unlabeled desA.
As shown in Fig. 2B and 2C, a strong shift of the desA promoter was observed in presence of the pure DmdR protein of either R. fascians or S. coelicolor. The mobility shift was removed by neutralizing the DmdR protein with anti-DmdR antibodies (Fig. 2B, lane 4) or by adding unlabeled excess desA probe (lane 3). The mobility shift required a divalent metal and was prevented by addition of the chelator 2,2-dipyridyl.
Constructions with the desA promoter in monocopy and multicopy vectors
The promoterless amy gene of S. griseus IMRU 3570 was subcloned as a 1.8 kb EcoRI-EcoRV band from plasmid pAM2PP [24] into the vector pTC191 (digested with SmaI-EcoRI) giving rise to plasmid pTCA. The promoter of the S. pilosus desA gene was subcloned from plasmid pTQ217 (a derivative of pTQ209) [19] as a 0.8 kb Ecl136II-SpeI DNA fragment upstream of the promoterless amy gene, giving rise to plasmid pTCDA. The correct orientation was confirmed by restriction endonuclease mapping of the construction. The construction with the incorrect orientation of the desA promoter with respect to the amy reporter gene was named pTCADi and was used as a negative control.
Both, the direct and the inverted desA-amy fusions, were subcloned as HindIII fragments into the multicopy Streptomyces coelicolor vector pIJ699 [25] and the low-copy pIJ2842 vector [26]. The constructions derived from the multicopy plasmid were named pUL99DA and pUL99ADi, respectively, whereas the constructions derived from the low-copy vector were designated as pUL42DA and pUL42ADi, respectively (Fig. 3).
Figure 3.
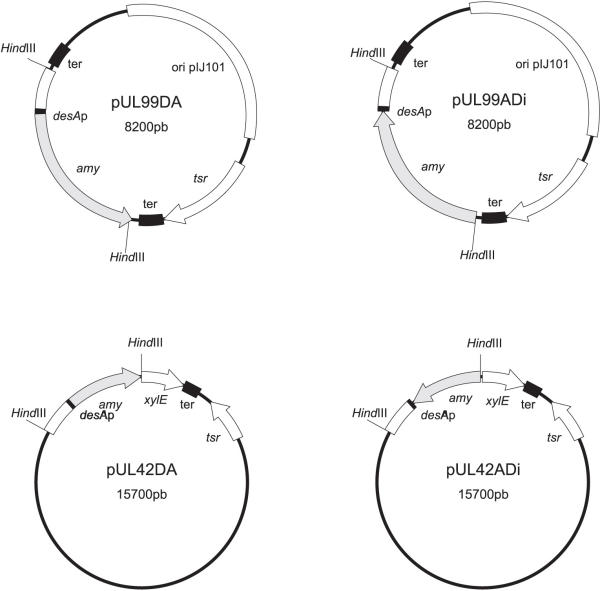
Physical map of the four vectors used to quantify iron regulation of the desA promoter. pUL99DA and pUL99ADi are multicopy vectors whereas pUL42DA and pUL42Adi are low copy number vectors; amy indicates the promoter-less amy gene; desAp corresponds to the desA promoter; xylE, catechol oxygenase gene; ter, transcriptional terminator; tsr, thiostrepton resistance gene (see Table 1).
Transformants with the pUL99DA and pUL42DA showed a strong expression of the amylase reporter gene when tested directly on colonies growing on plates of starch-containing LS medium with 2,2'-dipyridyl. Transformants with plasmids pUL99ADi and pUL42ADi that contained the desA promoter coupled in the inverted (incorrect) position to the amy gene did not show any expression of the amy gene and produced only the background halo of starch digestion due to the endogenous α-amylase of the S. coelicolor host (Fig. 4).
Figure 4.
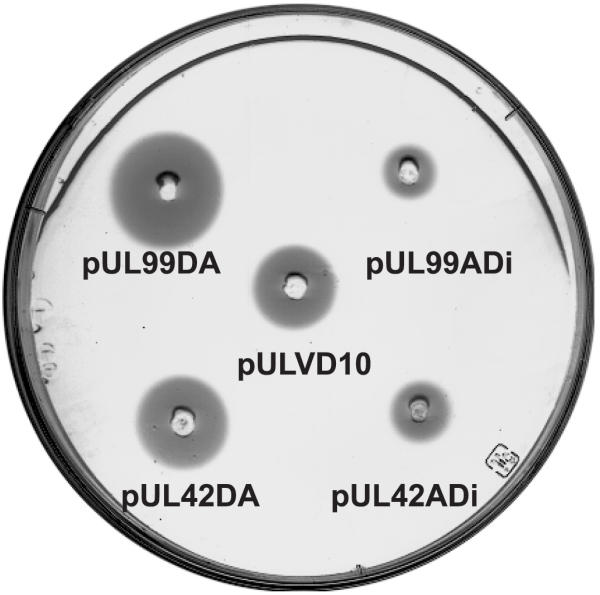
Direct test on agar plates of the reporter α-amylase expression ability of the desA promoter using round patches of growth of pUL99DA and pUL42DA transformants in LS medium containing 2,2'-dipyridyl as compared to transformants with pULVD10 in which the reporter amylase gene is under the control of the saf promoter. Control transformants with pUL99ADi and pUL42ADi are shown on the right. The plate was stained with I2 + IK; the figure is a negative of the plate photograph.
Expression of the desA-amy fusion is regulated by iron in multicopy and low-copy plasmids
To test the effect of iron on expression of the desA-amy fusion, the formation of amylase was quantified in S. coelicolor transformants containing plasmids pUL99DA or pUL42DA. S. coelicolor cells were grown in YEME + 34% sucrose collected and suspended in LS medium as indicated in Materials and Methods. To quantify the response to iron-depletion, iron in this medium was removed by addition of the chelating agent 2,2'-dipyridyl. Results showed that there was a seven-fold increase in the synthesis of the reporter enzyme in cells deprived of iron (Fig. 5A). When iron was complexed with the chelating agent, a large increase in the reporter enzyme activity was observed until 60 h of incubation while in cells growing in iron the amylase activity declined after 36 h.
Figure 5.
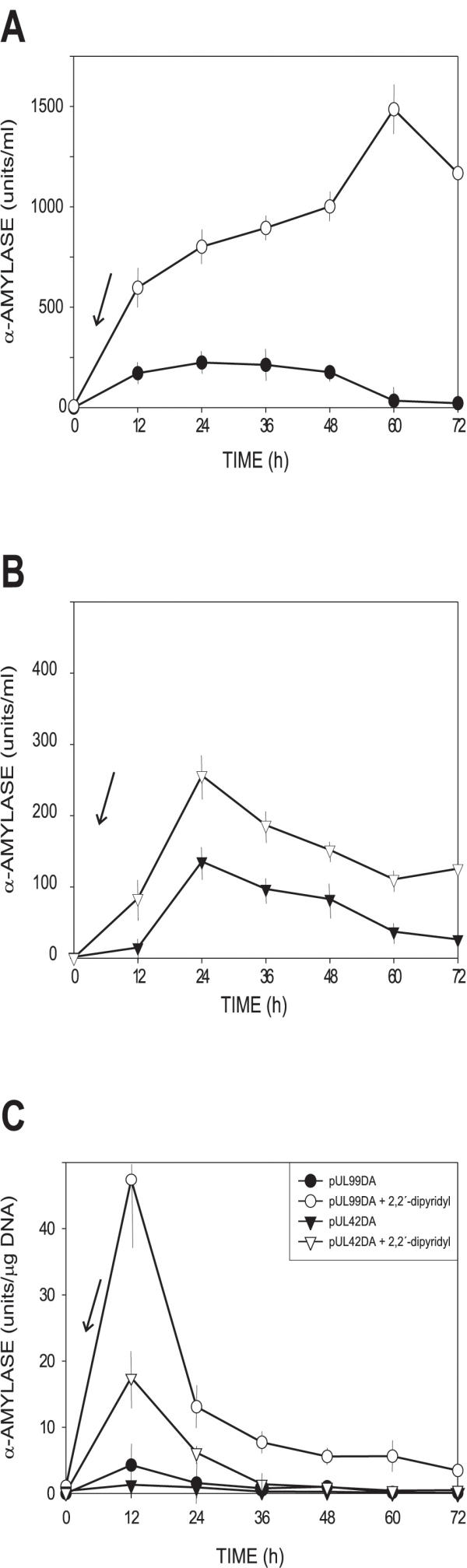
Iron regulation of expression of the reporter α-amylase gene coupled to the desA promoter in a multicopy transformant (pUL99DA) (panel A) and a low-copy number transformant (pUL99DA) (panel B) expressed as volumetric enzyme activity (panels A and B) or as specific activity (panel C). ○, ●, pUL99DA; ▼, ▽, pUL42DA. Closed symbols, control cultures in LS medium; open symbols, cultures induced with the iron-complexing agent 2,2'-dypirydyl added at time 0 (inclined arrows).
Similar results were also observed when the reporter α-amylase activity was expressed as specific activity per μg of DNA (dry weight can not be determined in starch-containing media). Although total amylase accumulated until the 60 h the highest specific rate of desA expression as measured by the specific rate of α-amylase peaked at 12 h following addition of the iron chelator (Fig. 5B).
Constructions containing the desA promoter in multicopy plasmids might, however, titrate the DmdR repressor. It was, therefore, of interest to test the effect of iron on low-copy plasmids (1 or 2 copies per genome).
Constructions carrying the desA promoter in the low-copy plasmid pUL42DA were also strongly regulated. Removal of iron with 2,2'-dipyridyl produced a two-fold increase in the expression of the reporter gene (Fig. 5B). The same effect was also observed when the reporter activity was expressed as specific amylase activity (Fig. 5C). In low-copy number transformants the reporter amylase activity decreased after 36 hours whereas in the high copy number transformants it accumulated in the culture until 60 h, reflecting a much higher ability to produce α-amylase in the transformants with the multicopy plasmids.
Taken together these results indicate that the desA promoter is regulated in the multicopy pIJ99DA transformant as well as in low-copy transformants. The DmdR repressor level is sufficient to keep most of the desA copies repressed under iron excess conditions even in transformants containing a high number of copies (about 50 copies per cell).
Iron-regulated versus non-regulated promoters as tools for gene expression
The iron-regulated desA promoter serves as an inducible system for expression of genes following iron removal with 2,2'-dipyridyl. Previously, we compared the strength of different Streptomyces promoters by coupling them to the amy reporter gene [27]. The best Streptomyces promoter available in our hands is the saf gene promoter that drives expression of the secretion activating factor (saf). The transcription initiation ability of the desA promoter was compared with that of saf promoter in plasmids containing the same replicon (from pIJ101).
Results of the comparative study (Fig. 6) showed that whereas the desA-mediated expression of the reporter gene was strongly regulated by iron, expression from the saf promoter was largely insensitive to iron starvation.
Figure 6.
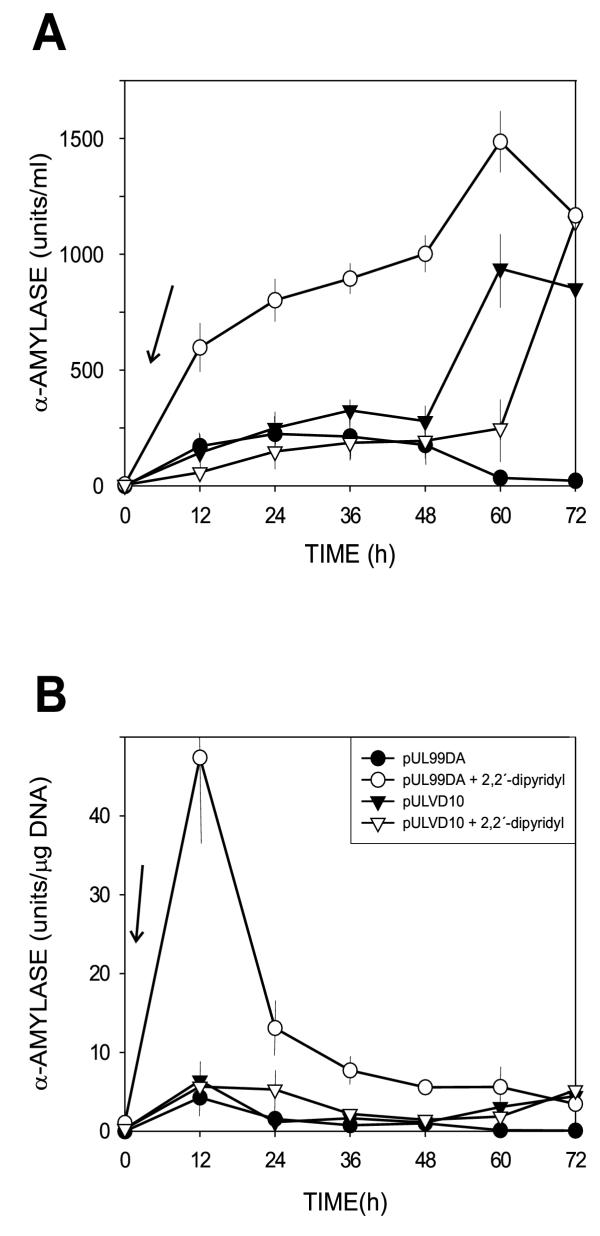
Expression of the reporter α-amylase gene from the iron-regulated desA promoter versus the non-regulated saf promoter. Experimental conditions were as in Fig. 5. ○, ●, pUL99DA; ▼, ▽, pULVD10. Closed symbols control cultures in LS medium; open symbols cultures induced by addition of the iron-chelator 2,2'-dipyridyl at time 0 (vertical arrows). Results are given as volumetric enzyme levels (panel A) or as specific enzyme production (panel B). The inclined arrows indicate the time of addition (0 h) of the 2,2'-dipyridyl inducer
The levels of amylase were about 50% higher in cultures expressing the reporter gene from the desA promoter under derepression conditions than in transformants expressing the reporter from the saf promoter (either under iron repressing or derepressing conditions).
Discussion
In bacteria and lower eurcaryotes iron is scavenged and transported into the cells by high-affinity iron-chelators (siderophores). When the concentration of iron in the environment is low, transcription of the siderophore-encoding genes is triggered. The specific transcriptional response of the genes encoding the siderophores and their receptors is an attractive subject because of its biotechnological interest [5].
In recent years considerable advances were made in the characterization of the DtxR repressor of C. diphtheriae that mediates iron-regulation. The regulatory protein DtxR binds to a 9 bp-interrupted palyndrome in the diphtheria toxin gene (see review by Tao et al. 1994). We have found previously that genes similar to dtxR that are generically named dmdR (for divalent metal dependent regulator) occur in Corynebacterium (formerly Brevibacterium) lactofermentum [22] in Rhodococcus fascians (J. Rincon and J.F. Martín, unpublished) and several Streptomyces species (Flores FJ and Martín JF, unpublished).
A few DmdR responsive operators occurring in the promoter regions of iron-regulated genes have been analyzed. Most of the information available refers to the tox gene of C. diphtheriae. The promoter of the desA gene of Streptomyces pilosus contains one of the consensus DmdR-binding sequences [20]. As shown in this article, purified DmdR repressor proteins from two different actinomycetes are able to interact with the 51 bp region containing the desA operator. The interaction required a divalent metal ion, either Fe2+, Mn2+ or Co2+. All the available evidence indicates that there is a common mechanism of iron regulation mediated by repressors of the DmdR family in corynebacteria and in higher actinomycetes.
There is a need of characterizing well-defined regulated promoters as tools for controlled gene expression in actinomycetes. Günter and coworkers [20] showed that the regulation of desferrioxamine B production is exerted at the transcriptional level. The concentration of iron in culture media can be regulated very simply by addition of the 2,2'-dipyridyl chelator [22] and, therefore, as shown in this article, expression from the desA promoter can be triggered at will by iron starvation at different times. There is no endogenous desA gene in most actinomycetes since this gene appears to be specific for secondary metabolism [19].
When the reporter activity was determined, we observed regulation both in low-copy number expression vectors and in multicopy vectors. It seems, therefore, that enough molecules of the DmdR iron regulator exist in S. coelicolor to bind the copies of the desA promoter in the multicopy vector. DNA binding to the desA promoter as shown by electrophoretic mobility shift assay occurred with the DmdR repressors of S. coelicolor, R. fascians (F. Flores and J.F. Martín, unpublished) and S. lividans [23]. The presence in several actinomycetes of a DmdR repressor (F.J. Flores and J.F. Martín, unpublished) makes this iron-regulated expression system very attractive for tailored expression of homologous or heterologous genes in a variety of actinomycetes.
Conclusions
- The iron-box of the iron-regulated desA promoter of S. pilosus has been characterized by the electrophoretic mobility shift assay (EMSA) technique using the purified DmdR repressor proteins of S. coelicolor and R. fascians that interact with the iron-box
- The iron-regulated desA promoters has been coupled to the reporter amy gene (encoding α-amylase) in low copy and high copy Streptomyces vectors.
- The desA promoter is strongly induced by addition of the iron-chelating agent 2,2'-dipyridyl.
- This iron-regulated desA promoter can be used for controlled expression of other transcriptionally-coupled genes in Streptomyces species. Expression from this promoter is switched off by iron and switched-on by 2,2'-dipyridyl.
Methods
Microorganisms, plasmids and culture conditions
Streptomyces coelicolor A3(2) was routinely used as a model actinomycete. S. coelicolor cultures were grown in YEME (yeast extract 10 g/l, malt extract 10 g/l) with 34% sucrose to obtain disperse growth [28] or in defined Lechevalier medium [29] supplemented with 10 g/l soluble starch (LS medium) for quantification of amylase production. Thiostrepton was added to the culture medium to maintain the plasmids in the transformants (final concentration 25 mg/ml in solid medium; 5 mg/ml in liquid media). All plasmids used in this work are listed in Table 1. E. coli DH5α was grown in TB medium [30] or in LA medium as indicated by Hannahan [31]. E. coli transformants were selected on LA plates containing ampicillin (100 μg/ml). 2,2'-dipyridyl was added, when required, at a final concentration of 250 μM.
Table 1.
Plasmids used in this work
| Plasmid | Description | Reference |
| pTC191 | A derivative of pUC19 containing identical restriction sites on both sides of the polylinker | [36] |
| pAM2PP | Contains the promoter-less amy gene (encoding the S. griseus α-amylase) | [24] |
| pTQ217 | A derivative of pTQ207 (Günter et al. 1993) containing the desA gene of S. pilosus encoding the enzyme lysine decarboxylase | T. Schupp, personal communication |
| pIJ699 | Bifunctional vector E. coli-Streptomyces of high copy number. The functional region in Streptomyces is flanked by transcriptional terminators. Allows positive selection of transformants | [25] |
| pULVD10 | A derivative of pIJ699 that contains the promoter-less amy gene expressed from the saf promoter (safp) of S. griseus | [27] |
| pIJ2842 | Low-copy number promoter-probe vector for Streptomyces promoters | [26] |
| pTCA | A derivative of pTC191 containing the amy gene without its promoter in a 1.8 kb DNA fragment | This work |
| pTCDA | A derivative of pTCD containing the desA promoter (desAp) coupled to the 5' end of the promoter-less amy gene | This work |
| pTCADi | A derivative of pTCD containing the desA promoter fused to the 3' end of the amy gene (negative control) | This work |
| pUL99DA | A derivative of the multicopy pIJ699 containing the transcriptional fusion desAp-amy in the correct orientation | This work |
| pUL99ADI | A derivative of pIJ699 containing the incorrect transcriptional fusion amy-desAp (non-functional) | This work |
| pIJ42DA | A derivative of the low-copy pIJ2842 containing the transcriptional fusion desAp-amy in the correct orientation | This work |
| pIJ42ADI | A derivative of the low-copy pIJ2842 containing the incorrect transcriptional fusion amy-desAp no functional | This work |
DNA procedures
Transformation conditions for S. coelicolor and plasmid DNA isolation from the transformants were as described by Hopwood et al. [32]. DNA digestion with restriction endonucleases, end-filling and removal of 5' or 3'-protruding ends were performed by standard procedures [33].
Preparation and labeling of the DNA probes
A 51 bp DNA fragment containing the 19 bp inverted repeat based on the nucleotide sequence of the desA gene [20] was synthesized by annealling two 47 bp oligonucleotides (with stagered ends) and subcloning the 51 bp DNA fragment in pBluescript KS+. The insert was sequenced to confirm the nucleotide sequence. After plasmid amplification the probe was scissed from the vector and isolated from the agarose gel by the Quiaex procedure (Quiagen) and labeled by end-filling with the Klenow fragment of the DNA polymerase and [32P]α-dCTP.
Electrophoretic mobility shift assays
The electrophoretic mobility shift assay was performed as described by Tao et al. [34] using as probes DNA fragments containing the desA regulatory region labeled by end-filling with Klenow DNA polymerase. The His-tagged DmdR protein of R. fascians was purified by filtration through a Ni 2+-NTA column after cloning the R. fascians dmdR in the expression vector pQE (Quiagen). The DmdR1 protein of. S. coelicolor was purified as a GST-fused protein by filtration through a Glutathion-Sepharose 4B affinity column (Amersham-Pharmacia Biotech). The dmdR1 gene was first cloned in a pGEX expression vector (Amersham-Pharmacia). Following elution of the GST-DmdR1 fused protein from the column, the DmdR1 protein was released by cleavage with thrombin as indicated by the manufacturer. The DmdR binding reaction was performed for 30 min at 30°C in a final volume of 30 μl containing 5 mM MgCl2, 40 mM KCl, 2 mM DTT, 125 mM MnCl2, 10% (v/v) glycerol, 1 μg of poli dI-dC, 8.5 μg BSA, purified DmdR protein (about 20 ng) and the desA labeled probe.
Quantification of the reporter α-amylase activity
The α-amylase activity was determined by quantifying the maltose released from soluble starch with 3,5-dinitrosalycilic acid [35]. All cultures in LS medium were inoculated with 1 ml of cells (O.D. = 1) grown in YEME + 34% sucrose washed twice with sterile saline solution. One unit of amylase was defined as the activity that forms one nmol of maltose per minute. The specific activity is given as units per μg of DNA in the cells.
Abbreviations
BSA, bovine seroalbumin; LA, Luria agar; TB, triptic broth; DTT, dithiothreitol; LS, Lechevalier starch.
Authors' contributions
JFM directed the research and designed the experimental work. FJF performed most of the experimental work in the article. JR helped with the EMSA assays. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by a grant of the Agencia de Desarrollo de Castilla y Leon (10-2/98/LE/0003). F.J. Flores received a fellowship of the Areces Foundation, Madrid (Spain). We thank T. Schupp for sending plasmid pTQ217.
Contributor Information
Francisco J Flores, Email: degffd@unileon.es.
Javier Rincón, Email: inbiotec@inbiotec.com.
Juan F Martín, Email: degjmm@unileon.es.
References
- Weinberg ED. The development of awareness of iron-withholding defense. Prospect Biol Med. 1993;36:215–221. doi: 10.1353/pbm.1993.0063. [DOI] [PubMed] [Google Scholar]
- Tai SS, Zhu YY. Cloning of a Corynebacterium diphtheriae iron-repressible gene that shares sequence homology with the AhpC subunit of alkil hydroperoxide reductase of Salmonella typhimurium. J Bacteriol. 1995;177:3512–3517. doi: 10.1128/jb.177.12.3512-3517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. Microbial envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- Crosa JH. Signal transduction and transcriptional and posttranscriptional control or iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1987;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Yong-Ho K, Yang ZK, Burlage RS. Measurement of iron-dependence of pupA promoter activity by pup-lux bioreporter. J Microbiol Biotechnol. 1997;7:352–355. [Google Scholar]
- Arahou M, Diem HG, Sasson A. Influence of iron depletion on growth and production of catechol siderophores by different Frankia strains. World J Microbiol Biotechnol. 1998;14:31–36. [Google Scholar]
- Bullen JJ, Rogers HJ, Griffiths E. Role of iron in bacterial infection. Curr Top Microbiol Immun. 1978;80:1–35. doi: 10.1007/978-3-642-66956-9_1. [DOI] [PubMed] [Google Scholar]
- Cox CD. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J Bacteriol. 1980;142:581–587. doi: 10.1128/jb.142.2.581-587.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Weaver LH, Matthews BW. Use of protein sequence and structure to infer distant evolutionary relationships. Chem Scripta. 1986;26B:251–255. [Google Scholar]
- Crichton RR, Charloteaux-Wauters M. Iron transport and storage. Eur J Biochem. 1987;164:485–506. doi: 10.1111/j.1432-1033.1987.tb11155.x. [DOI] [PubMed] [Google Scholar]
- Postle K. TonB and the gram-negative dilemma. Mol Microbiol. 1990;4:2019–2025. doi: 10.1111/j.1365-2958.1990.tb00561.x. [DOI] [PubMed] [Google Scholar]
- Hider RC. Siderophore mediated absortion of iron. Struct Bonding. 1984;58:25–87. [Google Scholar]
- Heinrichs D, Poole K. PchR, a regulator of ferripyochelin receptor gene (fpt A) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol. 1996;178:2586–2592. doi: 10.1128/jb.178.9.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol Chem. 1997;387:779–786. [PubMed] [Google Scholar]
- Bickel H, Bosshardt R, Gäumann E, Reusser P, Vischer E, Voser W, Wettstein A, Zähner H. Stoffwechsel-produkte von actinomyceten, über die isolierung und charakterisierung der ferroxiamine A-F, neuer wuchsstoff der sideramin-gruppe. Helv Chim Acta. 1960;43:2118–2128. [Google Scholar]
- Peter HH. Industrial aspects of iron chelators: pharmaceutical applications. In: Spik G, Montreuil J, Crichton RR, Mazurier J, editor. Proteins of Iron Storage and Transport. Amsterdam: Elsevier; 1985. pp. 293–303. [Google Scholar]
- Schupp T, Waldmeier U, Divers M. Biosynthesis of desferrioxamine B in Streptomyces pilosus : evidence for the involvement of lysine decarboxylase. FEMS Microbiol Lett. 1987;42:135–139. [Google Scholar]
- Schupp T, Toupet C, Divers M. Cloning and expression of two genes of Streptomyces pilosus involved in the biosynthesis of siderophore desferroxamine B. Gene. 1988;64:179–188. doi: 10.1016/0378-1119(88)90333-2. [DOI] [PubMed] [Google Scholar]
- Günter K, Toupet C, Schupp T. Characterization of an iron-regulated promoter involved in desferrioxamine B synthesis in Streptomyces pilosus : Repressor-binding site and homology to the diphtheria toxin gene promoter. J Bacteriol. 1993;175:3295–3302. doi: 10.1128/jb.175.11.3295-3302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Schiering N, Zeng H, Ringe D, Murphy JR. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- Oguiza JA, Tao X, Marcos AT, Martín JF, Murphy JR. Molecular cloning, DNA sequence analysis, and characterization of the Corynebacterium diphtheriae dtx R homolog from Brevibacterium lactofermentum. J Bacteriol. 1995;177:465–467. doi: 10.1128/jb.177.2.465-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günter-Seeboth K, Schupp T. Cloning and sequence analysis of the Corynebacterium diphtheriae dtxR homologue from Streptomyces lividans and S. pilosus encoding a putative iron represor protein. Gene. 1995;166:117–119. doi: 10.1016/0378-1119(95)00628-7. [DOI] [PubMed] [Google Scholar]
- Chary VK, de la Fuente JL, Liras P, Martín JF. amy. 1997. pp. 2977–2982. [DOI] [PMC free article] [PubMed]
- Kieser T, Melton RD. Plasmid pIJ699, a multicopy positive selection vector for Streptomyces. Gene. 1988;65:83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- Clayton M, Bibb MJ. Streptomyces promoter-probe plasmids that utilise the xyl E gene of Pseudomonas putida. Nucleic Acids Res. 1990;18:1077. doi: 10.1093/nar/18.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigal T, Gil JA, Daza A, García-González MD, Villadas P, Martín JF. Effects of replacement of promoters and modification of the leader peptide region of the amy gene of Streptomyces griseus on synthesis and secretion of α-amylase by Streptomyces lividans. Mol Gen Genet. 1991;231:88–96. doi: 10.1007/BF00293826. [DOI] [PubMed] [Google Scholar]
- Chater KF, Hopwood DA, Kieser T, Thompson CJ. Gene cloning in Streptomyces. Curr Top Microbiol Immun. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Martín JF, McDaniel L. Biosynthesis of candicidin by phosphate-limited resting cells of Streptomyces griseus. Eur J Appl Microbiol. 1976;3:135–144. [Google Scholar]
- Tartof KD, Hobbs CA. Improved media for growing plasmid and cosmid clones. Bethesda Res Lab Focus. 1987;9:12–14. [Google Scholar]
- Hannahan D. Studies on transformation of Escherichia coli. J Mol Microbiol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith DP, Ward JM, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Tao X, Boyd J, Murphy JR. Specific binding of the diphtheria tox regulatory element DtxR to the tox operator requires divalent heavy metal ions and a 9-base-pair interrupted palindromic sequence. Proc Natl Acad Sci USA. 1992;89:5897–5901. doi: 10.1073/pnas.89.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-González MD, Martín JF, Vigal T, Liras P. Characterization, expression in Streptomyces lividans, and processing of the amylase of Streptomyces griseus IMRU 3570: Two different amylases are derived from the same gene by an intracellular processing mechanism. J Bacteriol. 1991;173:5471–5478. doi: 10.1128/jb.173.8.2451-2458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente A, Cisneros E, Talavera A. Restriction end-converting vectors with repeated multiple cloning sites. Gene. 1994;139:83–86. doi: 10.1016/0378-1119(94)90527-4. [DOI] [PubMed] [Google Scholar]


