Abstract
Background
The generation of BAC/PAC contigs in targeted genome regions is a powerful method to establish high-resolution physical maps. In domestic animal species the generation of such contigs is typically initiated with the screening of libraries with probes derived from human genes that are expected to be located in the region of interest by comparative mapping. However, in many instances the available gene-derived probes are too far apart to allow the cloning of BAC/PAC contigs larger than a few hundred kb. High resolution physical mapping allows to estimate the sizes of gaps and to control the orientation of the individual sub-contigs, which helps to avoid errors during the assembly of smaller contigs into final Mb-sized contigs. The recently constructed porcine IMNpRH2 panel allowed us to use this approach for the construction of high-resolution physical maps of SSC 6q1.2.
Results
Two sequence-ready BAC/PAC contigs of the gene-rich region on porcine chromosome 6q1.2 (SSC 6q1.2) containing the RYRl gene were constructed. The two contigs spanned about 1.2 Mb and 2.0 Mb respectively. The construction of these contigs was monitored by the results provided by the mapping of 15 markers on the IMpRH7000rad and 35 markers on the IMNpRH212000rad radiation hybrid panels. Analyses on the IMpRH panel allowed us to globally link and orientate preliminary smaller contigs, whereas analyses on the high resolution IMNpRH2 panel allowed us to finally identify the order of genes and markers.
Conclusions
A framework map of 523 cR12000 was established covering the whole studied region. The order of markers on the framework 1000:1 RH map was found totally consistent with the data deduced from the contig map. The kb/cR ratio was very constant in the whole region, with an average value of 6.6 kb/cR. We estimate that the size of the remaining gap between the two contigs is of about 300 kb. The integrated physical and RH map of the investigated region on SSC 6q1.2 was used for a comparative analysis with respect to the syntenic regions on HSA 19q13.1 and MMU 7 and revealed a perfectly conserved gene order across the entire studied interval.
Background
Comparative genome analysis increases the knowledge of genome evolution and is especially important in livestock species where the currently available sequence information is very limited as compared to the vast amount of information available from the human and mouse genomes. Radiation hybrid mapping is seen as an efficient technique for the generation of high-resolution gene maps in different species and RH maps can be integrated in comparative mapping approaches to reveal the degree of synteny conservation between species [1].
Two RH panels have been reported for the pig: the 7 000 rad IMpRH panel [2] that provides medium-resolution global mapping information, and the 12 000 rad IMNpRH2 [3], that can be used to construct high-resolution local RH maps. Panels developed after a high level of cell irradiation (10 000 to 50 000 rads) are very useful for high resolution regional mapping studies but they require a characterization with a very large number of markers to be useful for genome-wide mapping studies [4].
The porcine RYRl gene region on SSC 6q1.2 is of special interest due to its economical importance. The porcine stress syndrome (PSS), which in pigs is caused by a single RYRl point mutation, is known to be associated with positive characteristics like increased muscling and increased lean meat content. Until now, it is not clear whether the RYRl mutation is also responsible for the positive carcass traits in stress susceptible pigs or whether these complex growth traits are influenced by other closely linked genes on SSC 6q1.2 [5-7]. Furthermore, this genomic region is also of special interest as it represents a GC-rich genomic region with a very high gene content. To investigate this genomic region we have previously reported the construction and analysis of a 1.2 Mb BAC/PAC contig [8].
In the present study, we report the construction of high-resolution framework and comprehensive RH maps of the RYRl gene region on the porcine chromosome 6q1.2 using the porcine IMpRH and IMNpRH2 panels as well as the comparison of the RH maps to an extended clone-based physical map of this region.
Results and Discussion
Construction of the BAC and PAC contig and analysis of end sequences
We previously reported the construction of a 1.2 Mb BAC/PAC contig on SSC 6q1.2 [8]. To extend the existing contig the porcine TAIGP714 PAC and RPCI-44 BAC libraries were screened with new probes either derived from end fragments of previously isolated porcine genomic clones or from human HSA 19q13.1 genes. Assembly of all 171 isolated BAC and PAC clones according to STS content, insert sizes and fingerprinting data resulted in the expansion of the existing 1.2 Mb contig [8] to 2.0 Mb and the generation of a new 1.2 Mb contig (Fig. 1). End sequences from all clones of the contig were generated and submitted to the EMBL database under accessions AJ514457-AJ514832. In total 292 end sequences from SSC 6q1.2 with an average read length of 708 bp totaling 207 kb of genomic survey sequences were generated. Thus, the BAC/PAC end sequences cover approximately 6 % of the studied genomic region. The end sequences contain an average GC content of 47 % exceeding the value of 41 % that is generally accepted as the average GC content in mammalian genomes [9]. The GC content analysis further confirms that SSC 6q1.2 is indeed closely related to HSA 19q13.1, which has a GC content of 46 % in the corresponding 4 Mb region. An analysis of repetitive elements revealed that 39.8 % of the end sequences consisted of repetitive DNA. Of the 39.8 % repetitive DNA, 20.5 % were SINE, 13.3 % were LINE, 2.4 % were of retroviral origin (LTRs), and 2.0 % represented DNA transposons. The predominance of SINEs is another typical hallmark of GC-rich and gene-rich genome segments [10]. The analysis of the end sequences also revealed three dinucleotide and one tetranucleotide microsatellite (AJ514594, AJ514613, AJ514706, AJ514795).
Figure 1.
Physical map of the isolated BAC/PAC contigs. STS markers are represented by vertical dotted lines, cDNA hybridization probes are represented as horizontal lines at the top, markers that are associated with genes are denoted in bold. The physical sizes covered by the different hybridization probes depend on the intron sizes of their respective genomic targets. BACs and PACs are indicated below the markers with their corresponding clone names, positions, and insert sizes.
The availability of the end sequences allowed the continuous verification of the contig assembly by comparative mapping. In BLAST searches against the human draft genome sequence, approximately 15 % of the BAC/PAC end sequences showed significant (E < 10-5) matches to HSA 19q13.1, which allowed the precise comparative mapping of 27 % of the tested BAC/PAC clones. Of the investigated clones, 73 % had no match in the human genome sequence, 23 % had matches with one end sequence, and 4 % had matches with both end sequences.
Physical mapping and comparative analysis
During the contig construction many gene-specific STSs were used, which allowed the unequivocal assignment of genes to individual clones. Further genes were localized by hybridization of heterologous cDNA probes to the individual BAC/PAC clones and BLAST analysis of the clone end sequences. Using these approaches, 33 genes in total were localized. Furthermore, the microsatellite SW193 was also localized by STS content analysis thus anchor in the physical clone-based map to the linkage map of this region [11].
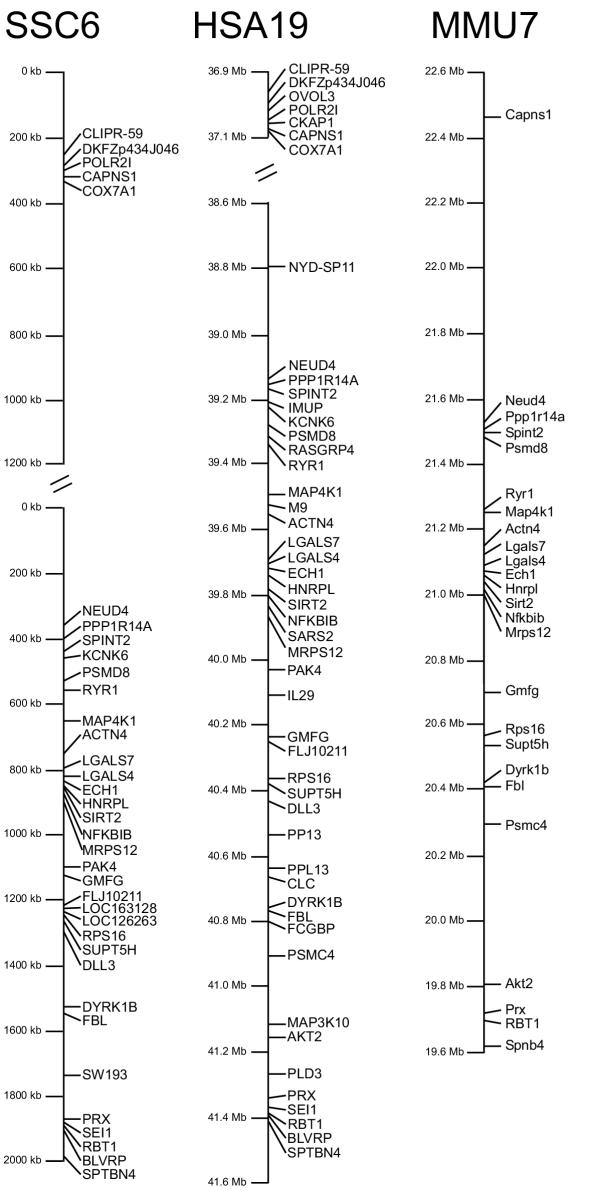
The gene assignments were compared with human and mouse maps and a comparative map for SSC 6q1.2, HSA 19q13.1 and MMU 7 was developed (Fig. 2). The gene order in this region of the pig genome corresponds exactly to the gene order of the NCBI HSA 19 map (http://www.ncbi.nlm.nih.gov build 31). The gene order of MMU 7 (http://www.ncbi.nlm.nih.gov MGSCv3) also corresponds exactly to the gene order of SSC 6 and HSA 19 but the orientation is inverted. The perfect synteny conservation between mouse and the two other species can only be observed since the latest update of the mouse maps as in the previous mouse genome assembly a major rearrangement of the gene order in this genome region was observed [8].
Figure 2.
Comparative maps of Sus scrofa chromosome 6q1.2, Homo sapiens chromosome 19q13 and Mus musculus chromosome 7. Comparative maps of Sus scrofa chromosome 6q1.2 (results from this study), Homo sapiens chromosome 19q13 (http://www.ncbi.nlm.nih.gov build 31), and Mus musculus chromosome 7 (http://www.ncbi.nlm.nih.gov MGSCv3). Gene order is perfectly conserved between the three species, however the gene order is inverted in the mouse with respect to the other two species. In the human map all known genes without hypothetical gene predictions are listed, while in the murine map only those genes are listed that have also been mapped in the pig. In the porcine map the position of the microsatellite SW193 is also indicated.
Whereas the gene order is perfectly conserved between human, mouse and pig, the physical distances between genes vary somewhat between the three species. Within the investigated region the gene-poor stretch between COX7A1 and NEUD4 accounts for the biggest part of these size deviations. The cloned region has a very uneven gene density. At the top and at the bottom of the map (Fig. 2) genes are clustered extremely dense with very short intergenic regions, while in the middle of the map, between the COX7A1 and the NEUD4 gene the gene content is actually very low.
RH mapping
In this study, we were able to build two comprehensive RH maps for SSC 6q1.2. On the 7000 rad IMpRH panel 15 STS markers were genotyped, while on the 12 000 rad IMNpRH2 35 STS markers were analyzed. Retention frequencies of markers ranged from 18.1 % to 32.8 % (average 22.9 %) on the IMpRH panel and from 27.8 % to 44.3 % with an average retention frequency of 37.5 % on the porcine IMNpRH2 panel.
During the building of these two contigs, we simultaneously analyzed data obtained on both IMpRH and IMNpRH2 panels using the Carthagene program. Intermediate rough analyses of RH data allowed us to monitor the construction of the contig. In particular it allowed us to orient a subcontig in the gene poor region from ITZ002 to ITZ004 as well as to estimate the size of remainmg gaps.
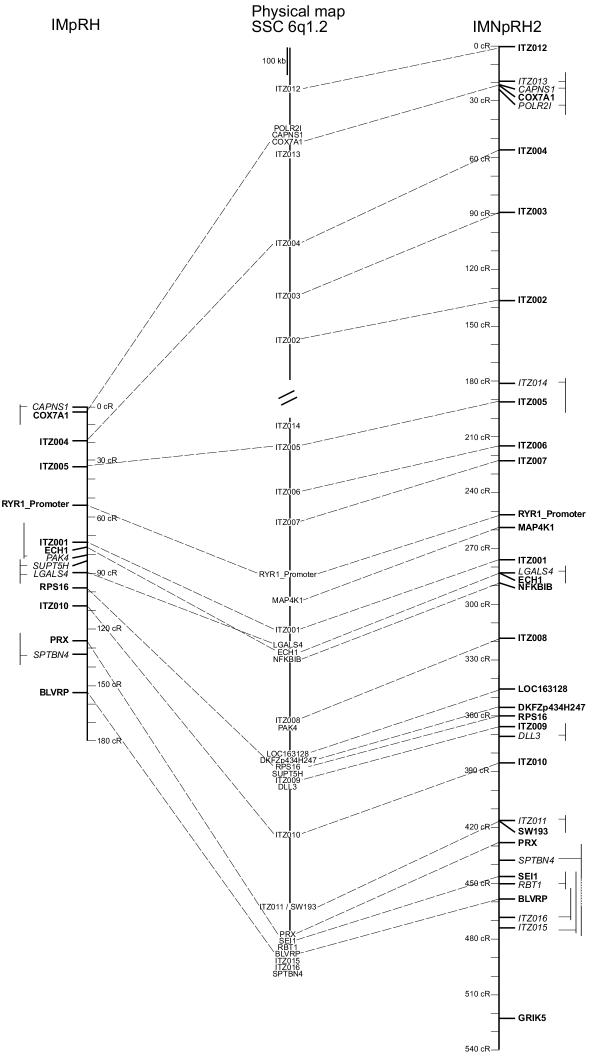
When the full RH data set was available for both panels, it appeared that at the scale of 10–100 kb, the degree of resolution of the IMpRH panel is not high enough, and furthermore the order of genes that could be determined on this panel is very sensitive to some small genotyping errors. To produce a final reference map we thus computed a 1000:1 framework map using only the 35 vectors produced on IMNpRH2 panel. The framework status of the map was tested by calculation of likelihood of maps produced after all local permutations in a slipping window of 6 markers, and by global local inversions. We confirmed that no altemate order could be identified with a difference of log likelihood of less than 3 compared to the proposed order. The framework map contained 24 of the 35 IMNpRH2 markers. Using this framework map comprehensive maps were produced on each panel. In order to avoid inflation of the map size, we chose to project additional markers at their most likely location, without altering the multipoint distance between framework markers (Fig. 3).
Figure 3.
Comparison of the clone-based physical map, RH7000 map and RH12000 mapofSSC 6q1.2. The RH maps represent comprehensive maps, where the markers of the 1000:1 framework map are highlighted in bold. Note that there is perfect agreement in the order of framework markers between the IMNpRH2 RH12000 map and the clone-based physical map. For the non-framework markers, altemate positions are indicated by thin vertical lines.
As shown in figure 3, the gene orders on the RH and physical maps are generally in good agreement. This agreement is perfect between the physical map and the 1000:1 framework RH map produced on IMNpRH2 panel. It demonstrates that at the 50–100 kb scale, fully accurate maps can be produced on this panel provided that 1000:1 framework maps are drawn.
Some minor discrepancies can be found when comprehensive maps are drawn. For instance, the location of SPTBN4 on the IMNpRH2 map seems incorrect. However a difference of log likelihood of only 1.57 is found between the maps constructed under the most likely order and the expected order. We thus think that our RH data do not sufficiently support the hypothesis of a very small rearrangement of this region. It should be pointed out, that even if additional markers are added at their most likely location on this kind of comprehensive map, their mapping does not affect the distance calculated between framework markers.
We also compared the resolution of both panels on the framework map established between COX7A1 and BLVRP. On IMpRH the distance is 146 cR7000, whereas the same fragment is 438 cR12000 long on IMNpRH2. In this region the ratio between the resolutions is thus 3.01, which is slightly higher than the value of 2.77 observed in the PRKAG3-RN region [3] and of 2.43 observed in a QTL region close to the centromere of SSC 7 [12]. In the gene rich region between RYRl and BLVRP, which is precisely mapped on the reported clone contig, a ratio of 6.6 kb / cR12000 (1370 kb / 207 cR12000) is observed on the IMNpRH2 panel.
The RH map allowed us to confirm the close link between the two contigs we produced. The distance between the extremity markers of the contigs (ITZ002 and ITZ014) was estimated at 43.2 cR12000. Considering a ratio of 6.6 kb/cR12000 in this region, we can estimate that the physical distance between both contigs could be around 285 kb, which is roughly similar to the 360 kb distance that would be estimated from the human-pig comparative map.
Conclusion
The IMNpRH2 panel allowed a highly accurate resolution of closely spaced markers and was very useful in evaluating the assembly of a clone contig. In most instances not only the order of markers but also the physical distances between markers could be very accurately estimated from the RH12000 map. During the contig building it helped us to orientate small sub-contigs, which were originally unlinked, and to estimate the size of remaining gaps. Combining analyses on both IMpRH and IMNpRH2 panels provides both the possibility to detect significant linkage between relatively distant markers on the IMpRH panel as well as to determine the accurate gene order on the higher resolving IMNpRH2 panel.
Methods
DNA library screening and chromosome walking
Library screenings were done as described [8]. Briefly, the TAIGP714 PAC library [13], http://www.rzpd.de was screened by PCR of hierarchical DNA pools. The porcine genomic BAC library RPCI-44 was screened by radioactive hybridization according to the RPCI protocols http://www.bacpac.chori.org.
DNA sequence analysis
End sequences of isolated BAC and PAC DNA were generated with a LICOR 4200L automated sequencer system. Further analyses were performed with the online tools of the European Bioinformatics Institute http://www.ebi.ac.uk/, BLAST database searches in the GenBank database of the National Center for Biotechnology Information NCBI and the RepeatMasker searching tool for repetitive elements (Smit, A.F.A. and Green, P. http://repeatmasker.genome.washington.edu/). Single copy sequences were used to design primer pairs for the chromosome walking using the programs GeneFisher and Primer3 http://bibiserv.techfak.uni-bielefeld.de/cgi-bin/gf_submit?mode=START, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi.
RH mapping
Prior to RH mapping of swine genomic inserts, each designed primer pair was tested for correct localization on SSC 6 on a somatic cell hybrid panel [14]. PCR was performed in a 22.5 μl reaction volume containing 25 ng of template DNA, 100 μM of each dNTP, 1.5 mM MgCl2, 10 pmol of each primer and 1 U Taq polymerase in the reaction buffer supplied by the manufacturer (QBiogene, Heidelberg, Germany). After a 5 min initial denaturation at 94°C, 38 cycles of 45 s at 94°C, 45 s at the annealing temperature of the specific primer pair, and 45 s at 72°C were performed in a MJ Research thermocycler (Biozym, Hess. Oldendorf, Germany). Finally an extension step at 72°C for 5 min was performed. Primer sequences and annealing temperatures are given in table 1. PCR products were separated on 1–2 % agarose gels. Gels were scored twice by independent investigators.
Table 1.
Primer sequences for RH mapping
| Primer | Forward sequence (5 '-3') | Reverse sequence (5 '-3') | Tm (°C) | PCR-Product (bp) | Retention IMpRH | Retention IMNpRH2 |
| BLVRB | GGT TAT GAG GTG ACA GTG | CCA AGA GCA CGA TGA TGG | 55 | 148 | 0.14 | 0.38 |
| CAPNSl | TGT ATT CCT GAA CGG GAG | TGC AAG AGA GGG CTA ATG | 54 | 272 | 0.31 | 0.38 |
| COX7A1 | CAC CTA CTG GAC GAA TCC | AGG TCC CGA GGT ATT ACA G | 59 | 388 | 0.29 | 0.38 |
| DLL3 | ACC CCA GAA TCC TCG TAC | AGA GCA GGA CAT AGC ATC | 55 | 395 | 0.39 | |
| ECHl | ACC AGG AGG TCC TGC TTG | ATG TAG TTG AGG CCC TCT G | 58 | 136 | 0.18 | 0.34 |
| GRIK5 | CAA CTT CCA GGC CCT GTC | TTG ATG AGC TCG CCA ACC | 60 | 174 | 0.31 | |
| ITZ001 | TAG CCT TTC CTG TGG AGG | ACA CAA AAG CAC ACA CCG | 56 | 423 | 0.19 | 0.32 |
| ITZ002 | AGC CTT CAC CCA GAA GTC | GAT GCA CAA GGA GCT GAG | 59 | 287 | 0.36 | |
| ITZ003 | CAT AGG GCT TTT CAC CAG | TTC TTG CCG AAT ATC AGA G | 54 | 297 | 0.28 | |
| ITZ004 | TTC ACT TTG GGT CTG CTG | ACC TAC CCA CTG CTA TGC | 55 | 311 | 0.24 | 0.26 |
| ITZ005 | TCC AGA CAA GTG AGA AAC AA | TTA ATT TAT AAT GCC TGG TCA | 55 | 320 | 0.24 | 0.37 |
| ITZ006 | ATC TGC GGA GAG GAA AAG | TCG TGT TTG TTG GAA TGT C | 54 | 201 | 0.35 | |
| ITZ007 | GTG ACT TGT AGA CCA CAG | CCT ACA GAG GGA GAA TCC | 55 | 346 | 0.35 | |
| ITZ008 | AGC TGA GAC CAA TGC CAT | ATA ATT GGG AGT TCT CGC | 53 | 122 | 0.36 | |
| ITZ009 | CTG CTC CTC ATT CCC ATG | CCG TCT TAT GCT TGA GTC | 55 | 187 | 0.40 | |
| ITZ010 | CAG ATT GGG ATG AAG CTC | TTA TCA ATC CCA ACA CAC C | 54 | 114 | 0.22 | 0.37 |
| ITZ011 | AGA CGG GAA ATT GAG ACC | TCC CTG TGG CAG TAA ATG | 58 | 345 | 0.37 | |
| ITZ012 | GTG GGG CCC TAT AAA GAC | CCG AGG GTC AAA TGT CTG | 56 | 223 | 0.38 | |
| ITZ013 | TTG GAG GGT TCA ACT ACG | GTA AAC CCG TTC ACG TTG | 57 | 197 | 0.38 | |
| ITZ014 | TGC CTG TTC ACG AAC CAC | TCC TTG TGT GGG CTA CAG | 59 | 100 | 0.32 | |
| ITZ015 | AAG AAA AGG AAA AGG TTT GG | TGT TCT CGC AAA CAG TGA G | 58 | 304 | 0.33 | |
| ITZ016 | AGA CAG GCT CCG ATG AAG | AGA AAG GCT TCC CTG CTG | 59 | 209 | 0.35 | |
| LGALS4 | GAT GTC GCC TTC CAC TTC | TGA TGA CCA GCT CGA AGG | 56 | 142 | 0.18 | 0.35 |
| LOC 163128 | TCC AAA GAG AAG GTG GTG | ATG AGG GCA TAG GAG AGC | 55 | 170 | 0.36 | |
| DKFZp434H247 | TTC TGC AGC TTC TTC TGC | ATC GAA GTC CTG TTG CTG | 54 | 145 | 0.41 | |
| MAP4K1 | CCT ACC CAC GCC TAT GC | CCA GCC AGC AGG AAA GC | 56 | 136 | 0.33 | |
| NFKBIB | CTG CAC CTG GCA GCC ATC | GCT GGA GCA GCA CGC AAG | 60 | 154 | 0.32 | |
| PAK4 | CAG CGA GTG TCC CAT GAG | CAT GGG TCA GCA GGA TGG | 58 | ~1600 | 0.19 | |
| POLR21 | ACC ACC CGT GCC AAA AG | CCG CGC ACT GTG TGA CT | 55 | 172 | 0.37 | |
| PPP1R14 | GCT GAG CAA GCT GCA GTC | GGT ACA GCT CCT CCA AGC | 58 | 164 | 0.32 | |
| PRX | GCC TCA GGT GAC CTT GTC | CCC ACA TCC AGC TCA AGC | 58 | 117 | 0.21 | 0.38 |
| RBTl | CCA TGG ATG AGA CTG AGC | GGC ACA GAA GAG GTT GTG | 56 | 218 | 0.33 | |
| RPS16 | CGC TGA TCA TCA CGA TGG | GCT TTT GGG CAA GGA ACG | 56 | 294 | 0.24 | 0.41 |
| RYR103/104 (RYR_Promoter) | TTC GTT TCT GCT TCG CC | CTC TCT CCT CCC ATT TC | 48 | 162 | 0.20 | 0.34 |
| SEIl | GAG CTG GAT GAT GCT GAG | GCT GTG ATG GAG CTT GAG | 56 | 184 | 0.32 | |
| SPTBN4 | ATA TCC TGC CCC AAG AAG | GAG GAG GTC GAC GTT TTG | 58 | 282 | 0.20 | 0.38 |
| SUPT5H | AGG AGC TTC CCC AGG AAG | TGG GTG AGG ATC GGG AAG | 58 | 121 | 0.18 | |
| SW193 | TGC CAT CCT TTC TTT CAT TAC G | TCA CTC TGA GGG GTC CTG AC | 62 | 101 | 0.37 |
To construct the RH map, 15 markers were mapped on the IMpRH panel [2] and 35 on the IMNpRH2 panel [3] according to the INRA protocols http://www.toulouse.inra.fr/lgc/pig/RH/IMpRH.htm. After genotyping the IMpRH panel the chromosomal assignments were performed with software available on the INRA WWW server and submitted to the IMpRH database http://imprh.toulouse.inra.fr/.
Statistical analysis of RH results
Vectors obtained on IMpRH and IMNpRH2 panels were analyzed with Carthagene software [15]. A framework map was built using buildfw option, which constructs a 1000:1 framework map by a stepwise locus adding strategy under the haploid model of fragment retention. The framework map was tested using a flips algorithm, which checks all local permutations in a window of 6 markers, and a greedy algorithm, which tries to improve the map by inversion of parts of the reference map. When the most likely order did not fit the expected order based on the human-pig comparative map, the likelihood of the two possible orders were calculated to determine the strength of the indication of a possible modification of gene order between both species. The final framework map was recomputed under a diploid model. Additional markers were mapped relatively to the framework map at their most likely location, projecting the markers on the map using the following formula (using the diploid model).
![]()
where Loc (M) is the location on the framework map of marker M mapped between the nth and n + 1th markers of the framework (respectively named Fwkn and Fwkn+1), Dmltpt(X,Y) and D2pt(X,Y) are the multipoint and two point distances between markers X and Y.
List of abbreviations
BAC bacterial artificial chromosome; cR centi Ray; HSA human chromosome; PAC Pl derived artificial chromosome; RH radiation hybrid; SSC porcine chromosome; STS sequence tagged site; MMU murine chromosome; IMpRH INRA Minnesota porcine Radiation Hybrid panel, IMNpRH2 INRA Minnesota Nevada porcine Radiation Hybrid panel 2
Authors' contributions
FM constructed most of the BAC/PAC contig, did the genotypings of the RH panels and drafted parts of the manuscript. DM analyzed the RH data and wrote the radiation hybrid mapping related parts of the manuscript. CD provided some initial help with the RH analysis. RV isolated several BACs and PACs of the final contigs. BB provided the TAIGP714 PAC library. AR and MY provided the IMpRH and IMNpRH2 panels and helped with analysis of RH data. TL conceptualized the investigation, coordinated the planning of the experiments and finalized the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
Acknowledgments
We thank H. Klippert, S. Neander, and D. Seinige for expert technical assistance. We would also like to thank P. de Jong and his lab for providing the RPCI-44 library and excellent supplementary information. This study was supported by a grant of the German Research Council DFG (Le 1032/7-1+2) to T.L.
Contributor Information
Flávia Martins-Wess, Email: flavia.martins@tiho-hannover.de.
Denis Milan, Email: milan@toulouse.inra.fr.
Cord Drögemüller, Email: Cord.Droegemueller@tiho-hannover.de.
Rodja Voβ-Nemitz, Email: Rodja.Christoph.Voss-Nemitz@tiho-hannover.de.
Bertram Brenig, Email: bbrenig@gwdg.de.
Annie Robic, Email: arobic@toulouse.inra.fr.
Martine Yerle, Email: yerle@toulouse.inra.fr.
Tosso Leeb, Email: Tosso.Leeb@tiho-hannover.de.
References
- Goldammer T, Kata SR, Brunner RM, Dorroch U, Sanftleben H, Schwerin M, Womack JE. A comparative radiation hybrid map of bovine chromosome 18 and homologous chromosomes in human and mice. Proc Natl Acad Sci USA. 2002;99:2106–2111. doi: 10.1073/pnas.042688699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerle M, Pinton P, Robic A, Alfonso A, Palvadeau Y, Delcros C, Hawken R, Alexander L, Beattie LB, Milan D, Gellin J. Construction of a whole genome radiation hybrid panel for high-resolution gene mapping in pigs. Cytogenet Cell Genet. 1998;82:182–188. doi: 10.1159/000015095. [DOI] [PubMed] [Google Scholar]
- Yerle M, Pinton P, Delcros C, Amal N, Milan D, Robic A. Generation and characterization of a 12,000 rads radiation hybrid panel for fine mapping in pig. Cytogenet Genome Res. 2002;97:219–228. doi: 10.1159/000066616. [DOI] [PubMed] [Google Scholar]
- Flaherty L, Herron B. The new kid on the block – a whole genome mouse radiation hybrid panel. Mamm Genome. 1998;9:417–418. doi: 10.1007/s003359900788. [DOI] [PubMed] [Google Scholar]
- Fujii J, Otsu K, Zorzato F, de Leon S, Khanna VK, Weiler JE, O'Brien PJ, MacLennan DH. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 1991;253:448–451. doi: 10.1126/science.1862346. [DOI] [PubMed] [Google Scholar]
- Zhao F, Ambady S, Ponce de Leon FA, Miller LM, Lunney JK, Grimm DR, Schook LB, Louis CF. Microsatellite markers from a microdissected swine chromosome 6 genomic library. Anim Genet. 1999;30:251–255. doi: 10.1046/j.1365-2052.1999.00502.x. [DOI] [PubMed] [Google Scholar]
- Bidanel JP, Milan D, lannuccelli N, Amigues Y, Boscher MY, Bourgeois F, Caritez JC, Gmand J, Le Roy P, Lagant H, Quintanilla R, Renard C, Gellin J, Ollivier L, Chevalet C. Detection of quantitative trait loci for growth and fatness in pigs. Genet Sel Evol. 2001;33:289–309. doi: 10.1051/gse:2001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Wess F, Voβ-Nemitz R, Drögemüller C, Brenig B, Leeb T. Construction of a 1.2-Mb BAC/PAC Contig of the Porcine GeneRYR1 Region on SSC 6q1.2 and Comparative Analysis with HSA 19q13.13. Genomics. 2002;80:416–422. doi: 10.1006/geno.2002.6846. [DOI] [PubMed] [Google Scholar]
- The intemational human genome sequencing consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Smit AFA. Interspersed repeats and other mementos of transposable elements in mammalian gcnomcs. Curr Opin GenetDev. 1999;9:657–663. doi: 10.1016/S0959-437X(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Keele JW, Smith TP, Beattie CW. A microsatellite linkage map of the porcine genome. Genetics. 1994;136:231–245. doi: 10.1093/genetics/136.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeure 0, Renard C, Yerle M, Faraut T, Riquet J, Robic A, Schiex T, Rink A, Milan D. Rearranged gene order between pig and human in a QTL region on SSC. Mamm Genome. 2003;14:71–80. doi: 10.1007/s00335-002-3034-1. [DOI] [PubMed] [Google Scholar]
- Al-Bayati H, Duscher S, Kollers S, Rettenberger G, Fries R, Brenig B. Construction and characterization of a porcine Pl-derived artificial chromosome (PAC) library covering 3.2 genome equivalents and cytogenetical assignment of six type 1 and type 11 loci. Mamm Genome. 1999;10:569–572. doi: 10.1007/s003359901046. [DOI] [PubMed] [Google Scholar]
- Yerle M, Echard G, Robic A, Mairal A, Dubut-Fontana C, Riquet J, Pinton P, Milan D, Lahbib-Mansais Y, Gellin J. somatic cell hybrid panel for pig regional gene mapping characterized by molecular cytogenetics. Cytogenet Cell Genet. 1996;73:194–202. doi: 10.1159/000134338. [DOI] [PubMed] [Google Scholar]
- Schiex T, Chabrier P, Bouchez M, Milan D. Boosting EM for Radiation Hybrid and Genetic Mapping. WABI'2001 (Workshop on Algorithms in Bioinformatics) 2001. p. LNCS 2149.