Abstract
Background
The storage and distribution of electrocardiogram data is based on different formats. There is a need to promote the development of standards for their exchange and analysis. Such models should be platform-/ system- and application-independent, flexible and open to every member of the scientific community.
Methods
A minimum set of information for the representation and storage of electrocardiogram signals has been synthesised from existing recommendations. This specification is encoded into an XML-vocabulary. The model may aid in a flexible exchange and analysis of electrocardiogram information.
Results
Based on advantages of XML technologies, ecgML has the ability to present a system-, application- and format-independent solution for representation and exchange of electrocardiogram data. The distinction between the proposal developed by the U.S Food and Drug Administration and ecgML model is given. A series of tools, which aim to facilitate ecgML-based applications, are presented.
Conclusions
The models proposed here can facilitate the generation of a data format, which opens ways for better and clearer interpretation by both humans and machines. Its structured and transparent organisation will allow researchers to expand and test its capabilities in different application domains. The specification and programs for this protocol are publicly available.
Background
Electrocardiogram (ECG) data are acquired, stored and analysed using different formats and software platforms. Medical informatics will fully exploit the benefits from its research only when data can be openly shared and interpreted. Therefore, there is a need to develop cross-platform solutions to support biomedical training, decision-making and telemedicine applications [1].
An important goal is to describe these data independently on the number of channels, instrumentation platform or type of experiments. Moreover, an ECG record should also include annotations relating to the acquisition protocols, patient information and analysis results. These data modelling tasks should consist of flexible and inexpensive tools to enhance pattern recognition capabilities.
The development of these systems will depend on the existence of information that clearly specifies domain terminologies, functional hierarchies and decision rules. The availability of such ontological representations [2] will allow the emergence of standards, which will facilitate the integration of information on a global communication infrastructure.
ECG data have been traditionally recorded using flat file formats, such as the MIT-BIH file library [3]. This type of data format lacks the information necessary to support a meaningful analysis, interoperability and integration of multiple resources. Different governmental, academic and private organisations have proposed minimum requirements for the representation and storage of biomedical information, including signals and images [4]. These efforts aimed to promote the application of standards for message exchange and data integration. In 1993, for example, the CEN/TC251 WG3 (Comité Européen de Normalisation European, Committee for Standardisation, Technical Committee 251) reviewed several data exchange formats for healthcare applications. It includes Abstract Syntax Notation (ASN.1) and Health Level Seven (HL7) [5]. The former defines norms to describe an electronic message based on different data types. One of the disadvantages of ASN.1 is that it does not fully support scalable solutions and query processing. HL7 has been a Standards Development Organisation affiliated to the ANSI (American National Standards Organisation) since 1997 and has become the standard for electronic exchange of historical and administrative data in health services worldwide. The next generation of the messaging standard (V3) has been under development since.
CORBAmed, the Healthcare Domain Task Force of the Object Management Group (OMG) [6], deals with interoperability problems between heterogeneous information systems. To facilitate the seamless and automated data exchange between numerous applications, a common interface architecture was developed that serves in a number of today's information systems. Liaisons have been established with other organisations such as HL7.
The Digital Imaging and Communications in Medicine (DICOM) standards committee supports the achievement of data compatibility between imaging systems and other healthcare information at different levels. This standard has been applied by many private organisations, which need to incorporate diverse bio-signals associated to medical imaging. The DICOM standard is a useful resource that also provides guidelines on how to represent ECG features [4].
More recently, the eXtensible Markup Language (XML) [7] has been suggested as a promising approach to representing biomedical data. Developed as a subset of SGML in 1996 to "be straightforwardly usable over the Internet" and published as a first recommendation by the W3C (World Wide Web Consortium) in 1998, XML soon became a ubiquitous syntax for data and data-exchange over the Internet. Since then, XML-based Markup Languages, specified as e.g. Document Type Definitions (DTD) or XML Schemas (XSD) have been emerging in unlimited numbers and in nearly every imaginable domain [8]. Advantages of XML syntax include platform-, vendor- and application independence as well as an easy-to-follow hierarchical data structure and wide support. "XML's greatest advantage is that it is a user-driven, open standard for exchanging data both over corporate networks and between different enterprises, notably over the Internet. XML's biggest potential lies undoubtedly in its ability to mark up mission-critical document elements self-descriptively" [9]. By following a strict separation of content and presentation information, XML technologies increase the re-usability of information in its purest way as access to the original (raw) data is always given. The use of XML syntax for the exchange of electronic patient records was shown in all aspects in Synapses [10] and SynEx [11] project implementations [12-14].
The U.S Food and Drug Administration (FDA) Centre for Drug Evaluation and Research has proposed recommendations for the exchange of time-series data. It includes a hierarchical structure for the representation of signals, including ECG data, which may be encoded as an XML file. This protocol focuses on the acquisition of multiple records from different subjects within a single file [15,16]. The HL7 committee has been actively cooperating with the World Wide Web Consortium (W3C) to define XML guidelines to represent medical information [17]. HL7 has endorsed the Clinical Document Architecture (CDA), which supports the generation and exchange of clinical messages [18]. Other XML-based initiatives for the representation and distribution of biomedical information are: The ASTM E31.25 subcommittee [19], the CEN/TC251 Task Force on XML Applications in Healthcare [20] and the Clinical Data Interchange Standards Consortium (CDISC) [21]. However, these efforts have not focused on ECG data. Some of them place a greater emphasis on the administrative and financial transactions associated with a clinical environment.
Recent advances include I-Med, which is an XML-based format for clinical data [22]. This project consists of a domain-independent interface for exchanging several types of medical information. Its major goal is to provide a unique platform for clinical transactions. These messages can include ECG records, which may be described by basic features, such as QRS duration and text-based interpretations. One major limitation of this solution is that it partially addresses important ECG data content-definitions.
This article introduces a markup language for supporting ECG data exchange and analysis (ecgML). It synthesises key recommendations specified by the initiatives presented above.
Methods
There is a need to harmonise the representation of digital ECG data originating from the full spectrum of devices along with annotations for events, and to include necessary associated information, such as patient identification, interpretation and other clinical data. The hierarchical data tree structures depicted in Figures 1 to 6 are proposed to address such concerns. Tables 1 to 8 describe the elements and attributes defined in this model. In this paper terms written in bold and italic prints represent either XML element or attribute names. Element names should be words concatenated with the first letter of each word capitalised (UpperCamelCase, http://searchwebservices.techtarget.com/sDefinition/0,290660, sid26_gci824363,00.html). Attribute names satisfy the same rule except for the first word (lowerCamelCase, http://searchwebservices.techtarget.com/sDefinition/0,,sid26_gci824366,00.html).
Figure 1.
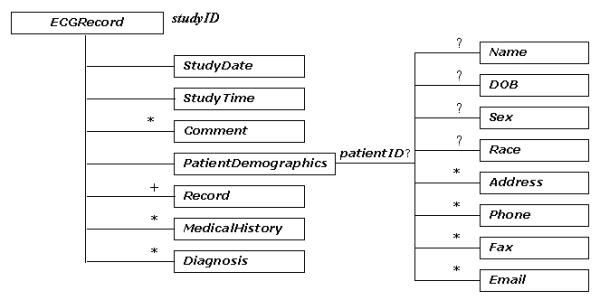
The tree diagram of ecgML: ECGRecord element
Figure 6.
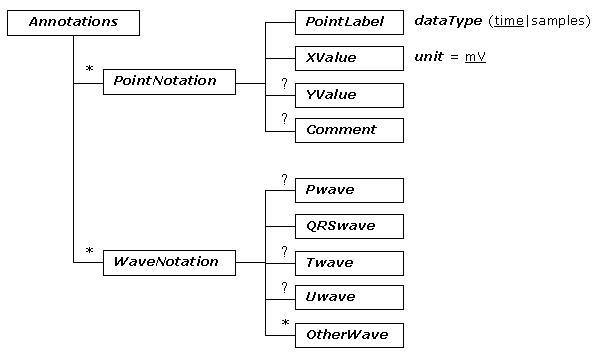
The tree diagram of ecgML: Annotations element
Table 1.
The description of ecgML: ECGRecord element
| ECGRecord | The root element for XML-based ECG record | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| studyID | Unique ID for an ECG record. | Required | string | ECG000001 |
| StudyDate | Study date. To be expressed as YYYY-MM-DD. | Required | date | 2002-10-22 |
| StudyTime | Study time. To be expressed as HH:MM:SS.SSS | Required | time | 12:01:00 |
| Comment | Comments about the ECG Record | Optional | string | Subject under stress 1 hour after dose. |
| PatientDemographics | Describes patient demographics | Required | See Table 2 | |
| Record | The details for ECG data. | Required | See Table 3 | |
| MedicalHistory | Description of the patient's clinical problems and diagnoses. | Optional | string | Ventricular ectopy |
| Diagnosis | For the latest diagnostic interpretation of the ECG. | Optional | string | Myocardial infarction |
Table 8.
The description of ecgML: Annotations element
| Annotations | Annotations for each ECG record. Based on FDA XML Data Format Specification (revision C). | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| PointNotation | A set of fiducial points with an X and Y position. | |||
| PointLabel | Name of the fiducial points. | Required | Based on DICOM waveform list | R peak |
| XValue | X position of notation. To be expressed as HH:MM:SS.SSS | Required | time or samples | 00:27:01 |
| YValue | Y position in mV of notation | Optional | float | 0.3 |
| Comment | Comment about the point notation | Optional | string | Paced beat |
| WaveNotation | Annotations for interval measurements. | |||
| Pwave | The annotations of P wave (onset, offset, peak, annotation, comment) | Optional | See Table 9 | Normal |
| QRSwave | The annotations of QRS wave (onset, offset, peak, annotation, comment) | Required | See Table 9 | PVC |
| Twave | The features of T wave (onset, offset, peak, annotation, comment) | Optional | See Table 9 | inverted |
| Uwave | The annotations of U wave (onset, offset, peak, annotation, comment) | Optional | See Table 9 | Normal |
| OtherWave | The annotations for other duration (onset, offset, peak, annotation, comment). | Optional | See Table 9 | |
Figure 2.
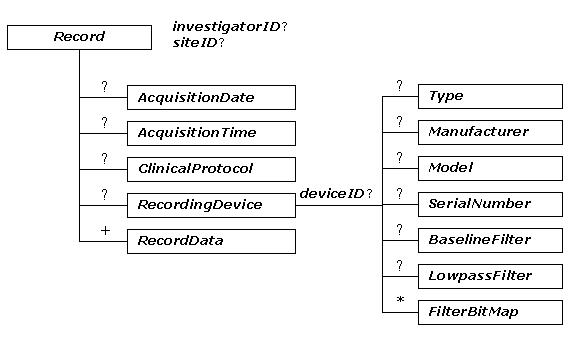
The tree diagram of ecgML: Record element
Figure 3.
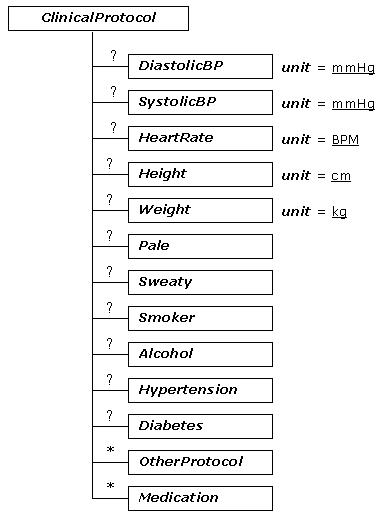
The tree diagram of ecgML: ClinicalProtocol element
Figure 4.
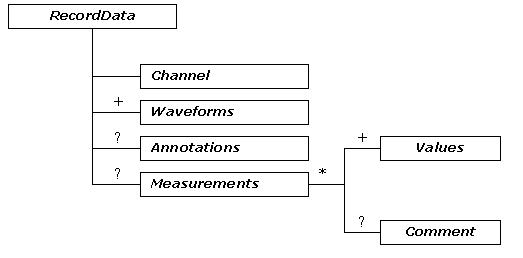
The tree diagram of ecgML: RecordData element
Figure 5.
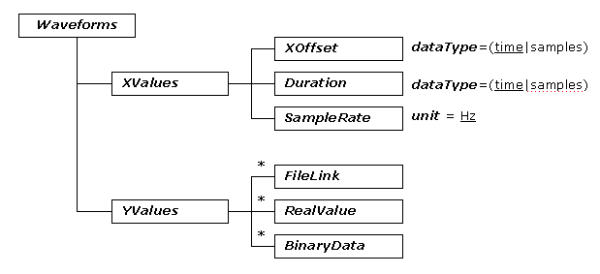
The tree diagram of ecgML: Waveforms element
Table 2.
The description of ecgML: PatientDemorgraphics element
| PatientDemographics | Describes patient demographics | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| patientID | Patient Data (Based on SCP-ECG). | Optional | string | 1532948 |
| DOB | Optional | Date | 1965-04-05 | |
| Sex | Optional | -Male -Female -Unknown -Unspecified | Male | |
| Race | Optional | string | African | |
| Phone | Phone number | Optional | string | 00442890368197 |
| Fax | Fax number | Optional | string | 00440289036 |
| Email address | Optional | string | hy.wang@ulst.ac.uk | |
| Address | Patient address. Based on HL7's PRA and I-Med recommendation | |||
| StreetAddress | Street address | Optional | string | 24 Shore Road |
| State | State or province | Optional | string | N. Ireland |
| PostalCode | Zip or postal code | Optional | string | BT37 0QB |
| City | City name | Optional | string | Belfast |
| Country | Country name | Optional | string | UK |
| Name | Patient name. Based on IMed DTD | |||
| FirstName | Patient firstname | Optional | string | Frank |
| MiddleName | Patient middle name | Optional | string | J |
| LastName | Patient lastname | Optional | string | Smith |
Table 3.
The description of ecgML: Record element
| Record | The element for the details of ECG data | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| investigatorID | A text description of the referring physician | Optional | string | Dr. John |
| siteID | A text description of the place where the ECG data was acquired. | Optional | string | Loyal Hosptial |
| AcquisitionDate | Acquisition date of the recording. To be expressed as YYYY-MM-DD | Optional | date | 2002-10-20 |
| AcquisitionTime | Acquisition time of the recording. To be expressed as HH:MM:SS.SSS | Optional | time | 11:05:32.00 |
| RecordingDevice | Description of the device that made the recording | Optional | See Table 4 | |
| ClinicalProtocol | Additional patient clinical information | Optional | See Table 5 | |
| RecordData | Ecg data for each channel. | Required | See Table 6 | |
Table 4.
The description of ecgML: RecordingDevice element
| RecordingDevice | Description of the device that made the recording. Based on FDA XML Data Format Specification (revision C) and SCP-ECG. | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| deviceID | ID number of recording device. | Optional | string | 35 |
| Type | Type of recording device. | Optional | string | 12-lead ECG |
| Manufacturer | Manufacture of recording device. | Optional | string | GE Medical Systems |
| Model | Model of recording device. | Optional | string | MAC 500 |
| SerialNumber | Serial number of recording device | Optional | string | 023001236 |
| BaselineFilter | High cut off in Hz. Based on SCP-ECG acquisition data. | Optional | float | 0.5 |
| LowpassFilter | Low cut off in Hz. Based on SCP-ECG acquisition data. | Optional | float | 120 |
| FilterBitMap | Other filters. Based on SCP-ECG acquisition data. | Optional | -60 Hz notch filter -50 Hz notch filter -Artifact filter -Baseline filter -Undefined | 60 Hz notch filter |
Table 5.
The description of ecgML: ClinicalProtocol element
| ClinicalProtocol | Additional patient clinical information | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| DiastolicBP | Diastolic blood pressure in mmHg. Based on SCP-ECG patient data. | Optional | unsignedInt | 72 |
| SystolicBP | Systolic blood pressure in mmHg. Based on SCP-ECG patient data. | Optional | unsignedInt | 120 |
| HeartRate | Heart rate in BPM. | Optional | unsignedInt | 75 |
| Height | Height in cm. Based on SCP-ECG patient data. | Optional | float | 172 |
| Weight | Weight in Kg. Based on SCP-ECG patient data. | Optional | float | 120 |
| Pale | Abnormal-looking skin on the face, or tongue | Optional | Yes/No | Yes |
| Sweaty | Abnormal reaction to heat | Optional | Yes/No | No |
| Smoker | Smoking? | Optional | string | A pack |
| Alcohol | Drinking alcohol | Optional | string | seldom |
| Hypertension | A disorder characterised by high blood pressure | Optional | Unknown/Yes/No | Yes |
| Diabetes | A disease marked by elevated levels of sugar in the blood. | Optional | Yes/No | No |
| OtherProtocol | Other clinical symptom | Optional | string | SOB(short of breath) |
| Medication | Drugs. Based on SCP-ECG drug information. | Optional | string | Digoxin |
Table 6.
The description of ecgML: RecordData element
| RecordData | Ecg data for each channel | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| Channel | Lead identifier | Required | Base on DICOM lead labelling scheme | Lead I |
| Waveforms | A description of ECG waveform | Optional | See Table 6 | |
| Annotations | A collection of annotations | Optional | See Table 7 | |
| Measurements | A collection of each channel measurements | Optional | See Table 8 | |
Table 7.
The description of ecgML: Waveforms element
| Waveforms | A 2-D plot with an X and Y axis is used to display the ECG waveform data. Refer to FDA XML Data Format Design Specification (revision C). | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| XValues | Describe the X-axis domain | |||
| XOffset | Offset from the StartTime. | Required | time | 00:23:10 |
| Duration | Duration of the each record. | Required | time | 00:00:10 |
| SampleRate | Sample rate in Hz for ECG records. | Required | unsignedInt | 500 |
| YValues | Describe the Y-axis domain. | |||
| unit | Name of base unit for Y-axis. | Required | Use UCUM when appropriate. | mV |
| FileLink | A reference to an external file storing ECG data. | Optional | string | |
| RealValue | A list of actual value of ECG data | |||
| From | Offset from the origin of the x axis to the beginning of waveform | Required | time or samples | 20:00:00 |
| To | Offset from the origin of the x axis to the ending of waveform | Required | time or samples | 21:00:04 |
| Data | The list of Y values separated by white space | Required | A list of float values | 1.25 2.23 3.2 |
| Comment | Comments on the format, e.g. separated by a specific delimiter | Optional | string | separated by white spaces |
| Binary Data | Binary representation of ECG data | |||
| encoding | Encoding scheme for embedding the binary data within XML document | Required | Base64/Hexidecimal | Base64 |
| From | Offset from the origin of the x axis to the beginning of waveform | Required | time or samples | 00:00:00 |
| To | Offset from the origin of the x axis to the ending of waveform | Required | time or samples | 12:00:04 |
| Scale | Scale factor to use to convert a binary data into a real value | Required | float | 1.0 |
| Data | Encoded binary ECG data | Required | string | A923B420 |
Table 9.
The description of ecgML: Measurements element
| Measurements | Measurements of each recorded lead | |||
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| Values | The each measurement. | Required | float | 20 |
| label | Based on SCP-ECG the lead measurements. | Required | string | P-duration |
| unit | Name of base unit for the each measurement. | Required | Use UCUM when appropriate. | ms |
| Comment | Comments on a measurement. | Optional | string | normal |
Table 10.
The description of ecgML: subelements for elements Pwave, QRSwave, Twave, Uwave and OtherWave
| Element/attribute | Description/Origin | Required | Values/Data Type | Example |
| Onset | Beginning value of the wave in time or samples | Required | Time/samples | 00:00:12.000 |
| Peak | Peak value of the wave in time or samples | Required | Time/samples | 00:00:12.600 |
| Offset | Ending value of the wave in time or samples | Required | Time/samples | 00:00:13.002 |
| Annotation | Annotation of the wave | Optional | string | Normal |
| Comment | Any comments on the annotation | Optional | string |
Each patient record starts with a root element ECGRecord, which is uniquely identified by its attribute studyID. The StudyDate and StudyTime elements represent the latest time record of the study of the ECG recording. Diagnosis contains a text version of the latest diagnostic interpretation of the ECG, while MedicalHistory is a description of medical history of patient's clinical problems and disgnoses. There are two main components for each record: one PatientDemographic and one or more Record components. It is worth noting that each record can have only one PatientDemographic element, which would be kept updated all the time; while multiple Record elements are allowed to be held in one patient record. This opens up every opportunity to keep track of the history of the patient's diagnoses.
PatientDemographic contains information of general interest concerning the person from whom the recording is obtained, such as demographic data (e.g. patientID, Name, etc.) and contact information (e.g. Address, etc.). This component is required in each record.
Record represents the physical storage for the basic content of an ECG recording. The AcquisitionDate and AcquisitionTime attributes specify the acquisition date and time for each record, which makes it possible to include multiple time-related ECG recordings within a file. investigatorID and siteID are used to identify who is responsible for the recording and where it is acquired. There are three main components: zero-or-one RecordingDevice, zero-or-one ClinicalProtocol, and one-or-more RecordDate. Such flexible structure allows each recording to have its own characteristics.
RecordingDevice is an optional element, which describes the device that generated the data. It should support the full spectrum of ECG devices, including standard 12-lead ECGs, Holter monitors, transtelephonic monitors and implanted devices. The main components in this section include deviceID, Type, Manufacturer, Model and a description of filtering technique used during the ECG acquisition (e.g. BaselineFilter and LowpassFilter).
ClinicalProtocol is an optional element, which may include information relating to a patient's clinical report. The unit attribute of each element is used to describe the measurement unit of each observation. Currently, this section only includes basic clinical dimensions, such as DiastolicBP and HeartRate. However, other variables can be easily added.
RecordData is a key ecgML element. There can be multiple RecordData elements within a file, which are identified by their Channel element names. The DICOM lead labelling format is recommended for this purpose. RecordData includes three main sub-components: Waveforms, Annotations and Measurements.
Based on the FDA-recommended PlotGroup format [16], Waveforms are represented by a series of values along two dimensions X, Y (XValues and YValues). Based on these values, a plot of voltage vs. time may be generated with a viewer program. The XValues (time) are evenly spaced. Xoffset represents the initial value. SampleRate represents the sampling frequency measured in Hz. The duration of a channel signal is represented by the element Duration. ecgML supports three formats to represent YValues: a RealValue element, a BinaryData element (associated with a specified encoding scheme, which may be base64 or hexadecimal), and a FileLink to refer to an external file.
The elements From and To, which are encoded into the elements BinaryData and RealValue, illustrate the beginning and ending values of the corresponding waveform. The Scale associated with BinaryData indicates how to convert the binary YValues into real values. The element Data in RealValue contains a list of float data separated by delimiters, representing the real value of each sample ECG data.
Annotations would typically be used to describe events specific to the corresponding channel. It defines a time point or interval, which can be used for performing the measurements. This consists of a collection of PointNotation and WaveNotation elements. Each PointNotation can be specified with a PointLabel (the name of the specific point, e.g. P wave onset), a XValue (time, expressed as HH:MM:SS.SSS format), YValue (amplitude in mV) and any relevant comment. WaveNotation includes descriptions for basic ECG waves, such as Pwave, QRSwave, Twave, Uwave, and other events that can be defined by the user (OtherWave). Wave descriptions are based on the following five elements: Onset (the beginning value), Peak (the peak value, for a T wave, it is possible to have two Peak values), Offset (the ending value), Annotation (annotation for the specified wave, such as "normal" or "abnormal"), and any comments on the annotation are given using the Comment element. The value of Onset, Peak, and Offset can be expressed as either time or sample values.
The Measurements element contains a list of Values (the measurements of each recorded channel). Each Values element may be associated with a label and a measurement unit.
There are different levels at which a record can define supplementary information. A Comment at the ECGRecord level can be used to indicate additional acquisition information, for example, place and technical conditions of the acquisition process. A Comment at the YValues level may typically be used to define the format of the representation of the YValues, e.g. which delimiter is used. A Comment at the Measurement level may be used to describe, for example, whether a measurement is a global average or an instantaneous value.
This research applies the DICOM recommendation for defining ECG channel names, fiducial point markers and waveform encoding details. Moreover, it applies the Unified Code for Units of Measure (UCUM) scheme for defining measurement units, such as cm for Height and mV for YValues [24] when appropriate.
This specification has been encoded into an XML-based data protocol. Additional files 1 and 2 are the DTD and XSD files for ecgML respectively. Additional file 3 is an ECG record, which has been generated using ecgML.
Results
Evaluation of the model
It is fundamental to demonstrate the system-, application- and format-independence of ECG data when using ecgML. Special importance should be given to illustrate the autonomy of content from its presentational scheme, e.g. printed graphs, tabular data to be imported into data mining systems for further analysis or audio files. Figure 7 illustrates the distinction separation of the five important components in XML publishing. Based on advantages of XML technologies, ecgML exhibits a remarkable advantage over existing systems where every information system has its own internal information-model and information is merged and intertwined with its representation format. Figure 8 exemplifies a scenario where the raw ECG data is kept in an ecgML data file and therefore independently from possible presentation information. Various XSLT transformations (stored as XSL files and applied on the fly, transparent to the user) convert the ecgML source into user- and/or application-specific data formats, such as MPEG (audio), MatLab (text) and SVG/PNG (graphics). The centralised storage of the ECG record and dynamic creation of data representations avoids redundancy.
Figure 7.
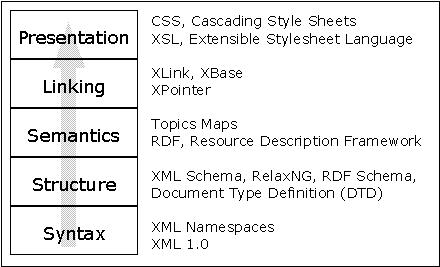
Publication Model
Figure 8.
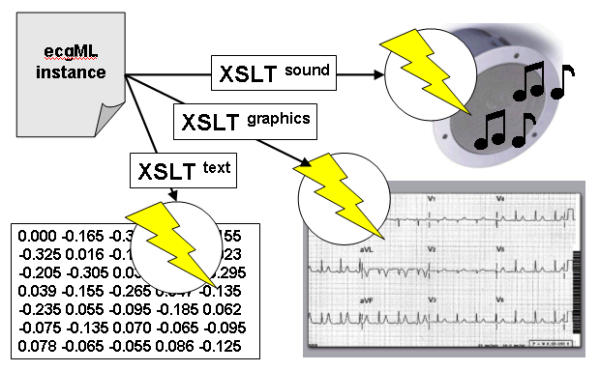
Dynamic transformation of ecgML data
The FDA, together with a number of other institutions, has developed and published an XML vocabulary [16] to represent collected time-series data. However, there are some significant differences between the FDA proposal and ecgML. The FDA proposal is intended to represent collected biological data, including ECG, electroencephalogram (EEG), or other time series data such as temperature, pressure and oxygen saturation. The main goal is to facilitate the submission of the biological data and to make sure that accuracy and consistency of the measurements made from the collected biological data is achieved. It is important for the FDA to view the biological data in an appropriate way. Thus, the data model (specified in a DTD) includes some presentation information, such as elements MinorTickInterval, MajorTickInterval and LogScale. On the other hand, the purpose of ecgMLis to develop anopen and transparent way of representing, exchanging and mining ECG data. Therefore, ecgML not only consists of basic components, which may be used to perform knowledge discovery in ECG data (e.g. ClinicalProtocol, Diagnosis and Measurements) but also follows the principle of separating content and presentation information, which will exhibit great advantages when using ecgML in combination with inter-media transformation.
Accompanying tools
A series of tools are being developed to assist users in exploiting ecgML-based applications. These include an XML-based ECG record generator, ECG parser and ECG viewer. The generator will automatically produce XML-based ECG records from existing ECG databases, e.g. the MIT-BIH database [3]. The ECG parser allows the user reading the ECG records and access their contents and structure, whereas the ECG viewer provides onscreen display of the required waveform data (Figure 9). It shows all annotation information of the individual waveform. The hierarchical structure of the XML-based ECG record is displayed. It can be expanded and shrunk at any level. This interface can also show individual episodes of the ECG waveform chosen from the ecgML structure. The viewer tool graphically locates boundaries (i.e. beginning, peak, and end) of the P, QRS and T waveforms for each selected QRS complex.
Figure 9.
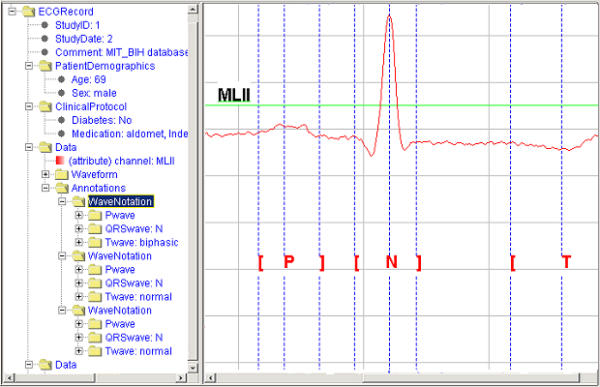
Screenshot of ECG viewer
Conclusions
ecgML will enable the seamless integration of ECG data into electronic patient records (EPRs) and medical guidelines. This protocol can support data exchange between different ECG acquisition and visualisation devices. Similarly, it may enable data mining using heterogeneous software platforms and applications. The data and metadata contained in an ecgML record may be useful to improve pattern recognition in ECG applications. It would also aid the implementation of automated decision support models such as case-based reasoning. Figure 10 illustrates the utilisation of map files to convert "raw" ecgML files into customised output formats, which will be imported into data mining systems for further analysis. ecgML may also be significant for problems such as future proof storage, context-sensitive (textual) search of patterns in ECG data, and its native inclusion into medical guidelines. Further research will address the following issues.
Figure 10.
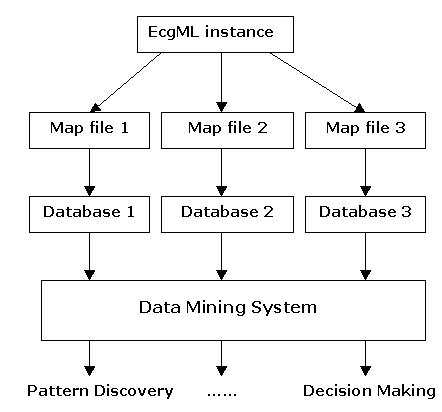
Converting XML-based ECG record into tabular data using map files. Notations for all tree diagrams are illustrated as follows. Lines of descriptive text outside an element box indicate attributes that the element should have. Default value is shown underlined.
• How does ecgML affect storage capacity?
• Does on-the-fly compression (as used by HTTP 1.1) make a difference in terms of transmission speed?
• Is it feasible to use ecgML in applications such as 24 hour monitoring?
• Does ecgML data contain all the significant information required for ECG analysis?
Competing interests
None declared.
Authors' contributions
HW co-designed and implemented ecgML (DTD and XSD files), developed support tools and drafted the manuscript. FA conceived the study, participated in the design of the model and drafted the manuscript. BJ helped to refine ecgML, brought expertise in XML and EPRs, and help to draft the paper. NB participated in the coordination of this study and contributed to the preparation of this manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
A DTD file for ecgML
An XML schema for ecgML
A sample file using ecgML
Contributor Information
Haiying Wang, Email: hy.wang@ulster.ac.uk.
Francisco Azuaje, Email: fj.azuaje@ulster.ac.uk.
Benjamin Jung, Email: benjamin.jung@cs.tcd.ie.
Norman Black, Email: nd.black@ulst.ac.uk.
References
- Värni A, Kemp B, Penzel T, Schlögl A. Standards for biomedical signal databases. IEEE Engineering in Medicine and Biology. 2001;20:33–37. doi: 10.1109/51.932722. [DOI] [PubMed] [Google Scholar]
- Ontology Homepage http://www.ontology.org/index.html
- Physionet Homepage http://www.physionet.org/
- NEMA's OFFICIAL DICOM WEB Page http://medical.nema.org/dicom.html
- Health Level Seven Homepage http://www.hl7.org
- CORBAmed Homepage http://www.acl.lanl.gov/OMG/
- Extensible Markup language (XML) http://www.w3.org/xml/
- XML Core Standards http://xml.coverpages.org/coreStandards.html
- Schroeter G. How XML is improving data exchange in healthcare http://www.softwareag.com/xml/library/schroeter_healthcare.htm
- Synapses Homepage http://www.cs.tcd.ie/synapses/public/
- SynEx Homepage http://www.gesi.it/synex/
- Jung B, Grimson J. Synapses/SynEx goes XML. In: Peter K, Blaz Z, Janez S, Marjan P, Rolf E, editor. In Proceedings of the Medical Informatics Europe '99 Conference (MIE99): August, 1999; Slovenia, Ljubljana. IOS Press; 1996. pp. 906–911. [Google Scholar]
- Jung B, Andersen EP, Grimson J. Using XML for Seamless Integration of Distributed Electronic Patient Records. In Proceedings of XML Scandinavia conference: May 2000; Gothenburg, Sweden. 2000.
- Grimson J, Stephens G, Jung B, Grimson W, Berry D, Pardon S. Sharing Health-Care Records over the Internet. IEEE Internet Computing. 2001;5:49–58. doi: 10.1109/4236.935177. [DOI] [Google Scholar]
- FDA application: Proposed Standard for Exchange ofElectrocardiographic and Other Time-Series Data http://www.fda.gov/cder/regulatory/ersr/ECGdata.htm
- FDA XML Data Format Design Specification http://www.cdisc.org/discussions/EGC/FDA_XML_Data_Format_Design_Specification_DRAFT_B.pdf
- Health Level Seven XML Patient Record Architecture http://xml.coverpages.org/hl7PRA.html
- HL7 to release first XML-based standard for healthcare http://xml.coverpages.org/hl7CDA-Ann.html
- ASTM, subcommittee E31.25 http://www.astm.org/COMMIT/COMMITTEE/E31.htm
- Dudeck J. TC 251 task force on XML application in healthcare. CEN/TC251 Task Force XML-Final Report. 1999. http://www.centc251.org/TCMeet/Doclist/TCdoc99/N99-067.doc
- Clinical Data Interchange Standards Consortium http://www.cdisc.org/
- I-Med Homepage http://www.hnbe.com/healthweb/imedpub/
- Schadow G, McDonald CJ. The Unified Code for Units of Measure http://aurora.rg.iupui.edu/~schadow/units/UCUM/ucum.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A DTD file for ecgML
An XML schema for ecgML
A sample file using ecgML


