Abstract
In (+) RNA coronaviruses, replication and transcription of the giant ∼30 kb genome to produce genome- and subgenome-size RNAs of both polarities are mediated by a cognate membrane-bound enzymatic complex. Its RNA-dependent RNA polymerase (RdRp) activity appears to be supplied by non-structural protein 12 (nsp12) that includes an RdRp domain conserved in all RNA viruses. Using SARS coronavirus, we now show that coronaviruses uniquely encode a second RdRp residing in nsp8. This protein strongly prefers the internal 5′-(G/U)CC-3′ trinucleotides on RNA templates to initiate the synthesis of complementary oligonucleotides of <6 residues in a reaction whose fidelity is relatively low. Distant structural homology between the C-terminal domain of nsp8 and the catalytic palm subdomain of RdRps of RNA viruses suggests a common origin of the two coronavirus RdRps, which however may have evolved different sets of catalytic residues. A parallel between the nsp8 RdRp and cellular DNA-dependent RNA primases is drawn to propose that the nsp8 RdRp produces primers utilized by the primer-dependent nsp12 RdRp.
Keywords: coronavirus, polymerase, replication, RNA, SARS
Introduction
Viruses of the plus-strand (+) RNA Coronaviridae family employ the largest multicistronic RNA genome composed of a single segment of 27–32 kb. This family includes the Coronavirus and Torovirus genera that together with the distantly related Arteriviridae and Roniviridae families form the Nidovirales order (Gonzalez et al, 2003; Draker et al, 2006 and reviewed in Lai and Holmes, 2001; Siddell et al, 2005). The coronavirus genome RNA is translated to express the two most 5′ open reading frames (ORFs), ORF1a and ORF1b, occupying approximately two-thirds of the genome. They encode major subunits of a poorly characterized membrane-bound complex that mediates the synthesis of the genome RNA (replication) and numerous subgenomic RNAs (transcription) (Gorbalenya, 2001; Thiel et al, 2001; Gosert et al, 2002; Prentice et al, 2004; Ziebuhr, 2005). ORF1a encodes the replicative polyprotein (pp) 1a, and ORFs 1a and 1b together encode pp1ab. Expression of the ORF1b requires a –1 ribosomal frameshift just upstream of the ORF1a stop codon. The two polyproteins pp1a and pp1ab are autocatalytically processed by at least two proteases to produce up to 16 end products called non-structural proteins (nsp) 1 to 16, as well as multiple processing intermediates (Thiel et al, 2003). The central and C-proximal regions of the polyproteins are processed at 11 conserved sites by the chymotrypsin-like proteinase known also as main proteinase (Mpro) that resides in nsp5 (reviewed in Ziebuhr et al, 2000).
The unique features of coronavirus RNA synthesis may be linked to the viral proteome bearing multiple enzymatic activities absent in other RNA viruses (Snijder et al, 2003). Replicase polyproteins of nidoviruses share a conserved array of functional domains (from the N- to C-termini): Mpro, RNA-dependent RNA polymerase (RdRp) (Cheng et al, 2005), zinc-binding domain-containing helicase (ZBD-HEL) (Ivanov et al, 2004b) and uridylate-specific endoribonuclease (NendoU) (Bhardwaj et al, 2004; Ivanov et al, 2004a). In coronaviruses, RdRp, ZBD-HEL and NendoU reside in nsp12, nsp13 and nsp15, respectively. In large nidoviruses, such as corona-, toro- and roniviruses, the conserved replicase polyprotein backbone is elaborated with additional enzymatic domains, including an exoribonuclease (ExoN) (Snijder et al, 2003; Minskaia et al, 2006) and a putative ribose-2′-O-methyltransferase (Feder et al, 2003; Snijder et al, 2003), which in coronaviruses, reside in nsp14 and nsp16, respectively. Despite recent progress in the characterization of replicase proteins, a large fraction of these proteins encoded in ORF1a (nsp1, nsp2, some nsp3 domains, nsp4, nsp6, nsp7, nsp8 and nsp10) remains functionally poorly characterized.
The crystal structure of a complex between the 197 a.a. nsp8 and the 83 a.a. nsp7 protein was recently determined for SARS coronavirus (SARS-CoV) (Zhai et al, 2005). It revealed a hexadecamer in which eight nsp7 molecules act as a mortar that holds the nsp8 octamer. RNA binding experiments with these subunits and the overall architecture of the complex suggest that the later encircles RNA. These authors have proposed that this complex provides a platform that might confer processivity to the synthesis of large RNAs in coronavirus nsp12 RdRp-mediated replication and transcription. This RdRp is phylogenetically clustered with RdRps of (+) RNA viruses whose genome RNAs have the 5′-end covalently linked to a viral protein genome-linked (VPg) (Koonin, 1991). For the well-studied poliovirus RdRp, covalent (oligo)nucleotide–VPg complex serves as a primer to direct the template-dependent synthesis of RNAs by RdRp (Paul et al, 1998). RdRps of the VPg-containing viruses share a conserved sequence motif G that was implicated in the recognition of primer–template RNA complex (Barrette-Ng et al, 2002; Gorbalenya et al, 2002; Thompson and Peersen, 2004). The motif G was also identified in RdRps of coronaviruses (Gorbalenya et al, 2002), and accordingly, the SARS-CoV RdRp was shown to be primer-dependent (Cheng et al, 2005). Since the 5′-end of genome RNA of coronaviruses is blocked by a cap rather than a VPg (Siddell et al, 2005), these data indicate that coronaviruses may have evolved a special mechanism to generate primers used in RNA synthesis.
Viruses and cellular organisms have evolved a number of mechanisms to produce RNA primers, which are either derived from pre-existing RNAs (Plotch et al, 1981; Mak and Kleiman, 1997) or synthesized on a template. In cellular organisms, template-derived RNA primers direct the DNA synthesis at both leading and lagging strands. A specialized, low-fidelity DNA-dependent RNA polymerase, known as primase, catalyzes the synthesis of short (4–15 nucleotides) RNA molecules to be used as primers by replicative DNA polymerases; these primers are subsequently removed by ribonucleases (Frick and Richardson, 2001). Like other and structurally unrelated template-dependent polynucleotide polymerases, primases catalyze the formation of internucleotidic phosphodiester bonds using a common phosphoryl transfer reaction.
In this report, we describe a new template-dependent oligonucleotide-synthesizing activity possessed by nsp8 of SARS-CoV. This protein recognizes a specific short sequence ubiquitous in the ssRNA coronavirus genome to catalyze the synthesis of complementary <6 nucleotides with a relatively low fidelity similar to that of known DNA-dependent RNA primases.
Results
The characterization of the SARS-CoV nsp8 (hereafter nsp8) protein described in this paper was initiated to test a possible modulating role of this protein for the poorly active nsp12 RdRp. Surprisingly, nsp8 alone was found to be able to catalyze template-dependent oligoribonucleotide synthesis. This observation prompted a detailed analysis of nsp8 properties related to this activity. The obtained results are described below.
Coronaviridae nsp8 forms an isolated protein family
Nsp8 protein is well-conserved among viruses of the Coronavirus genus with 17 and 66% positions being occupied by identical and similar, respectively, residues. Further database searches using single- and multiple-sequence queries and assisted by BLAST (Altschul et al, 1997) and HMMER (Bateman et al, 1999) programs identified no statistically significant similarity of nsp8 with other proteins. However in the profile searches, we observed an under-the-threshold hit with an equivalently located region in the ORF1a-encoded polyprotein of the Breda bovine torovirus (BToV-1) (Draker et al, 2006). In the torovirus pp1a/pp1ab polyproteins, this region is flanked by two potential 3CL cleavage sites, which may be recognized by Mpro (Figure 1 and AE Gorbalenya, personal communication). These data indicate that the identified region may be a very distant ortholog of nsp8. An alignment of coronavirus nsp8 and the putative BToV-1 ortholog identified eight absolutely conserved residues, which include Lys-58, Lys-82 and Ser-85 (Figure 1).
Figure 1.
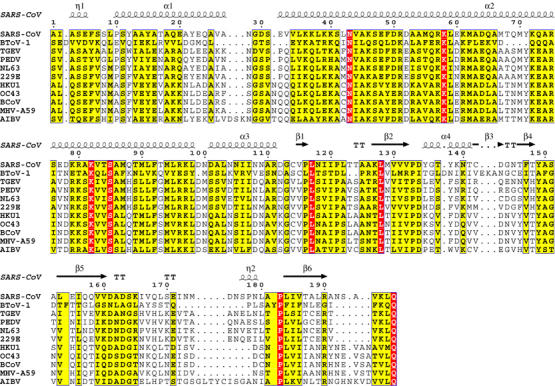
Sequence alignment of nsp8 proteins. The alignment of coronavirus nsp8 sequences was generated with the ClustalW program, version 1.82 (http://www.ebi.ac.uk/clustalw/). This alignment and individual nsp8 sequences were used to search sequence databases as described in Snijder et al (2003). Using results of these searches, the original alignment was extended to a distantly related (see text) torovirus sequence using the MUSCLE program. The resulting alignment was converted into this figure using the ESPript program, version 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Residues that are conserved in all or >70% sequences are boxed in red and yellow, respectively. Above the alignment, numbering and secondary structure elements (Zhai et al, 2005) for SARS-CoV nsp8 protein are depicted. National Center for Biotechnology Information (NCBI) Accession Numbers for replicase polyprotein sequences including nsp8: SARS-CoV, NP_828866; Breda torovirus (BToV-1), AY427798; Transmissible Gastroenteritis coronavirus (TGEV), NP_840006; Porcine epidemic diarrhea virus CV777 (PEDV), NP_839962; Human coronavirus NL63 (NL63), AAS58176; Human coronavirus 229E (229E), NP_835349; Human coronavirus HKU1 genotype A (HKU1), AAT98578; Human coronavirus OC43 (OC43), NP_937947; Bovine coronavirus (BCoV), NP_742135; Mouse Hepatitis virus strain A59 (MHV-A59), NP_740613; Avian Infectious Bronchitis Virus (AIBV), NP_740626.
The SARS-CoV nsp8 has RdRp activity
Purified nsp8 (Supplementary Figure 1A) was assayed for RdRp activity using homopolymeric RNA templates (poly(rA), poly(rU) and poly(rC)) in a filter binding assay (Dutartre et al, 2005). A weak polymerization with poly(rU) template and no activity using poly(rA) template were observed (data not shown). A strong activity using poly(rC) and oligo(rC15) template (Figure 2) was detected, with a linear polymerase activity after up to 4 h of incubation, in a template-dependent manner. The activity was found to be manganese-dependent, although magnesium but not Zn2+, Co2+ nor Ca2+ could also promote RdRp activity with a much lower efficiency (Supplementary Figure 1B). Nsp8 preparations are insensitive to rifampicin, a potent Escherichia coli RNA polymerase inhibitor. Nsp8 polymerase activity is free of contaminating or intrinsic terminal transferase (TNTase) activity. Indeed, a TNTase activity assay carried out with 5′-labeled oligo(rC15) and unlabeled guanine triphosphates as substrates shows no ladder-like products above the labeled oligo(rC15) template (data not shown). The RdRp activity was blocked by mutations at the most conserved positions of nsp8 (see below). Thus, the RdRp activity is catalyzed by nsp8 rather than a co-purified protein.
Figure 2.

Kinetics of RNA synthesis using nsp8 protein, NS5B polymerase of HCV and NS5 polymerase of Dengue virus and an oligo(rC15) RNA template. 1 μM of nsp8, 1 μM HCV NS5B and 400 nM of Dengue NS5 proteins were mixed with [α-32P]GTP (10 μM) and oligo(rC15) template (10 μM) to start the reaction (as described under ‘Materials and methods'). At various times (0, 1, 15 and 30 min), reaction aliquots were quenched by the addition of EDTA/formamide, and the RNA products were resolved on 14% polyacrylamide/7 M urea gel. Due to the likely formation of poly(rG) secondary structures, product size and abundance is not visually accurate for >8-mer products. M, RNA marker synthesized using T7 RNA polymerase and an appropriate template; each band product is indicated on the left.
Primer dependence of the nsp8 RdRp was investigated using poly(rC) or oligo(rC15) templates, which were annealed to labeled oligo(rG) primers of one of three sizes (G2, G6 or G15). Elongation of the labeled primers by nsp8 was not detected (not shown). An ability of the nsp8 RdRp to synthesize RNA in the primer-independent mode was compared to well-established primer-independent RdRps, the HCV NS5B and the Dengue virus (DV) NS5 (Ackermann and Padmanabhan, 2001; Kao et al, 2001). In this assay, an oligonucleotide of 15 consecutive cytidines (oligo(rC15) and GTP) were used as the template and the sole nucleotide substrate, respectively, to direct RNA synthesis. Figure 2 shows the synthesis of oligo(rG) products for each polymerase. The size pattern of products synthesized by nsp8 is similar to those that were produced using HCV NS5B and, to a lesser extent, DV NS5. In the reactions catalyzed by either nsp8 or HCV NS5B, a similar pattern of products up to 7-mers was found. The most prominent was the accumulation of pppGpG, which is a hallmark of a kinetic limitation in the initiation step of RNA synthesis. However and in contrast to NS5B and NS5, nsp8 was unable to catalyze the synthesis of the full-length complementary copies of the template. The seemingly full-length products are shorter and accumulated to a lesser extent than those produced by HCV NS5B or DV NS5. These results indicate that nsp8 is an RNA-directed, primer-independent RdRp acting in a distributive fashion.
Nsp8 is a sequence-specific RdRp
The nsp8 RdRp activity is dependent on the nucleobase of a homopolymeric template (poly(rA), poly(rU) or poly(rC)). A significant nsp8 RdRp activity was only detected using poly(rC) template. Since poly(rG) template generates strong secondary structures, it cannot be tested in a similar polymerase assay. To analyze whether guanines could be part of a template for nsp8, the heteropolymeric RNA template, 5′-UAU AAU GGA AAA-3′ (oligo 5), containing two internal guanines, was tested. No nsp8 RdRp activity was detected using this template. Based on these and other experiments (see below), we concluded that the template must contain at least one cytosine to promote RNA synthesis by nsp8. A 373-nt heteropolymeric RNA template identical to a part of the SARS-CoV genome was used to monitor RdRp activity, and the results are presented in Figure 3A. Nsp8 is able to synthesize only short RNA products (<6-mers), whereas HCV NS5B RdRp is able to synthesize both short and long products. The nsp8 heterogeneous product profile suggests various internal initiation sites (see below). Various short synthetic RNA oligonucleotides were then designed to refine relevant template requirements for an efficient synthesis of RNA products by nsp8 (Table I).
Figure 3.
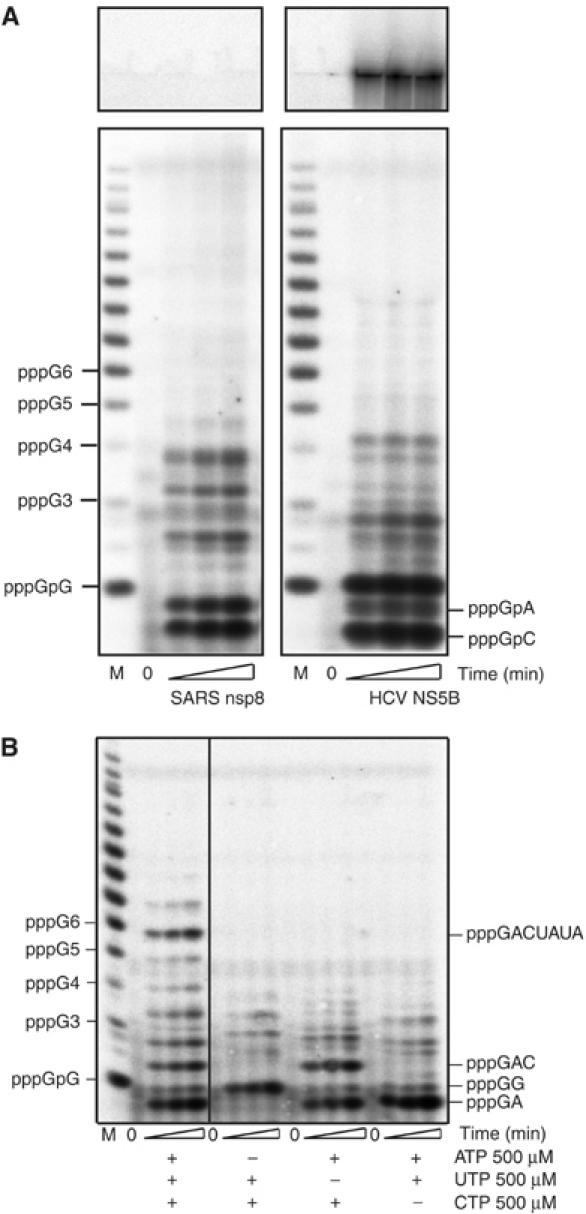
Nsp8 polymerase template requirements. (A) Time course of nucleotide incorporation using a purified 373 nt RNA template and 1 μM nsp8 (left panel) or 1 μM HCV NS5B (right panel) together with 500 μM ATP, UTP, CTP, and 10 μM [α32P]GTP. Reaction products are indicated on the side of the gel autoradiograph. Positions of the RNA markers (M) synthesized using T7 RNA polymerase are shown on the left. (B) Time course of nucleotide incorporation using RNA template 10 (5′-UAUAGUCCCAAA-3′) together with 1 μM nsp8 and 10 μM [α32P]GTP. Reactions were performed with all required nucleotides or lacking either ATP, UTP or CTP, as indicated.
Table 1.
Synthetic nucleic acid templates used in this study
| Oligo name | Sequence | Template usage efficiency (%) |
|---|---|---|
| Oligo 1 | 5′-AAAAAAAAAGUAUCC-3′ | 0 |
| Oligo 2 | 5′-UAUAAUCCGAAA-3′ | 35 |
| Oligo 3 | 5′-AAAAAAAAAAUUGCC-3′ | 0 |
| Oligo 4 | 5′-UAUAAGCCAAAA-3′ | 30 |
| Oligo 5 | 5′-UAUAAUGGAAAA-3′ | 0 |
| Oligo 6 | 5′-AAAACCUAAUAU-3′ | 0 |
| Oligo 7 | 5′-UAUAAACGAACUUUAA-3′ | 0 |
| Oligo 8 | 5′-AAAUCAAAA-3′ | 0 |
| Oligo 9 | 5′-AUAUCGUUUACAAAA-3′ | 0 |
| Oligo 10 | 5′-UAUAGUCCCAAA-3′ | 100 |
| Oligo 11 | 5′-AAGCCCAAA-3′ | 95 |
| Oligo 12 | 5′-UAUAAUCCAAAA-3′ | 100 |
| Oligo 13 | 5′-UAUAAUGCAAAA-3′ | 83 |
| Oligo 14 | 5′-UAUAAUCCCAAA-3′ | 80 |
| Oligo 15 | 5′-UAUAAUCCUAAA-3′ | 56 |
| Oligo 16 | 5′-UAUAAUUCUAAA-3′ | 69 |
| Oligo 17 | 5′-UAUUAAGCUAAA-3′ | 53 |
| All oligonucleotides are RNA except oligonucleotide 11, which is a single-stranded DNA template. RNA synthesis initiation sites are shadowed in gray. From this table and other experiments (not shown), the best priming site was found to be 5′-G/UCCNN-3′ (see text). | ||
| Oligo 10 is taken as a reference, and approximate relative template usage efficiencies are indicated on the right. |
Two sets of oligonucleotides pairs were designed, each containing two adjacent cytidines either internally or at the 3′-end of a template. Cytidines (underlined below) were placed downstream of either U (1st set—oligo 1: 5′-AAAAAAAAAGUAUCC-3′ and oligo 2: 5′-UAUAAUCCGAAA-3′) or G; 2nd set—oligo 3: 5′-AAAAAAAAAAUUGCC-3′ and oligo 4: 5′-UAUAAGCCAAAA-3′). With both sets, product synthesis was only observed using templates bearing internal CC (oligo 2 and oligo 4). Thus, nsp8 is unable to start the synthesis of a complementary sequence at the 3′-end of a linear RNA template and it requires a template cytidine to be flanked by at least two nucleotides from the 3′-end.
The specificity at the +2 and −1 positions relative to the template 3′-cytidine was then addressed. All the templates tested in this study and the results regarding the polymerase assay are shown in Table I. Sequences that include the cytidine-containing trinucleotides (5′–3′) -CCU-, -ACG-, -ACU-, -UCG-, -ACA-, -UCA- or -ACC- proved to be poor templates for nsp8 (Table I). Rational design of test sequences showed that the minimal sequences requirements are UCC, GCC, GCA, UCU and GCU in order to promote activity, with the optimal sequence being 5′-(G/U)CCNN-3′ (Figure 3B, and not shown). Synthesis starts on the 5′ C leaving one cryptic 3′ C. Figure 3B (left lanes) shows nsp8-dependent band products formation, using oligo 10 template (5′-UAUAGUCCCAAA-3′). Products accumulate over time, for each incorporation, confirming that nsp8 acts in a rather distributive fashion. Taken together, these results demonstrate that nsp8 is a sequence-specific RdRp.
As GTP is the required initiating (+1) nucleotide, we used the oligo 10 template (Table I) to measure Km(GTP) at position +1. An overall incorporation efficiency Vmax/Km of 0.65 × 10−5 min−1 μM−1 was measured for GTP, with Km(GTP) of 126±14 μM, a value comparable to that of other viral RdRps (Ranjith-Kumar et al, 2002; Castro et al, 2005; Selisko et al, 2006). This latter value is significantly lower than the intracellular GTP concentration (1–4 mM) (Hauschka, 1973), suggesting that the cellular GTP concentration is not rate-limiting for the nsp8-mediated RNA synthesis initiation reaction. The ATP and CTP incorporation efficiencies at the +2 position were on the same order of magnitude, 1.7 × 10−5 min−1 μM−1 and 0.65 × 10−5 min−1 μM−1, respectively.
We examined the nsp8 nucleotide insertion fidelity under conditions where the reaction mix assay specifically lacked one of the required NTP. Oligo 10 (Table I) was used with four different sets of nucleotides (Figure 3B). In the presence of the four NTPs, the synthesized product should correspond to 5′-GACUAUA-3′ (Figure 3B, left lanes). In contrast, when ATP, UTP or CTP are omitted, a full size product is not detected, indicating that in the absence of the required NTP, nsp8 does not significantly misincorporate a nucleotide (Figure 3B). Based on the observed rate of misincorporation, we could estimate a lower limit of 1 misincorporation per 10 nucleotides synthesized for nsp8 RdRp (data not shown, also see Figure 3 and Discussion). When ATP is omitted, a shift is observed from the usual dinucleotide product pppGA towards pppGG, indicating that nsp8 rather favors a shift in its initiating site over a misincorporation at the original site (Figure 3B).
Selective nsp8 inhibition using GTP analogs
The nsp8 protein may represent an attractive target against coronaviruses, several nucleotides analogs (3′-dGTP, ddGTP and 2′-O-methyl-GTP) were tested for their capability to inhibit nucleotide incorporation into RNA using a poly(rC) template. 3′-dGTP was found to efficiently inhibit nsp8 RdRp activity (Figure 4), whereas ddGTP and 2′-O-methyl-GTP were weak inhibitors (data not shown). Using increasing 3′-dGTP concentrations, most of the band products vanish, with no significant appearance of chain terminated products (Figure 4A). Comparatively, chain terminated products increasing over time are apparent when using HCV NS5B in a similar experimental setting (Figure 4B, asterisks) The fact that ladder-like product formation does not occur significantly using nsp8 suggests that most inhibition occurs at position +1 of the template.
Figure 4.
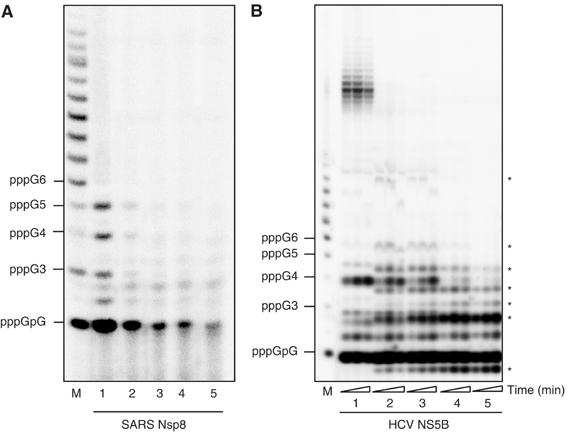
Inhibition of RNA synthesis by 3′-dGTP using a poly(rC) template. (A) Effect of increasing concentrations of 3′-dGTP on the initiation and full-length RNA product formation. The nucleoside analog was added to reaction mixtures containing 1 μM poly(rC), 10 μM [α32-P] GTP and 1 μM nsp8. Lanes 1–5 show the products of the reactions performed using the following concentrations of 3′-dGTP: 0, 100, 500 nM, 1 and 10 μM. Reactions were allowed to proceed for 60 min. M, RNA marker synthesized using T7 RNA polymerase and an appropriate template. (B) Same as in (A) using 10 μM oligo(rC15) as a template in conjunction with 1 μM HCV NS5B. Each chain terminated band product is indicated on the right by an asterisk (*). For each 3′-dGTP concentration (lanes 1–5: 0, 5, 10, 50 and 100 μM, respectively), the reaction was allowed to proceed for 30, 60 and 120 min.
Conserved nsp8 residues are critical for RdRp activity
In order to identify essential residues for nsp8 activity, alanine-scanning mutagenesis was applied to all conserved charged and selected polar amino-acid residues. In total, 14 mutants of nsp8 were generated (Figure 5), expressed in E. coli and purified to apparent homogeneity using affinity and exclusion chromatographies. The RdRp specific activity of these mutants was measured using [3−H]GTP incorporation into RNA using a poly(rC) template, and was expressed as a percentage of the activity of the wild-type nsp8 (Figure 5 and Supplementary Figure 3). The K58A, R75A, K82A and S85A mutations abolished or greatly reduced the RdRp activity. The nsp8 RdRp activity was most sensitive to replacement of either of the three residues, K58, K82 and S85, which are also conserved in the torovirus sequence. The D161A mutant remains substantially active. Although the nsp8 activity is metal-ion dependent, there are no essential and conserved acidic residues that may chelate a catalytic Mn2+ (see Discussion). Activities recorded with the nsp8 mutants are thus fully consistent with the alignment presented in Figure 1.
Figure 5.
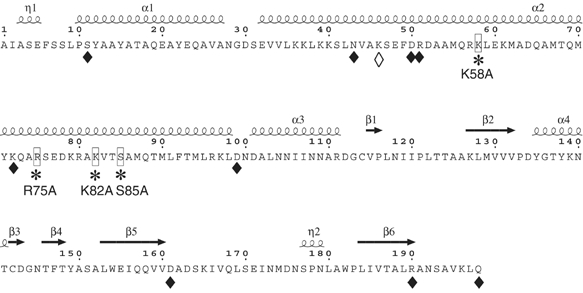
Alanine scanning mutagenesis of the nsp8 protein. Indicated amino acids were replaced by alanine using site-directed mutagenesis. One micromolar of nsp8 wt and 1 μM of Ala mutants were tested for polymerase activity by measuring [3H]GTP incorporation using a poly(rC) template. Nsp8 polymerase specific activity is represented as follows: empty lozenge (◊); 100% activity relative to nsp8 wt; black lozenge (⧫), between 12 and 40% activity and; asterisk (*), less than 4% activity (see Supplementary Figure 3 for precise values).
A structure- and activity-based model of nsp8 RdRp
The crystal structure of nsp8 in complex with nsp7 has been determined recently (Zhai et al, 2005). Nsp8 and nsp7 form a hexadecameric toron structure able to encircle and bind RNA as judged by both the presence of a strong positive charge in the inner channel and biochemical assays. Nsp7 is thought to play the role of a mortar, bringing cohesive force to the complex with no obvious role in RNA binding (Zhai et al, 2005). All conserved nsp8 residues, which are essential for the RdRp activity, are located on the second large alpha helix (Figure 1) with residues Lys-58 and Arg-75 surrounding, but slightly outside the channel. These residues were implicated in RNA binding. The essential residues map to a dimeric nsp8, which is part of two equivalent dimers in the hexadecameric nsp8/nsp7 complex.
Interestingly, the comparison of the folding of the head domain of nsp8 (corresponding to the most C-terminal 99 aa residues) shows similarity to a family of the RNA-binding domains (RBD) (Supplementary Figure 4) (Krissinel and Henrick, 2004). This family is characterized by the two ssRNA recognition motifs Rnp1 and Rnp2 (Maris et al, 2005). These motifs are composed of mainly hydrophobic and positively charged residues. Both the RBD and the head of nsp8 are folded into an α/β sandwich. The structure of the heterogeneous nuclear ribonucleoprotein D (hnRNP D), belonging to the RBD family, was solved in complex with ssRNA by NMR (Enokizono et al, 2005). Identical connectivity and sequential arrangement of secondary structural elements are also conserved in the RdRp palm subdomain (containing catalytic Asp residues in the GDD motif) (Hansen et al, 1997) (Supplementary Figure 5), providing evolutionary hints about a possible nsp8 origin (see Discussion).
Altogether, we can thus propose a model for a quaternary initiation complex involving two monomers of the nsp8 protein, an ssRNA template (5′-UAGC-3′) and the first two complementary nucleotides incorporated (GTP and CTP). We have superimposed the RNP motifs of nsp8 head domain onto the RNP motifs of hnRNP D. In this superimposition, the ssRNA is stacked by the hydrophobic surface of the two RNP motifs and points towards the inner part of the hexadecamer channel (Figure 6B). Thus, while an nsp8 molecule of the hexadecamer stacks the RNA template (Figure 6A, represented in black), a second is able to bind the nascent primer. Then, the two first NTPs incorporated (GTP in +1 and CTP in +2) are bound to two strictly conserved basic residues Arg-75 and Lys-82 present on the second nsp8 molecule (Figure 6A, represented in clear gray). The positioning of the GTP primer against the helix implies that triphosphated dimers and longer primers would not fit into the active site, consistent with the fact that they are not elongated.
Figure 6.
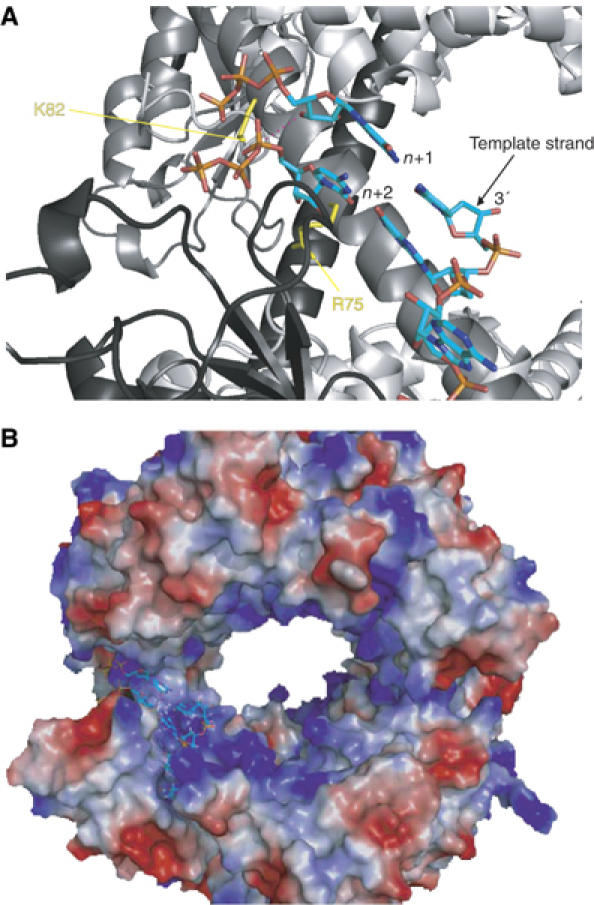
Model of two monomers of the nsp8 protein in complex with an ssRNA template (5′-UAGC-3′) and two nucleotides (GTP and CTP). RNA template, GTP and CTP are shown by a stick model. (A) The amino-acid residues related to null mutants, Lys-58 and Arg-75, are represented in yellow. The two first NTPs incorporated (GTP in +1 and CTP in +2) are indicated. Discontinuous purple line represents distance between GTP 3′-OH (incorporated nucleotide in position +1) and α-phosphate of CTP (incorporated nucleotide in +2 position), estimated to 3.8 Å. (B) The surface is colored according to the electrostatic potential nomenclature (blue, positive charge; red, negative charge). Images were generated using PYMOL.
Discussion
Many genetic and mechanistic features distinguish the coronavirus replication machinery among those encoded by other RNA viruses. We have now discovered a second RdRp in SARS-CoV, the first of this kind in RNA viruses, thus providing further evidence for the unprecedented sophistication of the replication complex in coronaviruses. In the context of other data accumulated in the field, the described nsp8 RdRp properties indicate that this enzyme may catalyze the synthesis of RNA primers for the primer-dependent nsp12 RdRp. Although primers are used in genome replication by numerous RNA viruses, coronaviruses may be unique in evolving a specialized RNA enzyme for primer synthesis (primase).
It was previously shown that the RdRp domain of coronaviruses is evolutionary clustered with RdRps of (+) RNA viruses that may use a protein (VPg-like) for priming RNA synthesis (Gorbalenya et al, 1989; Koonin, 1991). These RdRps are distinguished by the presence of the G sequence motif that was implicated in the primer/template recognition (Gorbalenya et al, 2002). Relevance of these observations for coronaviruses may be linked to the finding that the SARS-CoV nsp12 RdRp is active, in vitro, using a poly(rA)/oligo(dT)12−18 primer-template (Cheng et al, 2005). Since coronaviruses may not produce VPg, the identity of a primer and the mechanism of its generation have remained unresolved for these viruses. Besides VPg, cellular RNA molecules are recruited by some RNA viruses to prime polynucleotide synthesis in either replication or transcription of the genome. This strategy was adopted by influenza viruses and retroviruses, which either hijack a piece of a cellular mRNA (Plotch et al, 1981) or use a tRNA (Mak and Kleiman, 1997), respectively, as a primer. We propose that coronaviruses have evolved another strategy to produce primers, which may use the newly identified nsp8 RdRp as the primase.
In the DNA world, primases are ubiquitous and it may be instructive to compare the nsp8 RdRp with these enzymes. The general function of a DNA primase is to synthesize an RNA primer on a DNA template. In most viruses and cellular organisms that replicate their genomes through semidiscontinuous DNA synthesis, DNA-dependent DNA polymerase (DdDp) recognizes the primer/template complex to extend the primer-initiated synthesis of the complementary DNA strand (Frick and Richardson, 2001). In comparison to other template-dependent DNA and RNA polymerases, fidelity is of a less importance for DNA-dependent primases. Subsequent to the primer utilization in DNA synthesis, the RNA primer is removed and a gap in the nascent DNA is sealed with newly generated DNA by several enzymes including a high-fidelity DdDp.
In coronaviruses, the replication complex is yet to be characterized in sufficient details in vitro and in vivo (Ziebuhr, 2005). Recent analysis indicates that it is likely to include virus-encoded RNA processing enzymes ExoN (nsp14), EndoU (nsp15), and a putative ribose-2′-O-methyltransferase (nsp16) (Snijder et al, 2003; Bhardwaj et al, 2004; Ivanov et al, 2004a; Ziebuhr, 2005; Minskaia et al, 2006), which could excise primers synthesized by the low-fidelity nsp8 RdRp (with a misincorporation rate of 1/10 as a lower limit). This excision could be part of the methyl directed-mismatch repair activity that is worth further testing. Similarly to the E. coli and eukaryotic primases, the nsp8 RdRp exhibits a limited processivity. The prokaryotic DNA primases (i.e., from the E. coli DnaG family) generally recognize specific sequences on the template (Yoda et al, 1988), while eukaryotic primases are sequence-independent (Bullock et al, 1994). The SARS-CoV nsp8 revealed marked sequence preferences and, like cellular primases, RNA synthesis starts with a purine residue. Once the nsp8/RNA/nucleotides ternary complex has been formed, a rate-limiting step occurs before or during dinucleotide synthesis, a feature that is also common for cellular primases. In our hands, nsp8 and the purified nsp7–nsp8 complex exhibit comparable activities (not shown). The only noticeable biochemical difference between nsp8 and the nsp8–nsp7 complex, which may be the functional form of the nsp8 RdRp, is a relatively poor thermal stability of nsp8 (data not shown). Remarkably, this property was predicted upon examination of the crystal structure of the nsp8–nsp7 complex by Zhai et al (2005), who described nsp7 as a ‘mortar' protein.
Little is known about the initiation of RNA synthesis in coronaviruses although terminal sequences were implicated in the control of the process (Lai and Cavanagh, 1997). Depending on the polarity, plus or minus, and the size, genome or subgenome, single-stranded RNAs, partial single-stranded RNAs (known as replicative intermediates) and double-stranded RNAs (replicative forms) appear to serve as templates. The apparent complexity of the RNA synthesis may accommodate the postulated primase activity in different ways and the dissection of this aspect requires further analysis.
We note here that the sequence specificity of the nsp8 RdRp is not stringent and, potentially, this enzyme could initiate RNA synthesis at numerous internal places at the genome or its complement that would also be reminiscent of the modus operandi of cellular primases. In this way, the giant genome of coronaviruses could replicate much faster and, possibly, more accurately than it would otherwise using a single 5′-terminal primer. These properties could form a basis that has driven the origin of the primase in the coronavirus evolution.
The identification of nsp8 as a potential primase should facilitate developing functional assays for studying the replicase machinery in vitro. Our preliminary results show that in agreement with the reported results and the proposed model, purified nsp8 and nsp12 interact in GST-pull down experiments (Imbert et al, unpublished data). The 3- to 5-fold excess of the nsp8 synthesis relative to nsp12, due to downregulation of the latter by frameshifting (Brierley, 1995; Thiel et al, 2003), seems to be used to build the nsp7/nsp8 octamer complex containing four nsp8 subunits (Zhai et al, 2005). Consequently, equimolar stoichometric ratio between interacting nsp8 and nsp12 species may be maintained in the infected cell. The nsp8 has a unique bi-domain structure that is different from those of prokaryotic and eukaryotic primases. Its C-terminal domain has a fold also found both in a diverse family of RNA-binding proteins and the catalytic palm-subdomain of RdRps (Hansen et al, 1997; see Supplementary Figure 5). This finding indicates that the nsp8 RdRp and nsp12 RdRp may have originated from a common ancestor, possibly through a duplication during evolution leading to the emergence of the ancestral virus of the Coronaviridae family. Previously, duplications of PLpro and Mpro were implicated in the evolution of the coronavirus proteome (Ziebuhr et al, 2001).
The eight most conserved residues are distributed between the two domains of nsp8. Three polar and essential residues, Lys-58, Lys-82 and Ser-85, which may be part of the catalytic residues network for the phosphoryl transfer reaction, are located in the N-terminal domain. As in the case of the coronavirus Mn2+-dependent endonuclease nsp15, the metal-ion dependence in catalysis remains undefined and is unlikely to be promoted by acidic residues. However, our tertiary structure modeling analysis further suggests that the highly conserved Trp-182 in the C-terminal domain (head domain) is close to the α-phosphate of the +2 nucleotide and might be involved in Mn2+ coordination promoting metal-based nucleophilic activation at the phosphorus center. Indeed, such interaction between aromatic residues and cation, termed cation–π (Dougherty, 1996), have been described as a non-covalent bonding interaction relevant for molecular recognition and catalysis (Zaric et al, 2000). A role for Trp-182 in catalysis is consistent with the fact that in the putative torovirus ortholog, the residue is also an aromatic residue (Tyr). Regardless of the precise composition of the catalytic center of nsp8, it differs from that conserved in the nsp12 RdRp, impeding to determine undoubtedly if these two RdRps have been acquired independently or have diverged profoundly.
Materials and methods
Materials and reagents
RNA oligonucleotides were obtained from Dharmacon. DNA oligonucleotides were obtained from Invitrogen. A 373-nt template corresponding to nt 13905–14278 of the SARS-CoV genome (strain Frankfurt, GenBank Accession No. AY291315) was produced using an in vitro T7 transcription kit, and purified as described by the manufacturer (Ambion Inc.). Homopolymeric cytosine template (poly(rC)), 15-mers cytosine RNA oligonucleotide (oligo (rC15)), α-32P-labeled guanosine 5′-triphosphate (3000 Ci/mmol), α-32P-labeled cytosine 5′-triphosphate (3000 Ci/mmol), uniformly labeled [3H]GTP (5.20 Ci/mmol) and nucleosides 5′ triphosphate were purchased from Amersham Biosciences. Nucleosides analogs 3′-deoxy GTP, 2′-O-methyl-GTP and di-deoxy-GTP were purchased from Trilink, Inc. RNA molecular weight markers were synthesized as described in Dutartre et al (2005). HCV NS5B and NS5 Dengue polymerases were purified as in (Selisko et al, 2006).
SARS nsp8 plasmid constructions, E. coli protein expression and purification
The SARS-CoV nsp8 coding sequence was amplified by PCR from the cDNA prepared as previously described (Drosten et al, 2003). The cDNA was then subcloned in the pDest14 plasmid (Invitrogen) in a manner analogous as nsp9 described in Campanacci et al (2003). The ORF of the final construct (referred to as nsp8) encoded an N-terminally 6 His-tag. This construct was mutated using the QuikChange site-directed mutagenesis kit, according to the manufacturer's instruction (Stratagene). All constructions were verified by DNA sequencing (Millegen, France). Proteins expression and purification were performed as described (Campanacci et al, 2003). Proteins were homogenous as judged by SDS–PAGE (see Supplementary Figure 1A). They were concentrated to 5 mg/ml and stored in 50% glycerol at −20°C. Recombinant proteins were characterized by dynamic light scattering and circular dichroism spectra, which were undistinguishable from wt nsp8. Enzyme concentrations were determined using UVλ280 absorbance. No attempts were made to determine the proportion of active enzyme (enzyme active site concentration).
Nsp8-mediated steady-state incorporation of nucleotide using RNA templates
Polymerase activity was assayed by monitoring the incorporation of radiolabeled guanosine using either oligoribonucleotide or polycytosine (poly(rC)) templates. All indicated concentrations are final. The reaction was performed in an optimized polymerase buffer made of 50 mM Tris pH 7.5, 10 mM KCl, 4 mM MgCl2, 1 mM MnCl2, 10 mM dithiothreitol, 1% Triton X-100 containing 10 μM of [α-32P]GTP. The templates were either 10 μM RNA oligonucleotide, 1 μM poly(rC). Reactions were initiated by the addition of 1 μM purified nsp8 and incubated at 30°C. Aliquots were withdrawn over time from 10 s to 2 h and the reaction was stopped by the addition of EDTA/Formamide. Reaction products were separated using sequencing gel electrophoresis (14% acrylamide, 7 M urea in TTE buffer (89 mM Tris, 28 mM taurine, 0.5 mM EDTA)) and quantitated using photo-stimulated plates and a FujiImager (Fuji). In some instances, nsp8 activity was quantitated using a filter paper binding assay. Reactions were initiated by the addition of 1 μM nsp8 in polymerase buffer containing 1 μM poly(rC) template, 0.1 mM [3H]GTP (0.5 μCi), in the same buffer as above, incubated at 30°C, and stopped by spotting aliquots onto DE-81 paper discs (Whatman International Ltd). Filter paper discs were washed three times for 10 min in 0.3 M ammonium formate, pH 8.0, washed two times in ethanol, and dried. The radioactivity bound to the filter was determined using liquid scintillation counting. Under these conditions, the nsp8 specific activity was consistently in the vicinity of 62 c.p.m. min−1.
Steady-state kinetic studies
To determine the Km and Vmax for CTP incorporation by nsp8, the RNA 5′-UAUAAGCCAAAA-3′ template (10 μM) was mixed in polymerase buffer with 1 μM nsp8. The reaction was started by the addition of 10 μM [α-32P]GTP and increasing concentration of CTP (1, 5, 10, 50, 75, 100, 300 and 500 μM). To determine the Km and Vmax for the incorporation of ATP, RNA 5′-UAUAGUCCCAAA-3′ was incubated with 10 μM [α-32P]GTP and increasing concentration of ATP (1, 5, 10, 50, 75, 100, 300 and 500 μM). Finally, the Km and Vmax for GTP incorporation was determined using RNA 5′-UAUAGUCCCAAA-3′ template incubated with 10 μM [α-32P]CTP and increasing concentration of GTP (1, 5, 10, 50, 75, 100, 300 and 500 μM). The reactions were incubated at 30°C for 15, 30, 60 and 120 min. Aliquots were withdrawn during the time course of the reaction, and the reactions were quenched with EDTA/formamide. Products were separated using sequencing gel electrophoresis and quantified using photo-stimulated plates and a FujiImager (Fuji). Product formation was represented by the hyperbolic equation describing Vi dependence on NTP concentration
![]()
where Vmax and Km are the maximal velocity and the affinity constant of NTP incorporation by nsp8, respectively. Vmax and Km were determinated from curve-fitting using KaleidaGraph (Synergy Software).
Fold comparison
The fold comparison was performed using the SSM server (protein structure comparison service SSM at European Bioinformatics Institute, http://www.ebi.ac.uk/msd-srv/ssm) with a truncated form of nsp8 termed ‘head' domain (corresponding to the C-terminal 99 a.a.). The superimposition between the head of nsp8 and a member of the RBD, the C-terminal of CstF-64 (PDB code: 1p1t) (Perez Canadillas and Varani, 2003) shows a global RMSD of 2.8 Å. However, the RMSD calculated only for the two motifs is about 1 Å for RNP2 and 1.9 Å for RNP1 (Supplementary Figure 4). We also performed a structural alignment with different RNP members to check the correlation between the existence of the RNP motifs and the position in the structure. This alignment was carried out using MUSCLE (Edgar, 2004). Then, the alignment was analyzed and optimized with SeaView (Galtier et al, 1996), taking into account the secondary structure from the high-resolution models.
Modeling
The crystal structure of the nsp8 hexadecamer (Zhai et al, 2005) was submitted to 30 cycles of rigid body and 100 cycles of conjugate gradient with CNS (Brunger et al, 1998). Based on the structural alignment (Supplementary Figure 4), we have generated a superimposition between hnRNP D with its ssRNA (PDB: 1wtb) (Enokizono et al, 2005) and the head of one nsp8 monomer. Then, the position of the ssRNA was manually adjusted using Coot (Emsley and Cowtan, 2004) to avoid steric clash and to correct the direction of the ssRNA backbone to point out towards the central cavity of the hexadecamer. Another minimization cycle was performed taking into account the presence of the RNA template (as described above). To model the initiation state of nsp8, we docked the first two nucleotides base pair complementary (GTP in +1 and CTP in +2) to the ssRNA template. These two nucleotides are the first two to be incorporated in the nascent RNA chain. These two nucleotides of the new strand were manually docked on the top of the head respecting Watson and Crick base pairing dictated by the template. A last round of energy minimization was performed on this quaternary structure (primase/template/incorporated nucleotides).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Acknowledgments
We thank Hélène Dutartre for stimulating discussions and critical reading of the manuscript; Arnaud Gruez for helpful advice, Claire Debarnot and Sacha Grisel for excellent technical assistance. We acknowledge Eric J Snijder, Christian Cambillau, Valérie Campanacci and Sonia Longhi for materials and their help in the initial phase of the project. This work was supported by the Structural Proteomics in Europe (SPINE) Project of the European Union 5th framework research program (Grant QLRT-2001-00988) and subsequently by the Euro-Asian SARS-DTV Network (SP22-CT-2004-511064) from the European Commission specific research and technological development Programme ‘Integrating and strengthening the European Research area'.
References
- Ackermann M, Padmanabhan R (2001) De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem 276: 39926–39937 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrette-Ng IH, Ng KK, Mark BL, Van Aken D, Cherney MM, Garen C, Kolodenko Y, Gorbalenya AE, Snijder EJ, James MN (2002) Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J Biol Chem 277: 39960–39966 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL (1999) Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res 27: 260–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K, Guarino L, Kao CC (2004) The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol 78: 12218–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I (1995) Ribosomal frameshifting viral RNAs. J Gen Virol 76 (Part 8): 1885–1892 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Bullock PA, Tevosian S, Jones C, Denis D (1994) Mapping initiation sites for simian virus 40 DNA synthesis events in vitro. Mol Cell Biol 14: 5043–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanacci V, Egloff MP, Longhi S, Ferron F, Rancurel C, Salomoni A, Durousseau C, Tocque F, Bremond N, Dobbe JC, Snijder EJ, Canard B, Cambillau C (2003) Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr D 59: 1628–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C, Arnold JJ, Cameron CE (2005) Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective. Virus Res 107: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, Sarafianos SG, Ding J (2005) Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty DA (1996) Cation–pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271: 163–168 [DOI] [PubMed] [Google Scholar]
- Draker R, Roper RL, Petric M, Tellier R (2006) The complete sequence of the bovine torovirus genome. Virus Res 115: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Preiser W, Gunther S, Schmitz H, Doerr HW (2003) Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol Med 9: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutartre H, Boretto J, Guillemot JC, Canard B (2005) A relaxed discrimination of 2′-O-methyl-GTP relative to GTP between de novo and elongative RNA synthesis by the hepatitis C RNA-dependent RNA polymerase NS5B. J Biol Chem 280: 6359–6368 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M (2005) Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem 280: 18862–18870 [DOI] [PubMed] [Google Scholar]
- Feder M, Pas J, Wyrwicz LS, Bujnicki JM (2003) Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302: 129–138 [DOI] [PubMed] [Google Scholar]
- Frick DN, Richardson CC (2001) DNA primases. Annu Rev Biochem 70: 39–80 [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548 [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L (2003) A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol 148: 2207–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE (2001) Big nidovirus genome. When count and order of domains matter. Adv Exp Med Biol 494: 1–17 [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM (1989) Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res 17: 4847–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK (2002) The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC (2002) RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol 76: 3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Long AM, Schultz SC (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5: 1109–1122 [DOI] [PubMed] [Google Scholar]
- Hauschka PV (1973) Analysis of nucleotide pools in animal cells. Methods Cell Biol 7: 361–462 [DOI] [PubMed] [Google Scholar]
- Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J (2004a) Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA 101: 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J (2004b) Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 78: 5619–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Singh P, Ecker DJ (2001) De novo initiation of viral RNA-dependent RNA synthesis. Virology 287: 251–260 [DOI] [PubMed] [Google Scholar]
- Koonin EV (1991) The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol 72 (Part 9): 2197–2206 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Lai MM, Cavanagh D (1997) The molecular biology of coronaviruses. Adv Virus Res 48: 1–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM, Holmes KV (2001) Coronaviruses.In Field's Virology,Knipe DM, Howley PM (eds), Vol. 1, pp. 1163–1185. Lippincott Williams & Wilkins: Philadelphia, PA [Google Scholar]
- Mak J, Kleiman L (1997) Primer tRNAs for reverse transcription. J Virol 71: 8087–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J (2006) Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA 103: 5108–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E (1998) Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393: 280–284 [DOI] [PubMed] [Google Scholar]
- Perez Canadillas JM, Varani G (2003) Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J 22: 2821–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858 [DOI] [PubMed] [Google Scholar]
- Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR (2004) Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol 78: 9977–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Gutshall L, Kim MJ, Sarisky RT, Kao CC (2002) Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J Virol 76: 12526–12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selisko B, Dutartre H, Guillemot JC, Debarnot C, Benarroch D, Khromykh A, Despres P, Egloff MP, Canard B (2006) Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology 351: 145–158 [DOI] [PubMed] [Google Scholar]
- Siddell SG, Ziebuhr J, Snijder EJ (2005) Coronaviruses, Toroviruses, and Arteriviruses. In Hodder Arnold, In: Mahy BWJ, ter Meulen J (eds), pp 823–856 [Google Scholar]
- Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331: 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Herold J, Schelle B, Siddell SG (2001) Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol 75: 6676–6681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J (2003) Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84: 2305–2315 [DOI] [PubMed] [Google Scholar]
- Thompson AA, Peersen OB (2004) Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J 23: 3462–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Yasuda H, Jiang XW, Okazaki T (1988) RNA-primed initiation sites of DNA replication in the origin region of bacteriophage lambda genome. Nucleic Acids Res 16: 6531–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaric SD, Popovic DM, Knapp EW (2000) Metal ligand aromatic cation–pi interactions in metalloproteins: ligands coordinated to metal interact with aromatic residues. Chemistry 6: 3935–3942 [DOI] [PubMed] [Google Scholar]
- Zhai Y, Sun F, Li X, Pang H, Xu X, Bartlam M, Rao Z (2005) Insights into SARS-CoV transcription and replication from the structure of the nsp7–nsp8 hexadecamer. Nat Struct Mol Biol 12: 980–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J (2005) The coronavirus replicase. Curr Top Microbiol Immunol 287: 57–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J, Snijder EJ, Gorbalenya AE (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81: 853–879 [DOI] [PubMed] [Google Scholar]
- Ziebuhr J, Thiel V, Gorbalenya AE (2001) The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J Biol Chem 276: 33220–33232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5


